Abstract
Transposable elements are important sources of miRNA, long non-coding RNAs genes, and their targets in the composition of protein-coding genes in plants and animals. Therefore, the detection of expression levels of specific non-coding RNAs in various tissues and cells in normal and pathological conditions may indicate a programmed pattern of transposable elements’ activation. This reflects the species-specific composition and distribution of transposable elements in genomes, which underlie gene regulation in every cell division, including during aging. TEs’ expression is also regulated by epigenetic factors (DNA methylation, histone modifications), SIRT6, cytidine deaminases APOBEC3, APOBEC1, and other catalytic proteins, such as ERCC, TREX1, RB1, HELLS, and MEGP2. In evolution, protein-coding genes and their regulatory elements are derived from transposons. As part of non-coding regions and introns of genes, they are sensors for transcriptional and post-transcriptional control of expression, using miRNAs and long non-coding RNAs, that arose from transposable elements in evolution. Methods (Orbld, ncRNAclassifier) and databases have been created for determining the occurrence of miRNAs from transposable elements in plants (PlanTE-MIR DB, PlaNC-TE), which can be used to design epigenetic gene networks in ontogenesis. Based on the data accumulated in the scientific literature, the presence of 467 transposon-derived miRNA genes in the human genome has been reliably established. It was proposed to create an updated and controlled online bioinformatics database of miRNAs derived from transposable elements in healthy individuals, as well as expression changes of these miRNAs during aging and various diseases, such as cancer and difficult-to-treat diseases. The use of the information obtained can open new horizons in the management of tissue and organ differentiation to aging slow down. In addition, the created database could become the basis for clarifying the mechanisms of pathogenesis of various diseases (imbalance in the activity of transposable elements, reflected in changes in the expression of miRNAs) and designing their targeted therapy using specific miRNAs as targets. This article provides examples of the detection of transposable elements-derived miRNAs involved in the development of specific malignant neoplasms, aging, and idiopathic pulmonary fibrosis.
1. Introduction
Multicellular eukaryotes are characterized by the expression of a wide variety of non-coding RNAs (ncRNAs), the number of which is several times higher than the number of protein-coding genes [1]. In evolution, the emergence of ncRNAs was due to the protection of genomes from the expression of transposable elements using the RNA interference system (RNAi). This system includes the enzymes Dicer (ribonuclease III), RNA-dependent RNA polymerase (RdRP), and Argonaute-PIWI [2]. These enzymes process transposable elements’ (TEs) transcripts with the formation of small ncRNAs, which are then used by them as guides for silencing the expression of specific TEs [3]. Silencing is carried out due to the complementarity of the nucleotide sequences. In evolution, TEs became sources of regulatory sequences and a large number of protein-coding genes [4], which also become targets for RNAi since they contain complementary sequences for ncRNA.
The most known ncRNAs are miRNAs—single-stranded molecules (containing about 22 nucleotides) that regulate gene expression by binding to mRNA [5]. In addition, miRNA control is also possible at the transcriptional level due to RNA-directed DNA methylation (RdDM). This phenomenon has long been considered a specific feature of plants only [6]. However, recent studies have described RdDM in humans [7]. Since the RNAi system influences the formation of heterochromatin [8], and miRNAs are able to guide not only DNA methyltransferases, but also histone deacetylases to specific chromosomal loci [6,9], it can be argued that ncRNAs are drivers of epigenetic regulation. However, the higher-order control elements that regulate the features of ncRNA expression are species-specific transposable elements distributed in the genomes [10], which serve as key sources of ncRNA in plants [11] and animals [12]. In addition to epigenetic control using miRNAs, TEs create the global genomic network for in cis regulation of adjacent genes [3]. TEs make up 45% of the human genome [12,13].
TEs are classified into retroelements (REs) and DNA-transposons (which change their location by the “cut and paste” or “rolling circle” mechanism). Transposition of REs occurs by reverse transcription of their mRNA and insertion of cDNA into a new locus. REs are subdivided into long terminal repeats containing (LTR-REs) and not containing them (non-LTR-REs) [1]. Non-LTR-REs include the orders DIRS (Dictyostelium discoideum retroelements), PLE (Penelope-like), LINE (long interspersed nuclear elements), and SINE (short interspersed nuclear elements). Each of these orders is classified into several superfamilies (Figure 1) which differ in structural features and mechanisms of movement. For example, PLEs encode an endonuclease (EN—endonuclease), similar to group II introns, and reverse transcriptase (RT—reverse transcriptase), similar to eukaryotic telomerase [13]. Modern online resources are used to classify miRNAs and predict their potential targets, such as mirDeep (for animals), Shortstack, mirDeep-P, mirPlant, miRA, PIPmiR, miR-PREFeR, and miRCat2 (for plants) [14].

Figure 1.
Classification of retroelements.
In addition to miRNAs, noncoding RNAs also include tRNAs (73–93 nucleotides), small nuclear RNAs (150 nucleotides, denoted by the letter U, participate in splicing), small nucleolar RNAs (60–170 nucleotides, necessary for processing ribosomal RNA), Vault-RNA (100 nucleotides, regulate autophagy and apoptosis), Y-RNA (about 100 nucleotides, bind to the Ro60 protein), small NF90-associated RNAs (snaRs—117 nucleotides, involved in translation control), ribosomal RNAs (rRNAs), long non-coding RNAs (more than 200 nucleotides in size), and circular RNA (formed during splicing from exons or introns of mRNA genes) [5]. A common class of ncRNAs are small interfering RNAs (siRNAs) which are generated by the degradation of exogenous dsRNAs, transcribed from TEs or from other types of inverted repeats [2]. Animals are also characterized by the class of piRNAs, 21–35 nucleotides long, which are involved in the regulation of gene expression, antiviral response, and TEs silencing (by targeting histone modifications and DNA methylation) [15]. In humans, ncRNA designations are approved by the HUGO Gene Nomenclature Committee (HGNC). For each gene, www.genenames.org (accessed on 20 December 2022) provides information on its symbol, name, chromosomal localization, and links to key resources, such as Ensembl, NCBI Gene, and GeneCards [5]. The study of the relationship of TEs with miRNAs is promising for the use of ncRNAs as tools for correcting TEs dysregulation during aging, in cancer, and in various idiopathic diseases. To implement this, it is necessary to create an extensive, replenished universal online database that allows you to identify their relationship.
2. Differences of the Origin of miRNA from Transposons in Plants from Animals
Unlike in animals, plant miRNAs are completely complementary to their target mRNA sequences. Their binding, in most cases, causes mRNA cleavage [2]. Moreover, mRNAs can contain several regions that are complementary to miRNAs. Both stages of miRNA precursor cleavage are carried out in the nucleus using ribonuclease DCL1, after which the miRNA is transported into the cytoplasm by means of Hasty enzyme, which is homologous to the animal Exportin-5 protein [16]. Plants are characterized by a significant variety and number of specific small ncRNAs, which include tasiRNA (trans-acting short interfering RNA), nat-siRNA (natural antisense short interfering RNA), and hc-siRNA (heterochromatic small interfering RNA) [14]. TEs in plants during evolution become sources of both miRNA genes and protein-coding genes’ exons. Due to these processes, epigenetic regulators (miRNAs) and their targets (gene exons) are formed, and transposable elements (miRNA sources) form dynamic gene networks that control protein-coding genes expression. One of the mechanisms by which miRNAs originate from TEs is the formation of inverted repeats, which are transcribed into RNA hairpin structures processed by Dicer-like enzymes [17]. TE-derived miRNAs (TEDmiR) are involved in vital functions, such as stress responses, a barrier to hybridization in plants, and dynamic transformations of heterochromatin during ontogenesis [3].
For the first time in the world in 2007, a study of rice TEs revealed 21 different small ncRNAs formed from MITE (miniature inverted-repeat transposable elements), which are localized in introns and exons of protein-coding genes, EST regions (expressed sequence tag), and intergenic [18]. A total of 12 TEDmiRs in Arabidopsis and 83 TEDmiRs in rice, which also derived from MITE, were described the next year [19]. An analysis of the miRBase and Repbase Update allowed Lorenzetti et al. to create an online resource (http://bioinfotool.cp.utfpr.edu.br/plantemirdb; accessed on 20 December 2022) for the registration of miRNAs derived from TEs—PlanTE-MIR DB [11]. The main sources of miRNAs in plants are LTR-Res since they constitute the bulk of their genomes. For example, LTR-REs in Asparagus officinalis occupy 91% of the total DNA, in Hordeum vulgare—76%, in Allium cepa—58%, in Zea mays—55% [20]. In 2018, an article was published on the creation of the PlaNC-TE database (http://planc-te.cp.utfpr.edu.br; accessed on 20 December 2022), according to which, in 40 plant genomes, 14350 miRNAs originated from TEs [21].
The emergence of miRNAs from transposable elements is an important adaptive mechanism of plants, which is necessary for their survival. In the wheat genome, TEs occupy 85% of all nucleotides. Of these, the most prone to domestication into miRNA precursors are MITE. This mechanism of the miRNA generation plays a role in the development of wheat immune responses. Of the 48 miRNA families, 16 have been shown to be derived from TEs [22]. In different rice species, the miR812 family, derived from MITEs, is involved in immunity against fungal infections. These mechanisms involve many genes (such as ACO3, CIPK10, LRR) in the 3′- or 5′-UTR, of which MITEs are located [23]. In the tissues of Arabidopsis sporophytes, small RNAs of 21–22 nucleotides in length were identified, which are transcribed by RNA polymerase-IV from TEs genes. These ncRNAs were involved in the regulation of many plant genes [24]. In 2020, Marakli described 17 new TEDmiRs (in addition to those previously described in PlanTE-MIR DB) that are involved in purine, nitrogen metabolism, oxidative phosphorylation, and other critical plant functions [25]. The appearance of such articles makes it possible to expand the understanding of the role of TEs in the emergence of miRNAs and to create more global database systems for determining the mechanisms of epigenetic regulation of plant and animal ontogenesis.
3. The Role of Transposons in the Emergence of miRNA in Animals
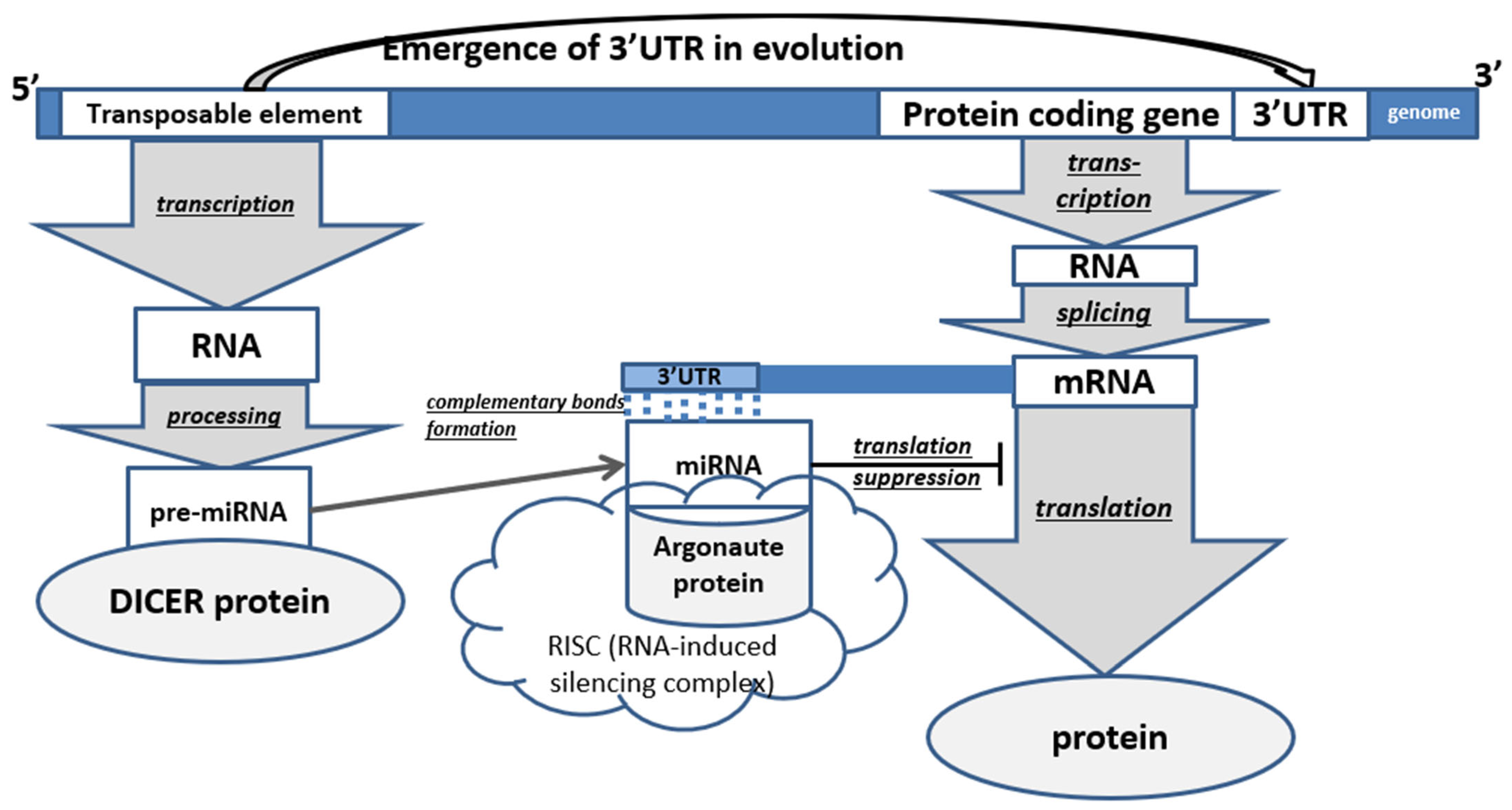
In animals, the main majority of all TEs are non-LTR retroelements (LINE and SINE). In humans, they occupy 35% of all DNA, in mice—28%, in Drosophila—17% [20]. miRNA precursor maturation (by cutting out a parts) is caused by specific enzymes; it initially occurs in the cell nucleus with the help of Drosha ribonuclease-III, after which the RNA is transported into the cytoplasm (with the help of Exportin-5) where it is acted upon by the DICER enzyme [16]. In animals, miRNAs interact with the 3′-UTRs of target mRNAs through partial base pairing (nucleotides 2 to 7 of miRNAs). Binding and interaction of microRNAs with the 3′-UTR lead to the repression of gene expression [2]. 3′-UTR of genes are characterized by the presence of TE residues in them, which form a mutually regulatory system since they become targets for miRNAs derived from their related TEs. As a result, a complex regulatory epigenetic gene network is formed that controls the development of the body (Figure 2) [26].
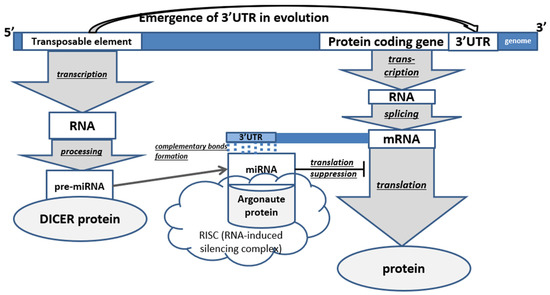
Figure 2.
Scheme of the role of transposable elements in post-transcriptional regulation of protein-coding genes in animals (PCG—protein-coding gene). The underlined words in the figure represent processes.
Evidence for the emergence of miRNAs from TEs in animals has been obtained in numerous studies. For the first time, back in 2005, Smalheiser and Torvik described a model for the formation of miRNAs from TEs sequences in mice, rats, and humans through the formation of hairpin DNA structures between two TEs [27]. In 2006, the results of the analysis of a miRNA cluster on human chromosome 19 were presented, according to which miRNAs are dispersed among Alu-retroelements (referred to as SINE). At least 30 different miRNAs were found to be complementary to Alu [28]. In 2007, 55 different TEDmiRs were described in humans [29]. In 2009, data on 7 TEDmiRs were published in the marsupial Monodelphis domestiva [30]. In the same year, 73 miRNAs transcribed from Alu or MIR in humans were characterized using computer modeling. The role of TEs was shown not only as sources of miRNAs, but also as regulators of their expression in time and space during the development of the organism. Retroelements Alu serve as the basis for the transcriptional regulation of certain miRNA genes [10]. Similar data were obtained in the study of piRNA and miRNA derived from TEs at the early stages of embryonic development. These ncRNAs affected the mRNA of genes involved in key pathways in the regulation of embryogenesis (including the Wnt and TGF-β genes) [31].
In 2010, data on the miR-1302 family, derived from MER53 transposons in humans, were published [32]. In 2011, an article was published about 226 TEDmiR in humans, 141 TEDmiR in mice, and 115 TEDmiR in rhesus monkeys. The authors noted a species-specific expansion of miRNA families, associated with evolutionary transpositions of certain TEs, with large segmental duplications of genomic loci [33]. In 2011, the results of the analysis of more than 15176 individual miRNAs in different animal species were described with the identification of 2392 TEDmiRs [34]. In the same year, a new approach was described for identifying miRNA targets. For this approach, the authors used the analysis of transcripts containing TEs—miRNA precursors. The method was named Orbld (Origin-based identification of miRNA targets). It helped identify targets for 191 TEDmiRs [35]. In 2012, the origin of 182 miRNAs, 788 siRNAs, and 4990 piRNAs from TEs was described in the silkworm [36]. In the same year, the mapping of all miRNA precursors from the miRBASE database, with the determination of the repetitive elements of the genomes overlapping these regions, was reported. The ncRNAclassifier method was developed to classify pre-ncRNAs arising from TEs, and 235 human TEDmiRs and 68 mouse TEDmiRs were described [37].
In 2013, genes of 1213 miRNAs in different eukaryotic genomes were studied, of which 1007 (83%) were derived from various TEs (467 from DNA transposons, 235 from LTR-RE, 186 from LINE, 119 from SINE). They identified primate-specific expansions in the miR-151, -378, -6130, -6127, -1260, -548, -4536, and -1273, including 45 human loci [38]. In 2014, using the RepeatMasker program, the GENCODE v.19 database was analyzed and 1900 TEDmiRs were discovered, of which 406 were previously described by other authors [39]. In bats, unlike other animals, DNA-transposons have the highest activity, in which are rich sources of most TEDmiRs. Among all miRNAs, TEDmiRs in bats account for 61%, which is a significant proportion compared to dogs (24%) and horses (17%) [40]. In 2016, data on the detection of 409 TEDmiRs in humans were published [41]. In 2016, an attempt was made to create an MDTE database (miRNAs derived from TEs) of miRNAs derived directly from TEs. Database address: http://bioinf.njnu.edu.cn/MDTE/MDTE.php (accessed on 20 December 2022). This database describes 1251 miRNAs derived from 30 TEs families in humans and 6 animal species (bull, house mouse, chicken, rhesus monkey, common chimpanzee, gray rat) [12]. However, at present, this database is not available, which indicates the relevance of creating a universal online database of miRNAs derived from TEs.
4. Prospects for the Creation of a Human Transposable Elements-Derived miRNA Database
It can be assumed that the majority of animal miRNAs evolved from TEs since transposons are characterized by high mutability during domestication, which causes difficulties in determining the belonging of TEs sequences [42]. Of greatest interest is the study of TEDmiRs in humans, which are associated with severe diseases, since this will reveal the key pathways of disease pathogenesis and, in the future, design targeted methods using ncRNAs targeting TEs. For example, in 2020, a bioinformatic analysis was carried out to find such TEDmiRs using the TransmiR v.2.0 database. A total of 51 specific miRNAs derived from TEs were identified, of which 34 are associated with various human pathologies [43]. Indeed, miRNAs are promising targets for the targeted therapy of various diseases, which is especially important for malignant neoplasms and idiopathic diseases (when the etiology and pathogenesis have not yet been established). In this regard, on the basis of the data presented in the scientific literature by various authors [12,29,32,33,35,37,39,41,43], data were collected on 467 miRNAs derived from transposable elements (Table 1). miRNAs are used to predict tumor formation and outcome. For this, appropriate bioinformation systems are used, such as OncomiR, an online resource for changes in miRNA regulation in malignant neoplasms, which is freely available at www.oncomir.org (accessed on 20 December 2022) [44]. Analysis of this resource using 467 miRNAs derived from transposable (Table 1) allowed me to identify 52 TEDmiRs, in which changes in the expression are characteristic of specific types of malignant tumors [44]. In order to find aging-associated microRNAs derived from transposons, a search was made for the association of 52 TEDmiRs, associated with cancer, with aging in the databases Scopus, WoS, and NCBI. I introduced phrases of specific miRNAs with the words “aging”, “change with age”, “senescence”, and “consenescence” into the search line.

Table 1.
Human miRNAs derived from transposons and their involvement in aging, carcinogenesis, and idiopathic pulmonary fibrosis.
Pathological activation of TEs is characteristic of both human aging and the development of malignant neoplasms, while aging is a risk factor for most types of cancer [45]. Therefore, the scientific literature was analyzed to search for an association with aging of the 52 TEDmiRs, the expression of which changes in malignant neoplasms. This would allow finding common epigenetic relationships between cancer and aging. In the long term, the results obtained could become the basis for a targeted effect on the mechanisms of aging in order to prevent the development of cancer. In my search, 16 of the 52 TEDmiRs (miR-151a, miR-192, miR-211, miR-28, miR-31, miR-335, miR-340, miR-378a, miR-450b, miR-487b, miR-495, miR-511, miR-576, miR-585, miR-708, miR-885) analyzed were found to be associated with aging (Table 1). Aging is characterized by a significant decrease in the level of miR-151a in the blood of healthy people [46], while the expression of miR-192 in the kidneys is significantly increased [47]. Comparison of centenarians with people from families with low life expectancy revealed a significant increase in miR-211 expression in centenarians, which was proposed to be used as a biomarker of aging [48]. A significant decrease in the level of miR-28 expression has been shown in the elderly [49]. Increased expression of miR-31 was revealed during replicative aging [50]. This miRNA is a target of histone deacetylators in both malignant neoplasms and aging [51]. The role of miR-335 was identified in human aging and in age-related neurological diseases [52]. Quantitative transcriptional reverse PCR analysis reveals the role of miR-340 in aging [53]. Estrogen-sensitive miR-378a is involved in the aging mechanisms of the human thymus, as confirmed in experiments on mice [54]. Disruption of miR-450b regulation in cellular senescence, caused by endogenous genotoxic stress, was found [55]. The involvement of miR-487b in the aging of skeletal muscle has been determined [56]. MiR-495 induces senescence of mesenchymal stem cells [57], and expression of miR-511 changes during aging of the nervous system [58].
More enrichment of miR-576 was found in blood plasma exosomes of the elderly compared with young people [59]. miR-487b can be used as a target for targeted therapy of aging-related muscle atrophy, which directly interacts with the long ncRNA MAR1 (muscle anabolic regulator 1) [56]. Oxidative stress contributes to aging and the development of cardiovascular and neurodegenerative diseases. It was found that miR-585 regulates the PARP-1 gene (poly- (ADP-ribose) polymerase 1), the product of which is involved in the repair of oxidatively damaged DNA. Overexpression of this miRNA increases DNA damage and suppresses cell survival [60]. As a result of the study of miRNA expression in Parkinson’s disease, it was proposed to use miR-885 as a biomarker of human aging and cellular senescence [61]. Experiments on mice have shown the role of miR-450b in aging [55], as well as a decrease in miR-511 expression during aging [58]. The study of 521 different miRNAs in 6 strains of mice with different average lifespans revealed a significant association of three miRNAs, including miR-708 [62], whose expression changes in specific human cancers [44].
An aging-associated disease, the etiology and exact pathogenesis of which is still unknown, is idiopathic pulmonary fibrosis, with an average global prevalence of 2–29 per 100,000 population [63]. Aging increases the risk of developing idiopathic pulmonary fibrosis from 4 per 100,000, for people aged 18–34, to 227.2 per 100,000, for people 75 and older. The average age of idiopathic pulmonary fibrosis patients is 66 years [64]. Survival with idiopathic pulmonary fibrosis is about 3 years, and available drugs only slow down the decline in lung function with little or no effect on mortality [65]. Therefore, it is important to search for the mechanisms of etiopathogenesis of idiopathic pulmonary fibrosis, the results of which could become the basis for creating criteria for the diagnosis and targeted treatment of the disease. The scientific literature was analyzed to look for a change in 467 TEDmiRs in idiopathic pulmonary fibrosis, with the result that identified 12 of these TEDmiRs that are associated with IPF [66,67,68,69,70,71]. Moreover, 9 of them (miR-31, miR-326, miR-335, miR-340, miR-374a, miR-487b, miR-493, miR-495, miR-708) are associated with cancer, and miR-31, miR-335, miR-340, miR-387b, miR-495, and miR-708 are associated with both cancer [44] and aging [50,51,52,53,56,57,62] (Table 1).
The limitations in the methodology are due to the need to search for articles in the Scopus, WoS, and NCBI databases, with their careful analysis and interpretation of the results, which are placed in a table in the form of conclusions. Limitations are due to the limited number of articles reporting associations of TEDmiRs with specific diseases and aging. Thus, when searching for aging-associated TEDmiRs, out of 467, only 52 were associated with malignant neoplasms and 12 with idiopathic pulmonary fibrosis. Prospects for creating a database of miRNAs derived from transposable and their role in the development of diseases are due to the possibility of designing targeted therapy. These miRNAs can be used as targets, and anti-miRNA, miRNA mimic, or peptides formed from their pri-miRNAs (called miPEP) can be tools for influencing them. Currently, scientific articles have already described works on the use of such molecules for the development of antitumor therapy. For example, the systemic delivery of anti-miRNA oligonucleotides (AMO) against miR-181a (oncomiR) in nude mice bearing chondrosarcoma xenografts prolonged survival from 23% to 45%, decreased tumor volume by 32% at day 38, restored RGS16 expression, and decreased MMP activity [72]. Anti-miRNA was applied against miR-21 for triple-negative breast cancer cells, which demonstrated high efficacy for tumor growth inhibition [73]. For pancreatic cancer cells, anti-miRNA-27a inhibited apoptosis and cell growth via Wnt/β-catenin pathway [74]. AMO against miRNA-221, with AMO-loaded exosomes, was described as an effective antitumor tool for colorectal carcinoma [75].
miPEP is translated from pri-miRNA, despite having a short reading frame. They are involved in carcinogenesis, like mature miRNAs formed by processing their pri-miRNAs. Since peptides are more stable molecules than miRNAs or AMOs, the use of miPEP is promising in anticancer therapy. For example, miR-200a encodes miPEP-200a and miR-200b encodes miPEP-200b, which suppresses epithelial-to-mesenchymal transition and thus, inhibits prostate cancer cells. MiR-200a and miR-200b are also implicated in epithelial-to-mesenchymal transition [76]. pri-miRNA-155 encodes miPEP155 that suppresses autoimmune inflammation [77]. It should be noted that some miPEP was shown to promote a positive autoregulatory loop. For example, miPEP133 (tumor-suppressor microprotein) is involved in the regulation of the expression of miR-34a [78].
Regulation of the expression of TEs is possible by means of miRNAs (derived from TEs since they are complementary to their sequences) at the transcriptional level. This is possible due to the phenomenon of RdDM [7]. This will increase the lifespan of people since the pathological expression of TEs is the cause of aging [79]. In addition, the use of miRNAs, complementary to specific TEs, will allow regulating their activity in cancer treatment since the role of pathological activation of TEs in carcinogenesis has been proven [80,81]. It is important to note that Tes are sources of lncRNAs that can serve as pri-miRNAs, with the ability to be translated on ribosomes to form peptides and be processed into miRNAs. Moreover, both formed functional molecules are characterized by participation in the same biological reactions. This indicates the importance of studying the relationship of TEs with lncRNAs and miRNAs. For example, lncRNA MIR22HG (activated in response to chemical stress) is transcribed into pri-miRNA-22, which is translated into a 9 kDa peptide involved in the antiviral response [82]. lncRNA MIR497HG is transcribed into pri-miR-497, which is further processed into two mature miRNAs: miR-497 and miR-195. At the same time, pri-miR-497 is translated into miPEP497 with an oncosuppressive function [83].
5. Conclusions
The identification of miRNAs derived from TEs is the basis for determining the regulatory mechanisms through which transposons exert global control over the functioning of genomes. It will allow designing possible ways of influencing physiological and pathological processes in the body, which is promising for the development of modern genetics and medicine. Therefore, it is necessary to create a universal, replenished online database of transposon-derived miRNAs. The scientific literature was analyzed and 467 specific transposon-derived miRNAs, which could form the basis for creating such an online database, were found. The analysis of the data presented in Table 1 made it possible to determine 52 different miRNAs derived from transposons, which are associated with specific malignant neoplasms. Moreover, it was found that 16 of these 52 miRNAs (miR-151a, miR-192, miR-211, miR-28, miR-31, miR-335, miR-340, miR-378a, miR-450b, miR-487b, miR-495, miR-511, miR-576, miR-585, miR-708, miR-885) are also associated with aging, 9 are associated with idiopathic pulmonary fibrosis, and 6 of them (miR-708, miR-495, miR-487b, miR-340, miR-335, miR-31) are associated with both malignant neoplasms and aging. Since TEs are involved in the global regulation of various body functions, my results can be further used to develop diagnostic algorithms for the diagnosis and targeted therapy of aging-associated diseases, such as malignant neoplasms and idiopathic pulmonary fibrosis. miRNAs derived from transposable elements or oligonucleotides antisense, as well as specific peptides formed during translation of pri-miRNAs, can be used as tools for such targeted therapy.
An analysis of the results presented in the table on the origin of miRNAs from transposons in humans showed that miRNAs are most often formed from LINE elements (108 miRNAs) and SINE elements (94 miRNAs) and less often from DNA transposons (64 miRNAs) and LTR-containing retroelements (53 miRNAs). Since, according to the results of the study (Table 1), the main sources of microRNAs in humans are LINE elements, we analyzed the scientific literature on the role of LINEs in the regulation of embryonic development, in which microRNAs play an important role [84,85,86]. In 2000, Wei et al. described the accumulation of multiple LINE1 insertions in human cell cultures [87]. In 2007, Garcia-Perez et al. revealed the accumulation of LINE1 insertions in human embryonic stem cells, which was accompanied by the suppression of the activity of specific genes required for cell differentiation. On the basis of the obtained data, the researchers suggested that TEs control the work of the genome during the growth and development of organisms [88]. Upon activation of LINE1, their proteins are used to mobilize SINEs. In 2011, Macia et al. reported the expression of several subfamilies of Alu elements in undifferentiated human embryonic stem cells. At the same time, activation of LINE1, located within protein-coding genes which indicates their role in the regulation of these genes, was mainly detected [89]. In addition to tissue cultures, consistent transpositions and activation of LINE1, Alu, and SVA have been identified in vivo during early embryogenesis, during tissue differentiation. These changes caused large-scale structural variations in the genomes of experimental animals. In 2004, Prak et al. showed, in transgenic mouse models, that LINE1 can move in vivo during early development [90]. In 2012, organ-specific and stage-specific changes in cell phenotypes were identified in the C57BL/6J mouse line due to structural transformations of their genomes, which were accompanied by changes in the transcriptional activity of certain ERs [91]. Experiments on two-celled mouse embryos have shown that LINE1 is required for the activation of global gene expression during early embryonic development [92]. LINE1 transcripts themselves function as lncRNAs, interacting with KAP1 and Nucleolin, stimulating rDNA gene expression and silencing other genes in a two-cell embryo by suppressing Dux (a transcription factor that controls the two-cell genetic program) [93].
Author Contributions
R.N.M.: research, processing the results, analysis of scientific literature on the topic, article writing. E.K.: developing the research design, article editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Ministry of Science and Higher Education of Russian Federation Megagrant (075-15-2021-595).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kannan, S.; Chernikova, D.; Rogozin, I.B.; Poliakov, E.; Managadze, D.; Koonin, E.V.; Milanesi, L. Transposable Element Insertions in Long Intergenic Non-Coding RNA. Front. Bioeng. Biotechnol. 2015, 37, 71. [Google Scholar] [CrossRef]
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef]
- Cho, J. Transposon-Derived Non-coding RNAs and Their Function in Plants. Front. Plant Sci. 2018, 9, 600. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C. The contribution of transposable elements to the evolution of regulatory networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar] [CrossRef]
- Seal, R.L.; Chen, L.L.; Griffiths-Jones, S.; Lowe, T.M.; Mathews, M.B.; O’Reilly, D.; Prierce, A.J.; Stadler, P.F.; Ulitsky, I.; Wolin, S.L.; et al. A guide to naming human non-coding RNA genes. EMBO J. 2020, 39, e103777. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Tian, J.; Mo, B. siRNA-mediated DNA methylation and H3K9 dimethylation in plants. Protein Cell 2013, 4, 656–663. [Google Scholar] [CrossRef]
- Chalertpet, K.; Pin-On, P.; Aporntewan, C.; Patchsung, M.; Ingrungruanglert, P.; Israsena, N.; Mutirangura, A. Argonaute 4 as an Effector Protein in RNA-Directed DNA Methylation in Human Cells. Front. Genet. 2019, 10, 645. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, U.; Srivastava, S.; Khatri, I.; Nanda, J.S.; Subramanian, S.; Arora, A.; Singh, J. Ablation of RNA interference and retrotransposons accompany acquisistion and evolution of transposases to heterochromatin protein CENPB. Mol. Biol. Cell 2017, 28, 1132–1146. [Google Scholar] [CrossRef]
- Samantarrai, D.; Dash, S.; Chhetri, B.; Mallick, B. Genomic and epigenomic cross-talks in the regulatory landscape of miRNAs in breast cancer. Mol. Cancer Res. 2013, 11, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.J.; Yi, X.; Zhao, X.W.; Zhao, Y.; Yin, J.Q. Alu-directed transcriptional regulation of some novel miRNAs. BMC Genom. 2009, 10, 563. [Google Scholar] [CrossRef]
- Lorenzetti, A.P.R.; de Antonio, G.Y.A.; Paschoal, A.R.; Domingues, D.S. Plant TE-MIR DB: A database for transposable element-related microRNAss in plant genomes. Funct. Integr. Genom. 2016, 16, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Qin, S.; Li, W.; Chen, L.; Ma, F. MDTE DB: A database for microRNAs derived from Transposable element. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016, 13, 1155–1160. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Morgado, L.; Johannes, F. Computational tools for plant small RNA detection and categorization. Brief. Bioinform. 2019, 20, 1181–1192. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zhoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Pashkovskiy, P.P.; Ryazansky, S.S. Biogenesis, evolution, and functions of plant microRNAs. Biochemistry 2013, 78, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, C.; Xia, J.; Jin, Y. Domestication of transposable elements into microRNA genes in plants. PLoS ONE 2011, 6, e19212. [Google Scholar] [CrossRef]
- Yao, C.; Zhao, B.; Li, W.; Qin, W.; Huang, B.; Jin, Y. Cloning of novel repeat-associated small RNAs derived from hairpin precursors in Oryza sativa. Acta Biochim. Biophys. Sin. 2007, 39, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Piriyapongsa, J.; Jordan, I.K. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 2008, 14, 814–821. [Google Scholar] [CrossRef]
- Kubiak, M.R.; Makalowska, I. Protein-coding genes’ retrocopies and their functions. Viruses 2017, 9, 80. [Google Scholar] [CrossRef]
- Pedro, D.L.F.; Lorenzetti, A.P.R.; Dominguez, D.S.; Paschoal, A.R. PlaNC-TE: A comprehensive knowledgebase of non-coding RNAs and transposable elements in plants. Database 2018, 2018, bay078. [Google Scholar] [CrossRef]
- Poretti, M.; Praz, C.R.; Meile, L.; Kalin, C.; Schaefer, L.K.; Schlafli, M.; Widrig, V.; Sanchez-Vallet, A.; Wicker, T.; Bourras, S. Domestication of High-Copy Transposons Underlays the Wheat Small RNA Response to an Obligate Pathogen. Mol. Biol. Evol. 2020, 37, 839–848. [Google Scholar] [CrossRef]
- Campo, S.; Sanuy, F.S.; Ramirez, R.C.; Gomez-Ariza, J.; Baldrich, P.; Campos-Soriano, L.; Soto-Suarez, M.; Segundo, B.S. A novel Transposable element-derived microRNA participates in plant immunity to rice blast disease. Plant Biotechnol. J. 2021, 19, 1798–1811. [Google Scholar] [CrossRef]
- Panda, K.; McCue, A.D.; Slotkin, R.K. Arabidopsis RNA Polymerase IV generates 21-22 nucleotide small RNAs that can participate in RNA-directed DNA methylation and may regulate genes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190417. [Google Scholar] [CrossRef]
- Marakli, S. In silico determination of transposon-derived miRNAs and targets in Aegilops species. J. Biomol. Dyn. 2020, 38, 3098–3109. [Google Scholar] [CrossRef]
- Spengler, R.M.; Oakley, C.K.; Davidson, B.L. Functional microRNAs and target sites are created by lineage-specific transposition. Hum. Mol. Genet. 2014, 23, 1783–1793. [Google Scholar] [CrossRef] [PubMed]
- Smalheiser, N.R.; Torvik, V.I. Mammalian microRNAs derived form genomic repeats. Trends Genet. 2005, 21, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polylmerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Piriyapongsa, J.; Marino-Ramirez, L.; Jordan, I.K. Origin and evolution of human microRNAs from transposable elements. Genetics 2007, 176, 1323–1337. [Google Scholar] [CrossRef]
- Devor, E.J.; Peek, A.S.; Lanier, W.; Samollow, P.B. Marsupial-specific microRNAs evolved from marsupial-specific transposable elements. Gene 2009, 448, 187–191. [Google Scholar] [CrossRef]
- Shao, P.; Liao, J.Y.; Guan, D.G.; Yang, J.H.; Zheng, L.; Jing, Q.; Zhou, H.; Qu, L. Drastic expression change of transposon-derived piRNA-like RNAs and microRNAs in early stages of chicken embryos implies a role in gastrulation. RNA Biol. 2012, 9, 212–227. [Google Scholar] [CrossRef]
- Yuan, Z.; Sun, X.; Jiang, D.; Ding, Y.; Lu, Z.; Gong, L.; Liu, H.; Xie, J. Origin and evolution of a placental-specific microRNA family in the human genome. BMC Evol. Biol. 2010, 10, 346–358. [Google Scholar] [CrossRef]
- Yuan, Z.; Sun, X.; Liu, H.; Xie, J. MicroRNA genes derived from repetitive elements and expanded by segmental duplication events in mammalian genomes. PLoS ONE 2011, 6, e17666. [Google Scholar] [CrossRef]
- Borchert, G.M.; Holton, N.W.; Williams, J.D.; Hernan, W.L.; Bishop, I.P.; Dembosky, J.A.; Elste, J.E.; Gregoire, N.S.; Kim, J.A.; Koehler, W.W.; et al. Comprehensive analysis of microRNA genomic loci identifies pervasive repetitive-element origins. Mob. Genet. Elem. 2011, 1, 8–17. [Google Scholar] [CrossRef]
- Filshtein, T.J.; Mackenzie, C.O.; Dale, M.D.; Dela-Cruz, P.S.; Ernst, D.M.; Tiwari, K.B.; Rubin, D.A.; Borchert, G.M.; Larson, E.D. Orbid: Origin-based identification of microRNA targets. Mob. Genet. Elem. 2012, 2, 184–192. [Google Scholar] [CrossRef]
- Cai, Y.; Zhou, Q.; Yu, C.; Wang, X.; Hu, S.; Yu, J.; Yu, X. Transposable-element associated small RNAs in Bombyx mori genome. PLoS ONE 2012, 7, e36599. [Google Scholar] [CrossRef] [PubMed]
- Tempel, S.; Pollet, N.; Tahi, F. NcRNAclassifier: A tool for detection and classification of transposable element sequences in RNA hairpins. BMC Bioinform. 2012, 13, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.T.; Cooper, E.A.; Favreau, C.J.; Howell, J.S.; Lane, L.G.; Mills, J.E.; Newman, D.C.; Perry, T.J.; Russell, M.E.; Wallace, B.M.; et al. Continuing analysis of microRNA origins: Formation from transposable element insertions and noncoding RNA mutations. Mob. Genet. Elem. 2013, 3, e27755. [Google Scholar] [CrossRef]
- Gim, J.; Ha, H.; Ahn, K.; Kim, D.; Kim, H. Genome-Wide Identification and Classification of microRNAs derived from repetitive elements. Genom. Inf. 2014, 12, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.N.; Vandeweqe, M.W.; Kern, C.; Schmidt, C.J.; Hoffmann, F.G.; Ray, D.A. Large number of novel miRNAs originate from DNA transposons and are coincident with a large species radiation in bats. Mol. Biol. Evol. 2014, 31, 1536–1545. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Jin, P.; Zhou, X.; Chen, L.; Ma, F. The Role of Transposable Elements in the Origin and Evolution of MicroRNAs in Human. PLoS ONE 2015, 10, e0131365. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.T.; Cardin, S.E.; Borcehrt, G.M. Burgeoning evidence indicates that microRNAs were initially formed from transposable element sequences. Mob. Genet. Elem. 2014, 4, e29255. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.E.; Huh, J.W.; Kim, H.S. Bioinformatics Analysis of Evolution and Human Disease Related Transposable Element-Derived microRNAs. Life 2020, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.W.; Chen, Y.; Chen, S.; Wang, X. OncomiR: And online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics 2018, 34, 713–715. [Google Scholar] [CrossRef]
- Pal, S.; Tyler, J.K. Epigenetics and aging. Sci. Adv. 2016, 2, e1600584. [Google Scholar] [CrossRef] [PubMed]
- Noren Hooten, N.; Fitzpatrick, M.; Wood, W.H., 3rd; De, S.; Ejogu, N.; Zhang, Y.; Mattison, J.A.; Becker, K.G.; Zonderman, A.B.; Evas, M.K. Age-related changes in microRNA levels in serum. Aging (Albany NY) 2013, 5, 725–740. [Google Scholar] [CrossRef] [PubMed]
- Sataranatarajan, K.; Feliers, D.; Mariappan, M.M.; Lee, H.J.; Lee, M.J.; Day, R.T.; Yalamanchili, H.B.; Choudhury, G.G.; Barnes, J.L.; Remmen, H.V.; et al. Molecular events in matrix protein metabolism in the aging kidney. Aging Cell 2012, 11, 1065–1073. [Google Scholar] [CrossRef]
- Smith-Vikos, T.; Liu, Z.; Parsons, C.; Gorospe, M.; Ferrucci, L.; Gill, T.M.; Slack, F.J. A serum miRNA profile of human longevity: Findings from the Baltimore Longitudinal Study of Aging (BLSA). Aging (Albany NY) 2016, 8, 2971–2987. [Google Scholar] [CrossRef]
- Zhang, T.; Brinkley, T.E.; Liu, K.; Feng, X.; Marsh, A.P.; Kritchevsky, S.; Zhou, X.; Nicklas, B.J. Circulating miRNAs as biomarkers of gait speed responses to aerobic exercise training in obese older adults. Aging (Albany NY) 2017, 9, 900–913. [Google Scholar] [CrossRef]
- Dellago, H.; Preschitz-Kammerhofer, B.; Terlecki-Zaniewicz, L.; Schreiner, C.; Grillari, J.; Wieser, M. High levels of oncomiR-21 contribute to the senescence-induced growth arrest in normal human cells and its knock-down increases the replicative lifespan. Aging Cell 2013, 12, 446–458. [Google Scholar] [CrossRef]
- Cho, J.H.; Dimri, M.; Dimri, G.P. MicroRNA-31 is a transcriptional target of histone deacetylase inhibitors and a regulator of cellular senescence. J. Biol. Chem. 2015, 290, 10555–10567. [Google Scholar] [CrossRef]
- Raihan, O.; Brishti, A.; Molla, M.R.; Li, W.; Zhang, Q.; Xu, P.; Khan, M.I.; Zhang, J.; Liu, Q. The Age-dependent Elevation of miR-335-3p Leads to Reduced Cholesterol and Impaired Memory in Brain. Neuroscience 2018, 390, 160–173. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, H.; Zhang, C.; Jing, Y.; Wang, C.; Liu, C.; Zhang, R.; Wang, J.; Zhang, J.; Zen, K.; et al. Investigation of microRNA expression in human serum during the aging process. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 102–109. [Google Scholar] [CrossRef]
- Guo, D.; Ye, Y.; Qi, J.; Tan, X.; Zhang, Y.; Ma, Y.; Li, Y. Age and sex differences in microRNAs expression during the process of thymus aging. Acta Biochim. Biophys. Sin. 2017, 49, 409–419. [Google Scholar] [CrossRef]
- Nidadavolu, L.S.; Niedernhofer, L.J.; Khan, S.A. Identification of microRNAs dysregulated in cellular senescence driven by endogenous genotoxic stress. Aging (Albany NY) 2013, 5, 460–473. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Li, J.; Guan, D.; Liang, C.; Zhuo, Z.; Liu, J.; Lu, A.; Zhang, G.; Zhang, B. A newly identified lncRNA MaR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia Sarcopenia Muscle 2018, 9, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, Y.; Liu, D.; Zhao, J.; Xu, J.; Ren, J.; Hu, Y.; Wang, Z.; Hou, Y.; Zhao, G. MiR-495 Promotes Senescence of Mesenchymal Stem Cells by Targeting Bmi-1. Cell Physiol. Biochem. 2017, 42, 780–796. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Sabbagh, J.J.; Blair, L.J.; Darling, A.L.; Wen, X.; Dickey, C.A. MicroRNA-511 Binds to FKBP5 mRNA, Which Encodes a Chaperone Protein, and Regulates Neuronal Differentiation. J. Biol. Chem. 2016, 291, 17897–17906. [Google Scholar] [CrossRef] [PubMed]
- Ipson, B.R.; Fletcher, M.B.; Espinoza, S.E.; Fisher, A.L. Identifying Exosome-Derived MicroRNAs as Candidate Biomarkers of Frailty. J. Frailty Aging 2018, 7, 100–103. [Google Scholar] [CrossRef]
- Dluzen, D.F.; Kim, Y.; Bastian, P.; Zhang, Y.; Lehrmann, E.; Becker, K.G.; Hooten, N.N.; Evas, M.K. MicroRNAs Modulate Oxidative Stress in Hypertension through PARP-1 Regulation. Oxid. Med. Cell. Longev. 2017, 2017, 3984280. [Google Scholar] [CrossRef]
- Behbahanipour, M.; Peymani, M.; Salari, M.; Hashemi, M.; Nasr-Esfahani, M.H.; Ghaedi, K. Expression Profiling of Blood microRNAs 885, 361, and 17 in the Patients with the Parkinson’s disease: Integrating Interatction Data to Uncover the Possible Triggering Age-Related Mechanisms. Sci. Rep. 2019, 9, 13759. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.P.; Buric, I.; George-Pandeth, A.; Flurkey, K.; Harrison, D.E.; Yuan, R.; Peters, L.L.; Kuchel, G.A.; Melzer, D.; Harries, L.W. MicroRNAs miR-203-3p, miR-664-3p and miR-708-5p are associated with median strain lifespan in mice. Sci. Rep. 2017, 7, 44620. [Google Scholar] [CrossRef]
- Zhao, J.; Ren, Y.; Qu, Y.; Jiang, W.; Lv, C. Pharmacodynamic and pharmacokinetic assessment of pulmonary rehabilitation mixture for the treatment of pulmonary fibrosis. Sci. Rep. 2017, 7, 3458. [Google Scholar] [CrossRef]
- Raghu, G.; Weycker, D.; Edelsberg, J.; Bradford, W.Z.; Oster, G. Incidence and prevalence of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2006, 174, 810–816. [Google Scholar] [CrossRef]
- Wyman, A.E.; Noor, Z.; Fishelevich, R.; Lockatell, V.; Shah, N.G.; Todd, N.W.; Atamas, S.P. Sirtuin 7 is decreased in pulmonary fibrosis and regulates the fibrotic phenotype of lung fibroblasts. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 312, L945–L958. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yang, L.; Wang, W.; Wang, J.; Wang, J.; Xu, Z. Discovery and validation of extracellular/ circulating microRNAs during idiopathic pulmonary fibrosis disease progression. Gene 2015, 562, 138–144. [Google Scholar] [CrossRef]
- Huang, C.; Xiao, X.; Yang, Y.; Mishra, A.; Liang, Y.; Zeng, X.; Yang, X.; Xu, D.; Blackburn, M.R.; Henke, C.A.; et al. MicroRNA-101 attenuates pulmonary fibrosis by inhibiting fibroblast proliferation and activation. J. Biol. Chem. 2017, 292, 16420–16439. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, Y.; Song, X.; Sun, W.; Zhang, J.; Liu, Y.; Li, H.; Meng, C.; Zhang, J.; Zheng, Q.; et al. Potential regulatory role of circular RNA in idiopathic pulmonary fibrosis. Int. J. Mol. Med. 2018, 42, 3256–3268. [Google Scholar] [CrossRef]
- Liu, G.; Friggeri, A.; Yang, Y.; Milosevic, J.; Ding, Q.; Thannickal, V.J.; Kaminski, N.; Abraham, E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010, 207, 1589–1597. [Google Scholar] [CrossRef]
- Wei, Y.Q.; Guo, Y.F.; Yang, S.M.; Li, J. MiR-340-5p mitigates the proliferation and activation of fibroblast in lung fibrosis by targeting TGF-β/p38/ATF1 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6252–6261. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Gu, L.N.; Qi, J.; Xia, Q.; Tian, L.; Jiang, W.; Cao, M. Construciton of potential idiopathic pulmonary fibrosis related microRNA and messenger RNA regulatory network. Chin. Med. J. 2021, 134, 584–586. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Chen, Y.; Yu, H.; Machan, J.T.; Alladin, A.; Ramirez, J.; Taliano, R.; Hart, J.; Chen, Q.; Terek, R.M. Anti-miRNA Oligonucleotide Therapy for Chondrosarcoma. Mol. Cancer Ther. 2019, 18, 2021–2029. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Xiong, G.; Guo, S.; Xu, C.; Xu, R.; Guo, P.; Shu, D. Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133. Mol. Ther. 2019, 27, 1252–1261. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, G.; Kong, D. miRNA-27a promotes the proliferation and inhibits apoptosis of human pancreatic cancer cells by Wnt/β-catenin pathway. Oncol. Rep. 2018, 39, 755–763. [Google Scholar] [CrossRef]
- Han, S.; Li, G.; Jia, M.; Zhao, Y.; He, C.; Huang, M.; Jiang, L.; Wu, M.; Yang, J.; Ji, X.; et al. Delivery of Anti-miRNA-221 for Colorectal Carcinoma Therapy Using Modified Cord Blood Mesenchymal Stem Cells-Derived Exosomes. Front. Mol. Biosci. 2021, 20, 8, 743013. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Morsalin, S.; Rao, V.N.; Reddy, E.S.P. Decoding of Non-Coding DNA and Non-Coding RNA: Pri-Micro RNA-Encoded Novel Peptides Regulate Migration of Cancer Cells. J. Pharm. Sci. Pharmacol. 2017, 3, 23–27. [Google Scholar] [CrossRef]
- Niu, L.; Lou, F.; Sun, Y.; Sun, L.; Cai, X.; Liu, Z.; Zhou, H.; Wang, H.; Wang, Z.; Bai, J.; et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci. Adv. 2020, 6, eaaz2059. [Google Scholar] [CrossRef]
- Kang, M.; Tang, B.; Li, J.; Zhou, Z.; Liu, K.; Wang, R.; Jiang, Z.; Bi, F.; Patrick, D.; Kim, D.; et al. Identification of miPEP133 as a novel tumor-suppressor microprotein encoded by miR-34a pri-miRNA. Mol. Cancer 2020, 19, 143. [Google Scholar] [CrossRef]
- Gorbunova, V.; Seluanov, A.; Mita, P.; McKerrow, W.; Fenyo, D.; Boeke, J.D.; Linker, S.B.; Gage, F.H.; Kreiling, J.A.; Petrashen, A.P.; et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature 2021, 596, 43–53. [Google Scholar] [CrossRef]
- Burns, K.H. Transposable elements in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef]
- Jansz, N.; Faulkner, G.J. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef]
- Razooky, B.S.; Obermayer, B.; O’May, J.B.; Tarakhovsky, A. Viral Infection Identifies Micropeptides Differentially Regulated in smORF-Containing lncRNAs. Genes 2017, 8, 206. [Google Scholar] [CrossRef]
- Prel, A.; Dozier, C.; Combier, J.P.; Plaza, S.; Besson, A. Evidence That Regulation of Pri-miRNA/miRNA Expression Is Not a General Rule of miPEPs Function in Humans. Int. J. Mol. Sci. 2021, 22, 3432. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wang, S.; Wang, Z. Role of microRNAs in embryo implantation. Reprod. Biol. Endocrinol. 2017, 15, 90. [Google Scholar] [CrossRef] [PubMed]
- McJunkin, K. Maternal effects of microRNAs in early embryogenesis. RNA Biol. 2018, 15, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Muraoka, M.; Okada, H.; Toyoda, A.; Ajima, R.; Saga, Y. The RNA helicase DDX6 controls early mouse embryogenesis by repressing aberrant inhibition of BMP signaling through miRNA-mediated gene silencing. PLoS Genet. 2022, 18, e1009967. [Google Scholar] [CrossRef]
- Wei, W.; Morrish, T.A.; Alisch, R.S. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal. Biochem. 2000, 284, 435–438. [Google Scholar] [CrossRef]
- Garcia-Perez, J.L.; Marchetto, M.C.; Muotri, A.R.; Coufal, N.G.; Gage, F.H.; O’Shea, K.S.; Moran, J.V. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 2007, 16, 1569–1577. [Google Scholar] [CrossRef]
- Macia, A.; Munoz-Lopez, M.; Cortes, J.L.; Hastings, R.K.; Morell, S.; Lucena-Aguilar, G.; Marchal, J.A.; Badge, R.M.; Garcia-Perez, J.L. Epigenetic control of retrotransposons expression in human embryonic stem cells. Mol. Cell. Biol. 2011, 31, 300–316. [Google Scholar] [CrossRef]
- Prak, E.T.; Dodson, A.W.; Farkash, E.A.; Kazazian, H.H., Jr. Tracking an embryonic L1 retrotransposition event. Proc. Nat. Acad. Sci. USA 2003, 100, 1832–1837. [Google Scholar] [CrossRef]
- Lee, K.H.; Chiu, S.; Lee, Y.K.; Greenhalgh, D.G.; Cho, K. Age-dependent and tissue-specific structural changes in the C57BL/6J mouse genome. Exp. Mol. Pathol. 2012, 93, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Jachowicz, J.W.; Bing, X.; Pontabry, J.; Boskovic, A.; Rando, O.J.; Torres-Padilla, M. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 2017, 49, 1502–1510. [Google Scholar] [CrossRef] [PubMed]
- Honson, D.D.; Macfarlan, T.S. A lncRNA-like Role for LINE1s in Development. Dev. Cell 2018, 46, 132–134. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).