Abstract
Soil salinization inhibits plant growth and seriously restricts food security and agricultural development. Excessive salt can cause ionic stress, osmotic stress, and ultimately oxidative stress in plants. Plants exclude excess salt from their cells to help maintain ionic homeostasis and stimulate phytohormone signaling pathways, thereby balancing growth and stress tolerance to enhance their survival. Continuous innovations in scientific research techniques have allowed great strides in understanding how plants actively resist salt stress. Here, we briefly summarize recent achievements in elucidating ionic homeostasis, osmotic stress regulation, oxidative stress regulation, and plant hormonal responses under salt stress. Such achievements lay the foundation for a comprehensive understanding of plant salt-tolerance mechanisms.
1. Introduction
Soil salinization is a major adverse environmental stress that seriously restricts plant growth and development and affects crop yields [1]. Globally, one-third of agricultural land (~950 million hectares worldwide, including 250 million hectares of irrigated land) is affected by salt stress [2]. Plants are classified as halophytic or glycophytic according to their salt tolerance. Most crops are glycophytic and sensitive to high salt concentrations in the soil. Improper use of fertilizers, excess irrigation, and industrial pollution are the primary causes of widespread soil salinity [3,4], which poses a serious threat to agricultural productivity and food security for both humans and livestock. Developing crops that can grow normally in salinized soils is fundamental to solving this problem.
The basic mechanisms of plant salt tolerance have been well studied through the continuous development of research methods and technological innovations and the unremitting efforts of researchers. Many resulting findings elucidating how plants resist and adapt to salt stress at different levels have been applied to agricultural production. High concentrations of Na+ and Cl− in the soil result in both osmotic and ionic stress, which decrease plants’ capacity to take up water and nutrients [5,6,7]. Roots are the frontline organs making contact with salt in the soil. They must adapt to the soil salinity to maintain plant growth and uptake of nutrients and water. Salt stress significantly reduces root mass and modifies the distribution of root system architecture components, differentially affecting the growth rate of the primary root and lateral roots and inhibiting lateral root formation. Aboveground tissues are also limited by salt stress, but the molecular mechanisms behind this are less well studied than those for roots. Plants have evolved a series of mechanisms to mitigate the effects of stressful conditions on growth. Therefore, understanding plant tolerance mechanisms is essential for effectively utilizing saline–alkali land and improving crop yields.
High levels of salinity induce ionic stress, osmotic stress, and oxidative stress. Plants employ various physiological and biochemical responses to help maintain ionic homeostasis, proper osmotic potential, and reactive oxygen species (ROS) homeostasis [8].
Great effort has been devoted to understanding the process of salt stress sensing, yielding substantial breakthroughs; however, knowledge of this process remains limited, and it is unclear whether there is a specific Na+ receptor. Ionic homeostasis is disrupted under salt stress. Excluding excess Na+ is a fundamental approach underpinning plant tolerance of salt stress and has been the focus of considerable research. The SALT OVERLY SENSITIVE (SOS) pathway plays a central role in excluding excess Na+ from plant cells. Identification of regulators of the SOS pathway has confirmed the central position of this signaling pathway in the salt-tolerance mechanism. A high level of potassium absorption and a high K+/Na+ ratio in plant cells are also important for ionic homeostasis [9,10,11,12]. Several K+ transporters, such as Arabidopsis K+ TRANSPORT 1 (AKT1) and HIGH-AFFINITY K+ TRANSPORTER 1 (HKT1), participate in the regulation of the salt stress response. Osmotic homeostasis helps plants maintain proper cellular morphology and ensures the availability of water. High concentrations of soluble salt repress the water potential at the root surface, thereby decreasing water uptake by plants [13] and leading to water deficit and osmotic stress. Ionic and osmotic stresses can also cause secondary stresses in plants, especially the production of toxic ROS, which severely damage cellular structures and reduce the bioactivity of macromolecules [14,15,16]. Various plant hormone signaling pathways respond to salt stress, helping plants to survive under high-salinity conditions by balancing stress tolerance and plant growth. In this review, we briefly summarize how plants respond to salt stress at various levels.
1.1. Salt Stress Sensing
Stress sensing is the process by which components (molecules/proteins) known as “sensors” [17] detect changes in the external environment and launch appropriate responses. During saline stress sensing, the calcium signaling response, i.e., the “secondary signaling process” of salt stress, occurs within a timescale of seconds, and excessive Na+ starts to be excluded from the root tissue 10 min after exposure to high salinity [18]. These findings suggest that salinity-induced ionic stress is rapidly perceived by plants, initiating the regulation of ionic homeostasis. The detrimental effects of salinity on plant performance often take several hours to manifest after NaCl exposure, suggesting that plants employ different processes for long-term saline-stress sensing.
Sodium ions are initially perceived by sensors localized on the plasma membrane (PM). Saline stress induces ionic stress that results in changes to the calcium status of the cytosol [8]. Thus, the initiation of salt stress signaling transduction is always associated with the regulation of Ca2+ channels. Glycosyl inositol phosphorylceramides (GIPCs) are a class of lipids abundant in the PM that collectively function as a monovalent-cation sensor by binding with monocations and regulating ionic-stress-induced Ca2+ signaling. Biosynthesis of GIPC is regulated by monocation-induced Ca2+ increases 1 (MOCA1), a glucuronosyltransferase involved in GIPC biosynthesis. The moca1 mutant is sensitive to salt stress and displays a defect in the production of Ca2+ spikes induced by excessive Na+ [19]. GIPC is thought to be an ionic-specific sensor of salt stress; however, GIPC can also bind K+ and Li+ [19]. Whether plants have a more specific sodium sensor needs further research.
AtANNEXIN4 (AtANN4) encodes a putative calcium-permeable transporter in Arabidopsis thaliana (Arabidopsis) that interacts with SOS3-LIKE CALCIUM-BINDING PROTEIN8 (SCaBP8) and the kinase SOS2 under salt stress to induce a calcium signal at the beginning of salt stress and initiate the SOS pathway, a conserved salt-stress-tolerance signaling pathway [20,21]. SCaBP8 and activated SOS2 form a complex that phosphorylates ANN4 to repress its calcium-permeable transporter activity, thereby creating a specific calcium signal that functions in the long-term salt stress response [20]. This suggests that the long-term salt stress response is different from the short-term stress response at the molecular level.
Increasing evidence indicates that changes in the plant cell wall participate in sensing salt stress [22,23]. Root tissue cells display a radial swelling when plants are treated with high salinity for 6–8 h [24,25], with the integrity of the plant cell wall changing under high-salinity treatment. In Arabidopsis, the PM-localized leucine-rich repeat receptor kinase (LRR-RK) Male Discoverer 1-Interacting Receptor-Like Kinase 2/Leucine-Rich Repeat Kinase Family Protein Induced by Salt Stress (MIK2) [23], as well as FEI1 and FEI2 (two additional leucine-rich repeat receptor kinases), function in perceiving changes in cell wall integrity as well as in the salt stress response [26]. In Arabidopsis, FERONIA (FER), a PM-localized receptor-like kinase, is required for the recovery of root growth when plants are subjected to high salinity. Pectin polysaccharide in the cell wall interacts with the extracellular domain of FER. High salinity weakens the cell wall and disrupts its integrity. FER perceives these changes and regulates calcium channels to mediate calcium signaling, thereby maintaining root cell morphology during growth under salt stress [25]. The content and composition of cell wall lignin are affected when plants are exposed to biotic and abiotic stresses [27]. SHORT ROOT IN SALT MEDIUM 3 (RSA3)/MURUS3 (MUR3)/KATAMARI1 (KAM1) encodes a xyloglucan galactosyltransferase involved in cell wall biosynthesis in Arabidopsis. KAM1 maintains endomembrane organization and prevents damage to plant cells under salt stress by decreasing cellular ROS accumulation and regulating the expression of stress-related genes [22]. These observations indicate that cell wall integrity status and changes in cell wall components under salt stress trigger downstream signaling, stimulating plant responses and initiating salt-tolerance mechanisms to regulate root growth and morphogenesis for adapting to stress conditions. These processes are typically initiated long after the beginning of salt stress, further confirming that the early response of plants to salt stress is different from their response to long-term salt stress and that plants employ different mechanisms for sensing short-term vs. long-term salt stress.
Saline stress induces both ionic stress and osmotic stress, with half of the stress response genes induced by salt stress also being induced by osmotic stress [28]; this suggests that osmotic perception probably contributes to the process of sensing salt stress. Rice (Oryza sativa) REDUCED HYPEROSMOLALITY-INDUCED CALCIUM INCREASE 1 (OSCA1) is a channel responsible for increases in internal calcium ion concentration induced by a stimulus in plants and is therefore considered to be the osmotic sensor. The osca1 mutant shows defective Ca2+ signaling under osmotic stress in root cells and guard cells, as well as decreased regulation of transpiration and inhibited root growth under osmotic stress [29,30]. Pei and colleagues recently demonstrated that OSCA1 functions as an osmotic-specific sensor [31]. Calcium signaling in the osca1 mutant is reduced more greatly under osmotic stress treatment than under NaCl treatment [30]. These findings confirm that salt-induced osmotic sensing differs mechanistically from ionic stress sensing. However, the osca1.1 mutant is hypersensitive to both sorbitol and NaCl, demonstrating that the osmotic sensor OSCA1.1 indeed contributes to the salt stress response of plants through salinity-induced osmotic perception [32].
1.2. Ion Homeostasis Regulation
Maintaining ionic homeostasis in cells is critical for plants in adapting to the presence of excess ions during salt stress. An appropriate ratio of K+/Na+ is required to maintain low sodium and high potassium levels in the cytoplasm, which prevent cellular damage and nutrient deficiency under salt stress [8,33]. Ionic homeostasis is related to ion transport. Sodium ions are transported into plant cells in a non-selective and non-active manner. No specific transporters responsible for Na+ influx have been identified yet in plant cells; rather, the uptake of Na+ into the cytoplasm is considered to occur passively. Excessive Na+ in the extracellular space induces the generation of an electrochemical potential across the PM, which promotes the movement of Na+ across the PM into the cytoplasm. Membrane potential differences (PDs) exhibit characteristic noise under salinity treatment. Upon salt stress, depolarization of membrane PDs to an excitation threshold sets off successive action potentials, leading to further losses of K+ and Cl− [34].
Sodium ions are probably transported into the cytoplasm via the K+ transport systems. K+ UPTAKE 1 (KUP1) is a dual-affinity K+ transporter responsible for K+ uptake in Arabidopsis [9]. Na+ competes with K+ for uptake by KUP1, suggesting that the influx of Na+ and K+ might occur via similar mechanisms. The low-affinity K+ transporters AKT1 and HKT1 both mediate the movement of Na+ across the PM [33,35,36]. Non-selective cation channels (NSCCs), voltage-independent cation channels (VICs), and non-selective outward-rectifying conductance (NORC) are also involved in the influx of Na+ and K+ into plant cells [35,37].
Unlike Na+ uptake, Na+ efflux is an active process. A PM-localized ATPase activated through Na+ was identified in the unicellular chrysophyte Heterosigma akashiwo [38,39]. PM H+-ATPases power Na+ exclusion in land plants [40,41]. In Arabidopsis, Na+/H+ EXCHANGER1 (NHX1) is a tonoplast-membrane-localized Na+/H+ antiporter responsible for transporting sodium ions from the cytosol into the vacuole [42]. HKT1s are a class of Na+ and K+ transporters with a stronger affinity for Na+ [43]. The xylem-parenchyma-localized transporter AtHKT1;1 is responsible for unloading Na+ from xylem vessels to repress the transport of excess Na+ from root to shoot tissues [44,45,46].
Na+ exclusion from cells is the main approach used by plants to resist salt stress. The Ca2+-dependent Na+ efflux pathway SOS was identified using genetic screens of Arabidopsis sos mutants subjected to salt stress and is the most powerful Na+ efflux pathway in plants (Figure 1). The SOS pathway consists of SOS3, SCaBP8, SOS2, and SOS1. SOS3 and SCaBP8 are helix E–loop–helix F (EF-hand) Ca2+-binding proteins responsible for sensing and decoding calcium signals in the cytosol stimulated by excessive salinity [21,47,48,49]. SOS3 functions primarily in roots, whereas SCaBP8 functions primarily in shoots [21,49]. Both proteins interact with SOS2, the most critical serine/threonine protein kinase in the SOS pathway, activating its kinase activity. Further, SOS2 is recruited to the PM by SOS3 and SCaBP8 for phosphorylation of the Na+/H+ antiporter SOS1 [21,50,51,52,53]. SOS1 is the most important determinant of Na+ transport out of the cytoplasm to the apoplast, which is promoted by a PM H+-ATPase-generated proton gradient [52,54]. The C-terminus of SOS1 is an intracellular domain containing an autoinhibitory domain, while the N-terminus is a transmembrane domain. The transport activity of SOS1 is autoinhibited by intermolecular interaction between these two domains [55]. SOS2 phosphorylates the 1136th and 1138th serines in the C-terminal autoinhibitory domain of SOS1, which relieves the autoinhibition of SOS1, resulting in SOS1 activation [56,57].
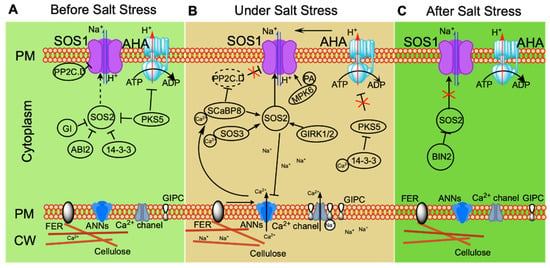
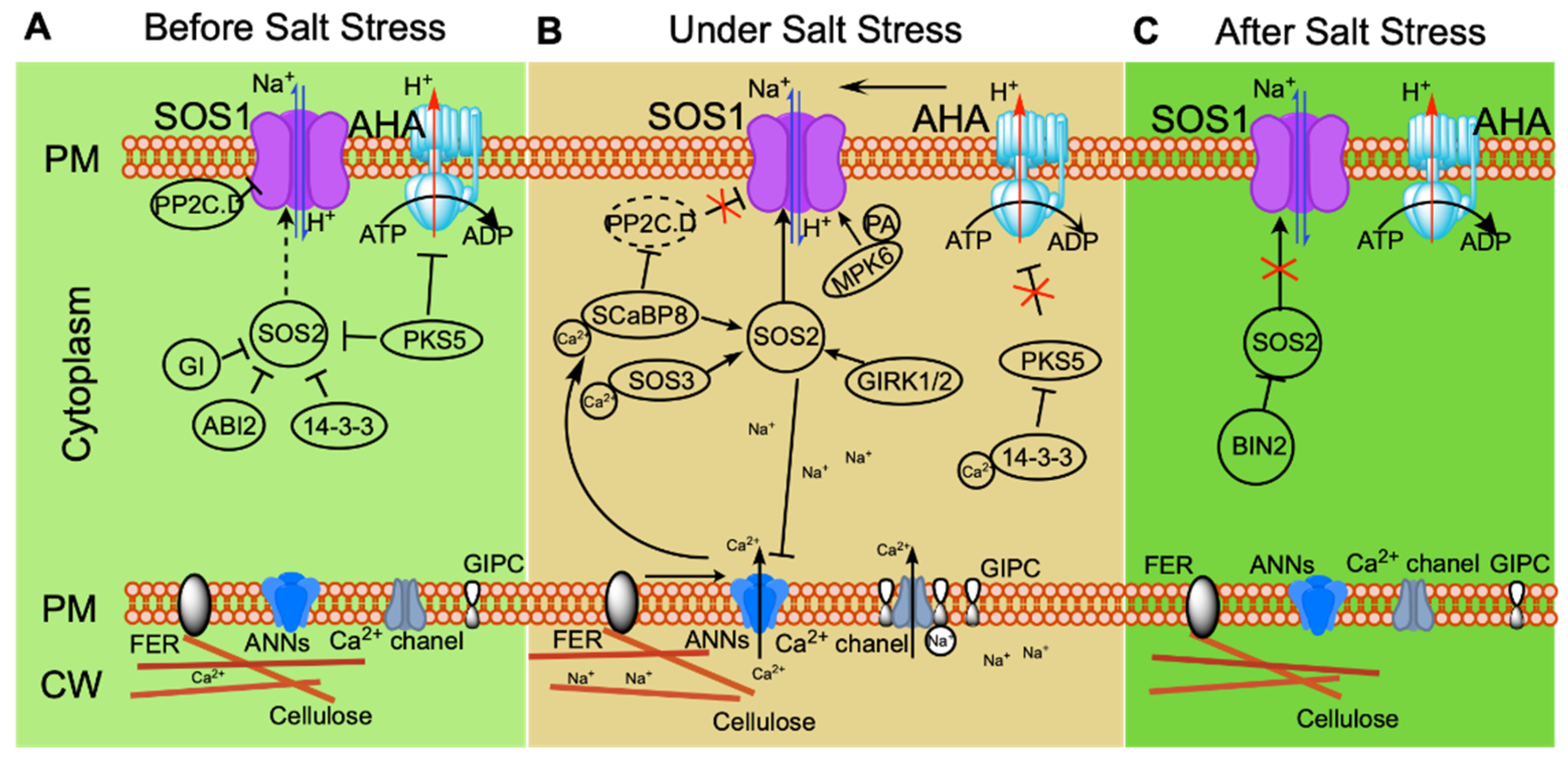
Figure 1.
Ionic-stress signaling pathways that maintain ionic homeostasis and thereby help plants to adapt to salt stress. Under non-stress conditions (before salt stress (A)), plasma membrane (PM) H+-ATPase activity is repressed by PKS5; SOS2 kinase activity is repressed by PKS5, 14-3-3, ABI2, and GI; and SOS1 activity is inhibited by clade D PP2C (PP2C.D). Under salt stress (B), GIPC binds Na+, inducing an increase in calcium signaling. FER perceives changes in the cell wall under long-term salt stress and mediates calcium signaling. The calcium receptors SOS3 and SCaBP8 bind Ca2+, interacting with and activating SOS2, which then phosphorylates SOS1 to activate its Na+/H+ antiporter activity. Salt stress induces PA accumulation, which promotes the kinase activity of MPK6. MPK6 then phosphorylates SOS1 to enhance the activity of SOS1. At the same time, SCaBP8 inhibits PP2C.D to relieve the inhibition of SOS1 by PP2C.D. ANN modulation of calcium signaling under salt stress positively regulates SCaBP8-activated SOS2; under long-term salt stress, SOS2 phosphorylates ANN4 and represses its Ca2+ channel activity, creating a specific calcium signal for long-term stress. After salt stress (C), BIN2 phosphorylates SOS2 and inhibits its kinase activity, helping plants to recover from stress.
Regulation of the SOS pathway has been the subject of extensive research, showing among other things that SOS2 plays an irreplaceable role in the pathway. In Arabidopsis, SOS2 kinase is induced specifically by sodium, but not by potassium or mannitol [51]. Structural analysis suggests that the N-terminus of SOS2 is a kinase domain, while its C-terminus is a regulatory domain that interacts with the kinase domain to cause self-inhibition. A 21-amino-acid sequence in the regulatory domain of SOS2, named the FISL motif (owing to the presence of the conserved residues A, F, I, S, L), is responsible for the binding of SOS2 to SOS3. The kinase activity of SOS2 requires coordinated release of the self-inhibitory FISL motif and the activation loop [58,59]. Under normal conditions, SOS2 kinase activity is inhibited through phosphorylation by PROTEIN KINASE SOS2-LIKE 5 (PKS5). 14-3-3 proteins repress SOS2 activity, and this repression is enhanced by the phosphorylation of SOS2 by PKS5 [60,61]. GIGANTEA (GI) also functions as a repressor of SOS2 kinase activity through interaction with SOS2. When plants experience saline stress, inhibition of SOS2 activity is released by degradation of GI and 14-3-3 through a 26S proteasome pathway [61,62]. Ca2+ signaling is stimulated under salt stress. This is perceived by 14-3-3 proteins that interact with PKS5 and suppress its kinase activity, further relieving the repression of SOS2 kinase activity by 14-3-3 and PKS5 and initiating Na+ efflux in the SOS pathway [60,61]. When salt stress is removed, SOS2 activity is inhibited through phosphorylation by BRASSINOSTEROID-INSENSITIVE 2 (BIN2), a protein kinase functioning as a negative regulator in the brassinosteroid signaling pathway; plants attenuate stress responses and focus on growth [63]. The protein phosphatase ABSCISIC ACID-INSENSITIVE 2 (ABI2) also inhibits SOS2 activity through an unknown molecular mechanism; abi2-1, a dominant-negative mutant of ABI2, is insensitive to NaCl compared with the wild type [64]. GEMINIVIRUS REP INTERACTION KINASE1 (GRIK1) and GRIK2 function as positive regulators of SOS2 kinase activity by phosphorylating the activation loop of SOS2 to activate its kinase activity [65]. These regulations of SOS2 illustrate the important role of SOS2 at the center of the SOS pathway.
In addition, SOS2 also functions in other pathways. However, the mechanism regulating SOS2 kinase activity and adjusting it between salt stress and non-salt-stress conditions is not clearly understood. Salt stress induces ROS stress [1], which in turn modulates the transcription level of SOS1 [66], indicating that the SOS pathway is modulated by ROS signals [1,67]. SOS2 interacts with CATALASE 2 (CAT2) and CAT3, two crucial regulators of H2O2 signaling, suggesting that SOS2 serves as a node connecting H2O2 signaling and salt stress responses [68]. Ethylene-insensitive 3 (EIN3) is phosphorylated by SOS2, and ethylene-related gene expression is thereby enhanced; therefore, salt stress and ethylene signaling are linked by SOS2 [69]. In summary, SOS2 functions as a connector linking the SOS pathway with other signaling pathways.
Various regulators of SOS1 have also been identified in recent years. Salt stress increases phosphatidic acid (PA) accumulation in the PM. PA binds to MITOGEN-ACTIVATED PROTEIN KINASE 6 (MPK6) and activates its kinase activity; the C-terminus of SOS1 is phosphorylated by PA-activated MPK6, in turn activating the Na+/H+ antiporter activity of SOS1 [70]. Two Clade D protein phosphatases 2C, D6 and D7 (PP2C.D6 D7), interact with SOS1 to repress its activity in a process dependent on their phosphatase activity under non-stress conditions. SCaBP8 perceives salinity-induced calcium signaling and inhibits the phosphatase activity of PP2C.D6 and D7 under salt stress, simultaneously regulating the subcellular localization of PP2C.D6, which mediates PP2C.D6 moving to the cytoplasm from the plasma membrane in turn to release SOS1 activity [71]. Identification of more and more regulators controlling the activity of the SOS pathway at different stages of stress confirms that the SOS pathway plays a central role in the regulating of ionic homeostasis (Figure 1).
In conclusion, in the absence of salt stress, the SOS pathway is maintained in a low-activity state. Maintenance of this state depends on the repression of SOS2 kinase activity by PKS5, 14-3-3, GI, and ABI2; inhibition of SOS1 by PP2C clade D; and suppression of PM H+-ATPase by PKS5 (Figure 1A). When plants experience salt stress, GIPC binds to Na+, increasing calcium signaling. Calcium receptors SOS3 and SCaBP8 bind the intracellular Ca2+ and interact with and activate SOS2, which phosphorylates SOS1 to activate its Na+/H+ antiporter activity. MPK6 is activated by PA induced in response to salt stress, further phosphorylating SOS1 and promoting its activity. Simultaneously, SCaBP8 relieves the inhibition of SOS1 by repressing the protein phosphatase activity of PP2C.D. Inhibition of H+-ATPase (AHA) by PKS5 is relieved by 14-3-3, creating a proton gradient across the PM that drives Na+/H+ antiporter SOS1 activity. FER perceives changes in the cell wall under salt stress and mediates calcium signaling for long-term stress. ANNEXINs (ANNs) modulate calcium signaling under salt stress, promoting activation of SOS2 activity by SCaBP8; SOS2 phosphorylates ANN4 and represses its Ca2+ channel activity, creating a specific calcium signaling cascade for long-term salt stress (Figure 1B). After salt stress is relieved, BIN2 phosphorylates SOS2 to inhibit its kinase activity, helping plants to recover from stress (Figure 1C). The capacity of plants to exclude Na+ needs to be appropriately regulated at different stages of stress for the maintenance of ionic homeostasis.
The SOS pathway is involved in potassium uptake and is crucial for regulating Na+/K+ homeostasis in plants [49]. AKT1 is an important potassium channel contributing to K+ influx transport. Salt stress decreases the transport activity of AKT1 [72]. SCaBP8 interacts with the C-terminus of AKT1 to inhibit its K+ transport activity [73]. The Arabidopsis sos1 mutant shows significantly reduced transcript levels of AtAKT1, AtHKT1;1, and STELAR K+ OUTWARD RECTIFIER (AtSKOR; encoding the single outward Shaker K+ channel) in root tissues compared with the wild type, as well as elevated accumulation of Na+ in root cells and decreased K+ uptake and transport, ultimately leading to suppression of plant growth [74]. Salinity stress also influences K+ levels by regulating another potassium transporter, TONOPLAST-LOCALIZED K+ CHANNEL1 (TPK1) [75]. CALCIUM-DEPENDENT PROTEIN KINASE (CDPK) phosphorylates TPK1 and activates K+ influx under salt stress [76]. Nonetheless, how plants actively regulate potassium uptake, including whether SOS signaling pathway elements directly regulate potassium uptake under salt stress, remains unclear.
1.3. Osmotic Homeostasis
Osmotic stress induced by salinity leads to various transient biophysical changes, such as shrinkage of the PM, decreases in turgor pressure in the cell, and physical changes to the cell wall [77]. Many proteins play essential roles in regulating salt-induced osmotic stress responses [8,78]. The osmotic-stress-stimulated Ca2+ channel OSCA1 is considered to be a hyperosmotic stress sensor [29,30]; however, the osca1 mutant shows no significant phenotypic differences from the wild type under salt stress. Therefore, whether OSCA1 is involved in salt-induced osmotic stress responses needs to be further researched.
The plastidial EXCHANGE ANTIPORTER 1 (KEA1), KEA2, and KEA3 regulate Ca2+ signaling induced by rapid hyperosmotic stress, and kea mutants are defective in Ca2+ signaling [79]. Therefore, KEA1/2/3 are thought to function as sensors of osmotic stress that regulate increases in Ca2+ levels under stress conditions. MscS-LIKE8 (MSL8), a PM-localized mechanosensitive (stretch-activated) ion channel in Arabidopsis pollen, is a sensor of hyperosmotic-stress-induced membrane tension. MSL8 promotes pollen survival under the hyperosmotic shock of rehydration [80]. The Ca2+-responsive phospholipid-binding copine protein BONZAI1 (BON1) plays a critical role in osmotic stress regulation by positively regulating Ca2+ signaling. Defects in BON1 disrupt Ca2+ signaling in the cytosol in response to osmotic stress [81]. Whether these osmotic sensors participate in salt-stress-induced osmotic stress sensing and regulation requires further investigation.
MAPK cascades are involved in signal transduction in response to both salt and osmotic stress. Under salt stress, PA-activated MPK6 phosphorylates SOS1, improving its Na+/H+ antiporter activity in Arabidopsis [69]. PA-mediated activation of MAPK signaling cascades under hyperosmotic stress caused by high salt and desiccation has also been observed in Asterochloris erici [82]. The MKK4-MPK3 and MKKK20-MPK6 pathways participate in the osmotic stress responses. mkk4 and mkkk20 mutants exhibit increased sensitivity to salt stress and increased water loss under dehydration conditions compared with the wild type [83,84]. Under osmotic stress treatments, mRNA levels of MAPKKK, MAPKK, and MAPK genes are upregulated, increasing the biosynthesis and accumulation of osmolytes [85,86].
Water uptake capacity is destroyed by salt stress, leading to dehydration and changes in cell turgor and thus to osmotic stress. Under high-salinity conditions, endogenous abscisic acid (ABA) levels increase to mediate stomatal closure and further regulate osmotic homeostasis [87]. ABA functions as a key link between salt stress and osmotic stress responses. SUCROSE NON-FERMENTING1-RELATED PROTEIN KINASES (SnRKs) are the central components regulating the ABA signaling pathway, and almost all SnRK2s are activated by osmotic stress. ABA binds to its receptors and participates in relieving the inhibition of SnRK2.2/3/6 kinase activity by clade A PP2Cs, initiating an ABA-responsive element (ABRE)-binding protein/ABRE-binding factor (AREB/ABF) signaling pathway to regulate osmotic stress and drought stress tolerance [8,79,88]. The SnRK2-AREB/ABF regulatory network modulates starch degradation in leaves through β-AMYLASE1 (BAM1)/α-AMYLASE3 (AMY3), which is also crucial for osmotic stress tolerance in plants [89]. SnRKs function in osmotic stress regulation in an ABA-independent manner. VARICOSE (VCS) is an mRNA decapping activator that belongs to the osmotic-stress-activated subclass I SnRK2s, which are involved in the regulation of mRNA populations under osmotic stress [88].
ABA also participates in the salt stress response through other regulators. FER functions in response to cell wall defects induced by long-term salt stress. ABI2, an important protein phosphatase that functions in ABA signaling, dephosphorylates FER, which senses defects in the cell wall. ABA regulates the kinase activity of FER via ABI2 [90], and, at high levels, helps prevent decreases in photosynthesis resulting from salt stress [91]. In all studies demonstrating crosstalk between the salt stress response and ABA signaling, high salinity increased endogenous ABA levels. Whether a direct interaction or a regulatory relationship exists between the central components of the salt stress and ABA signaling pathways remains unclear.
Common osmotic response pathways (both long-term and short-term) result in the biosynthesis and accumulation of compatible osmolytes, which can correct the cellular osmotic potential in cells and stabilize proteins and cellular structures and morphology. This is an adaptive strategy that is not specifically induced by salt stress. Compatible osmolytes prevent water loss to resist short-term osmotic stress and increase cellular turgor and cellular expansion to cope with long-term osmotic stress [1,92,93,94,95,96]. Many compatible osmolytes biosynthesized under salt stress also accumulate under other stresses, such as drought and cold stresses, and their biosynthesis is partly species- and tissue-specific [1,97,98,99,100].
Salt stress induces the accumulation of several types of osmolytes, including charged metabolites, polyols, sugars, and complex sugars. Charged metabolites include β-alanine betaine, choline-O-sulfate, glycine betaine, hydroxyproline, dimethyl sulfonium propionate (DMSP), proline, and putrescine [101,102,103,104]. Polyols include glucosylglycerol, glycerol, myoinositol, ononitol, pinitol, mannitol, and sorbitol [105,106,107,108,109,110,111]. Sugars include fructose and sucrose as well as the complex sugars trehalose, fructans, and raffinose. Ions such as K+ also function in osmotic regulation [111,112,113,114,115,116,117,118]. These osmolytes also act as signals of ABA accumulation under salt stress, regulating target gene expression [119].
1.4. Regulation of Oxidative Stress Responses
Oxidative stress is a secondary stress stimulated by salt stress in plants, with salt stress rapidly inducing the accumulation of toxic ROS and oxidative damage [8]. Although high concentrations of ROS are damaging to plants, ROS function as signaling molecules at low concentrations [1,120]. Plant cells perceive high ROS levels and rapidly initiate their elimination by scavenging ROS and inducing adaptive responses [11,121,122] (Figure 2). In plant cells, ROS comprise free radicals and non-radicals. The free radicals include superoxide radical (O2•−), hydroxyl radical (OH•), alkoxyl radical (RO•), and peroxyl radical (ROO•); non-radical ROS are singlet oxygen (1O2), hydrogen peroxide (H2O2), hypochlorous acid (HOCl), and excited carbonyl (RO*). The predominant ROS in plant cells are 1O2, H2O2, O2•−, and •OH [123,124].
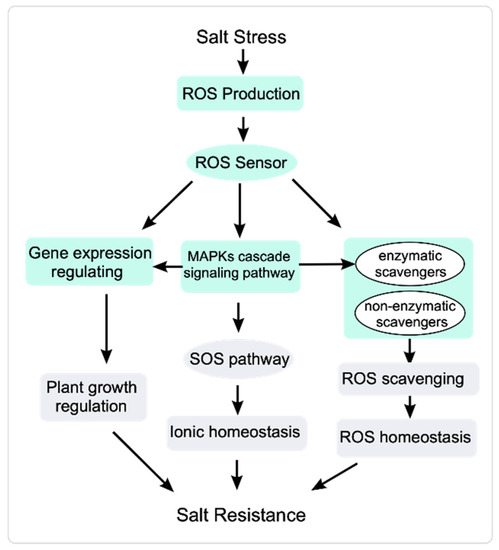
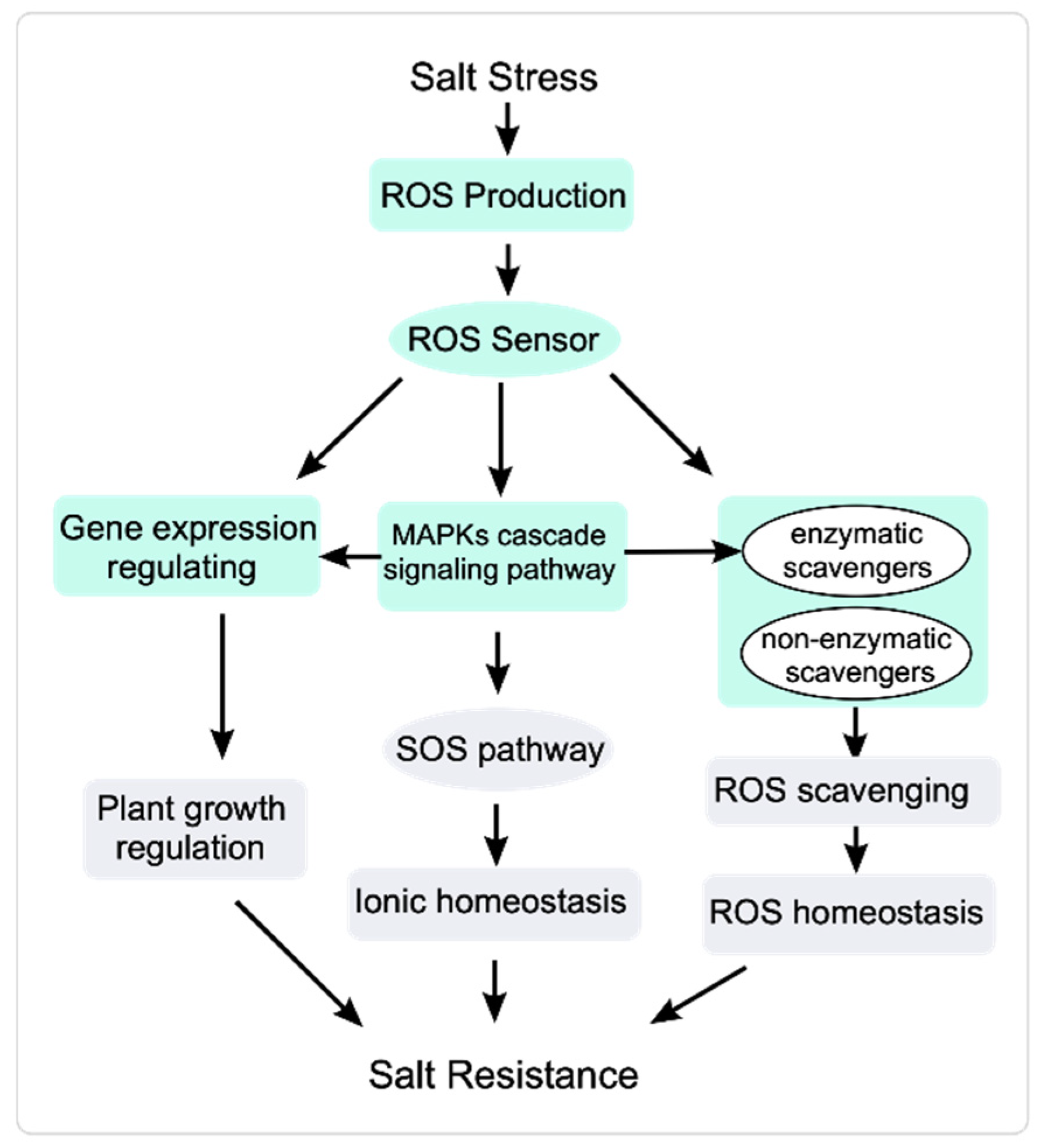
Figure 2.
ROS signal transduction response to salt stress. Salt stress induces a rapid increase in ROS accumulation. Sensors perceive the elevated ROS and transduce the ROS signal to stimulate plant responses. MAPK signaling cascades receive ROS signals and regulate the activity of the SOS pathway and ROS scavengers to modulate ionic homeostasis and ROS homeostasis, respectively. MAPKs also regulate gene expression to modulate plant growth under salt stress.
MAPKs are involved in regulating ROS homeostasis [125]. The receptor-like kinase SALT INTOLERANCE 1 (SIT1) activates MAPK3 and MAPK6 and regulates the homeostasis of ROS and ethylene to inhibit plant growth and even promote death during the salt stress response [126]. Salt stress increases ZmMPK5/17 levels and regulates oxidative homeostasis to help maize (Zea mays) plants cope with salt stress [127,128]. In Arabidopsis, MPK3/6 positively regulate salt stress responses by phosphorylating HEAT SHOCK FACTOR A4A (HSFA4A) and modulating ROS homeostasis [129]. ROS scavenging is important for regulating ROS homeostasis. MAPKs also activate ROS scavengers and regulate the expression of ROS-responsive genes to regulate oxidative homeostasis [130]. Transcriptome analysis of mekk1, mkk1/2, and mpk4 mutants shows that the MEKK1-MKK1 and MKK2-MPK4 regulatory pathways modulate the activity of ROS scavenging enzymes to help maintain ROS homeostasis [131] (Figure 2).
ROS levels must be strictly regulated to avoid their destructive effects on plant cells, so antioxidants are employed to modulate ROS metabolism [1]. Plant cells initiate rapid regulatory mechanisms to scavenge ROS when high ROS levels are sensed under environmental stress [11]. Many enzymatic and non-enzymatic antioxidant scavengers help to prevent damage induced by ROS in plants under salt stress [132,133,134]. Enzymatic ROS scavengers include ascorbate peroxidase (APX), catalase (CAT), polyphenol oxidase (PPO), dehydroascorbate reductase (DHAR), peroxidase (POX), glutathione peroxidase (GPX), peroxiredoxins (PRXs), thioredoxin (TRX), glutathione peroxidase (GR), glutathione S-transferase (GST), monodehydroascorbate reductase (MDHAR), and superoxide dismutase (SOD) [132,135,136,137,138,139,140]. Ascorbate (AsA), glutathione (GSH), alkaloids, carotenoids, flavonoids, phenolic compounds, non-protein amino acids, and tocopherol function as non-enzymatic scavengers [141,142,143,144,145,146,147] (Figure 3).
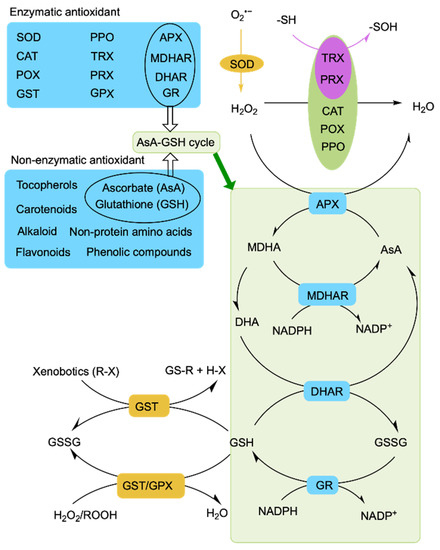
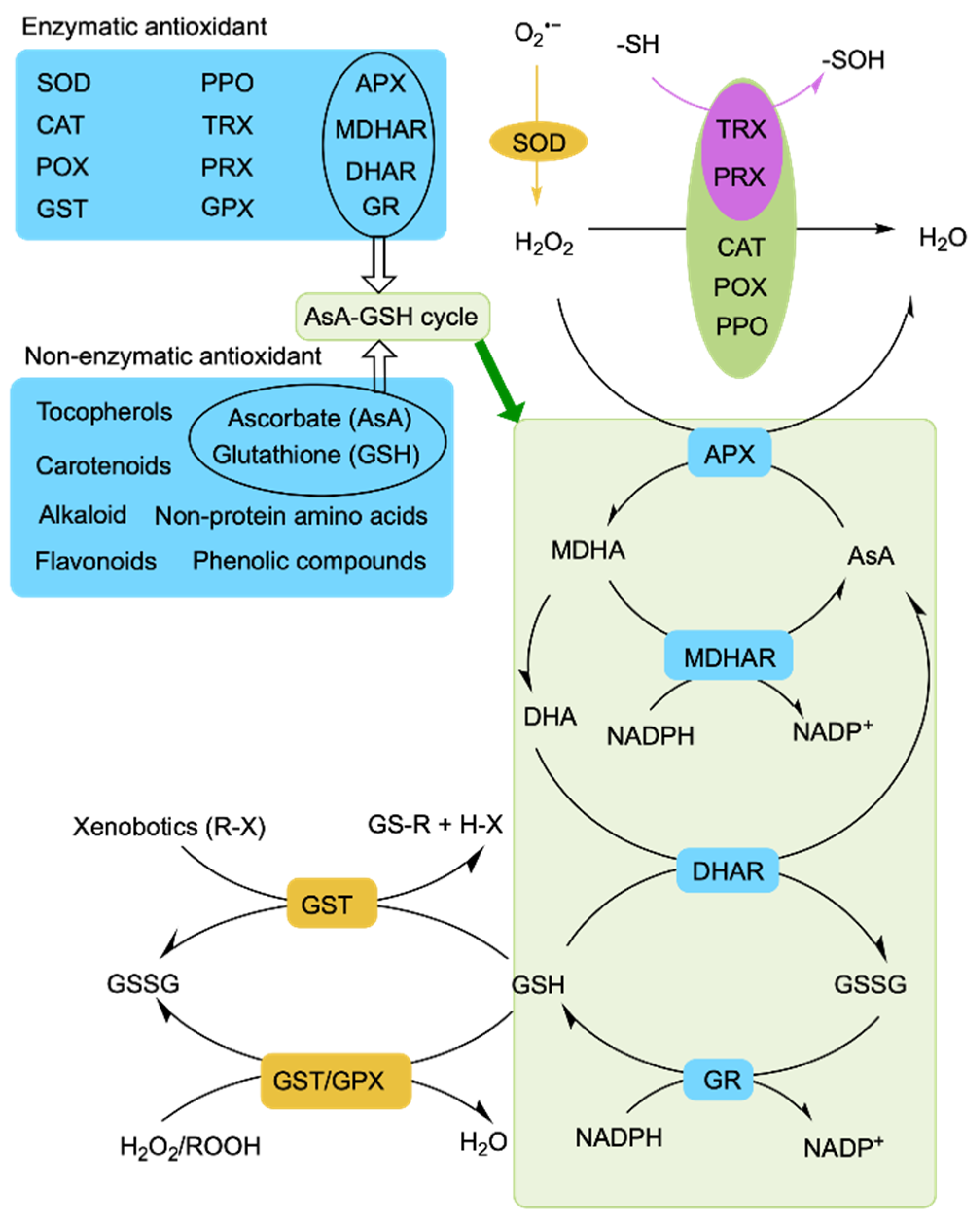
Figure 3.
Outline of antioxidant defense mechanisms in plants. SOD, superoxide dismutase; CAT, catalase; POX, peroxidase; AsA, ascorbate; DHA, dehydroascorbate; GSSG, oxidized glutathione; GSH, reduced glutathione; APX, ascorbate peroxidase; MDHA, monodehydroascorbate; MDHAR, monodehydroascorbate reductase; DHAR, dehydroascorbate reductase; GR, glutathione reductase; GST, glutathione S-transferase; GPX, glutathione peroxidase; PPO, polyphenol oxidase; PRX, peroxiredoxin; TRX, thioredoxin; NADPH, nicotinamide adenine dinucleotide phosphate; O, oxygen; H2O2, hydrogen peroxide; O2•−, superoxide radical; R, aliphatic, aromatic, or heterocyclic group; X, sulfate, nitrite, or halide group; ROOH, hydroperoxides; -SH, thiolate; -SOH, sulfenic acid.
Among antioxidant enzymes, SOD plays an important role as the first line of defense and converts O2•− into H2O2. The H2O2 is further converted into H2O and O2 by the H2O2 enzymes CAT, APX, and GPX [148] (Figure 3). CAT is an important enzyme that rapidly decomposes H2O2 into H2O and O2 without any reducing equivalent [149]. NCA1 (NO CATALASE ACTIVITY1) encodes a protein that can interact with CAT2 and enhance its catalase activity, thereby promoting CAT2 regulation of ROS homeostasis through scavenging cellular hydrogen peroxide under abiotic stress [132]. The nca1-3 mutant is hypersensitive to various abiotic stresses including saline stress. Heat shock protein (Hsp) 17.6CII, a chaperone protein, also activates CAT2 together with NCA1 to increase the abiotic stress tolerance of Arabidopsis [134]. In rice, the receptor-like kinase Salt Tolerance Receptor-like cytoplasmic Kinase 1 (STRK1) phosphorylates the CAT CatC to increase salt tolerance [148]. The AsA-GSH cycle comprises ASA, GSH, and four antioxidant enzymes (APX, DHAR, GR, and MDHAR), playing a significant role in the regulation of ROS homeostasis through detoxifying H2O2 [150]. Alkaloids can inhibit the oxidation induced by H2O2 and scavenge free radicals [151]. Tocopherol mainly scavenges 1O2 and •OH to maintain photosynthesis and protect chloroplasts [152]. Carotenoids, flavonoids, and phenolic acids regulate ROS homeostasis by scavenging free radicals [153,154,155,156]. All of this evidence suggests that plants actively engage in regulatory mechanisms to cope with salt-stress-induced oxidative stress and transduce stress signals by controlling ROS homeostasis.
1.5. Phytohormonal Responses to Salt Stress
Besides transporting excess Na+ out of cells, plants must also adjust their growth to adapt to salt stress or any other adverse environmental conditions. These growth adjustments require regulation by various plant hormones. In addition, salt stress elicits the responses of several plant hormones directly.
Auxin plays crucial roles in several aspects of plant development, such as shoot and root architecture, maintenance of meristem activity, and leaf morphogenesis. Many studies have demonstrated that auxin levels decrease under salt stress [157,158,159,160], but the processes regulating auxin biosynthesis under saline stress appear to be complex. Endogenous auxin biosynthesis is regulated by the YUCCA family of flavin monooxygenases (YUC). Expression of CsYUC10a and CsYUC11 decreases in cucumber (Cucumis sativus) under salt stress. However, the expression of YUC6 in poplar and potato and CsYUC11 in Arabidopsis enhances tolerance of salt stress [160,161,162], pointing to a complicated regulation process for auxin biosynthesis under saline stress. TRANSPORT INHIBITOR RESPONSE 1 (TIR1), encoding an auxin receptor, and AUXIN SIGNALLING F-BOX (AFB) are also downregulated under salt stress [163], leading to changes in the expression patterns of the auxin transporter genes AUXIN-RESISTANT1 (AUX1), PIN-FORMED1 (PIN1), and PIN2 [164]. These results illustrate how plants adjust their auxin response and auxin transport to adapt to saline stress, possibly as a means of balancing plant growth and stress tolerance.
Brassinosteroids (BRs) are plant hormones that promote plant growth. The receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1) recognizes the presence of BR through a series of phosphorylation cascade reactions among BR-related kinases. The key transcription factors BRASSINAZOLE RESISTANT 1 (BZR1) and BRI1-EMSSUPPRESSOR 1 (BES1) in the BR signaling pathway are then released to initiate expression of BR-responsive genes [165]. Increasing evidence indicates that BRs are involved in salt stress tolerance: The salinity tolerance of plants is enhanced by the exogenous application of BRs [166,167], and mutants of BR-biosynthesis-related genes and positive regulators of BR signaling are more sensitive to salt stress than wild-type plants [168,169,170,171]. These findings suggest that BRs positively regulate plant tolerance to salt stress. BIN2, a negative regulator of BR signaling, directly interacts with and phosphorylates SOS2, inhibiting its kinase activity and thereby downregulating the SOS pathway during the recovery stage after salt stress [63]. This suggests that BR signaling undergoes direct crosstalk with the SOS pathway via their central elements to modulate responses to different stages of salt stress.
Ethylene levels rise under salt stress, with salt stress upregulating the 1-Aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) ethylene biosynthesis genes to different degrees. Treatment with the ethylene precursor ACC enhances salt tolerance in plants [163,172,173]. Ethylene signaling is also involved in the salt stress response. Most mutants of positive regulators of ethylene signaling, such as etr2 (ethylene response 2), ein2 (ethylene insensitive 2), and ein3/eil1 (ein3-like 1), display increased sensitivity to salt stress, whereas ctr1 (constitutive triple response 1), a mutant of a negative regulator of ethylene signaling, is insensitive to salt stress [171]. These findings demonstrate that both ethylene and the ethylene signaling pathway promote salt tolerance in plants.
Salicylic acid (SA) at appropriate levels also promotes salt tolerance in plants. Oxidative toxicity and osmotic stress are both alleviated by the exogenous application of SA, but this positive effect requires a specific dosage range. Seed germination under salt stress is enhanced by the application of less than 50 μM SA and repressed by the application of more than 100 μM SA [174,175].
Gibberellin (GA) also functions as a growth-promoting hormone in plants. However, plants appear to actively downregulate GA signaling and retard their growth to aid survival under salt stress. Salt stress decreases the levels of DOMINANT SUPPRESSOR OF KAR2 (OsDSK2a), a positive regulator of GA signaling, which reduces active GA levels and slows plant growth [176]. The overexpression of genes encoding negative regulators of GA accumulation, such as CYP71D8 (CYP, Cytochrome P450) in rice and PtCYP714A3 in poplar (Populus trichocarpa), promotes salt tolerance by inhibiting plant growth [177,178]. This implies that GA plays a crucial role in defending plants from salt stress by modulating the balance between growth and stress tolerance.
In summary, studies into the roles of plant hormones in plant responses to salt stress have revealed that balancing plant growth and stress tolerance is crucial for plant survival under salt stress. Cooperative regulation by different plant hormones is essential for maintaining this balance.
1.6. Photosynthesis under Salt Stress
Salinity stress destroys the photopigment system and decreases chlorophyll production, leading to severe inhibition of photosynthesis [179]. Both nitrogen and magnesium ions (Mg2+) are essential for chlorophyll biosynthesis [180], and salt stress seriously reduces their uptake [181,182], inhibiting chlorophyll biosynthesis. A decrease in stomatal number and an increase in closed stomata under high salt concentrations leads to decreases in CO2 absorption, further inhibiting photosynthesis [183,184]. Stomatal density in the adaxial part of phyllodes is significantly lower in Acacia auriculiformis under seawater-induced salinity stress compared with that in salt-free control plants [183]. Moreover, salt stress significantly inhibits photosystem II (PSII), which is crucial for light energy conversion and photosynthetic efficiency [185].
The ultrastructure of chloroplasts is affected by salt stress. Salinity stress causes swollen chloroplasts in rice mesophyll cells [186]. In wheat (Triticum aestivum), the granum thylakoids of chloroplasts are loosely arranged with a slim spindle shape under 200 mM NaCl compared with non-stress conditions [187]. Salinity stress also results in the swelling of thylakoids and the accumulation of starch granules in Thellungiella salsuginea, which helps maintain osmotic equilibrium [188]. Increasing chloroplast number helps halophytes to overcome stomatal limitations induced by salinity stress [189].
Photosynthetic pigments harvest light energy for conversion to chemical energy in the photosystem. Salt stress damages photosynthetic pigments. For example, contents of chlorophyll a, chlorophyll b, and total chlorophyll are reduced in Acacia auriculiformis plants under seawater treatment [190]. In Nicotiana benthamiana, intermediates of chlorophyll biosynthesis such as Mg-photoporphyrin-IX, protoporphyrin-IX, and protochlorophyllide show reduced abundance under salinity stress [191].
Parameters determining photosynthetic efficiency, including the quantum yield of PSII, activity of PSII, maximum quantum efficiency of PSII photochemistry (Fv/Fm), electron transport (QY), photochemical quenching (qp), non-photochemical quenching (NPQ), and linear electron transport rate (ETR), are significantly altered under salt stress [192]. Many key elements of photosynthesis are inhibited by salinity stress. Salt affects the amount of PSI proteins, with a significant decrease in the amount of PsaA protein observed under NaCl treatment and decreases in the amounts of PsaF and PsaK seen when PEG is applied in combination with salt [193]. Salt stress reduces the activity of ribose 5-phosphate by 55% [194], whereas severe salt stress decreases the regeneration of ribulose 1,5-bisphosphate and carboxylase/oxygenase (RuBisCo), resulting in impaired PSII electron transport [195]. Glyceraldehyde 3-phosphate dehydrogenase phosphoglycerate kinase and phosphofructokinase are also inhibited by salt stress [196].
In conclusion, salt stress leads to lower stomatal conductance and reduced photosynthetic rate, chlorophyll content, and activity of key enzymes, significantly reducing the photosynthetic capacity of plants.
1.7. Transcription Factors in Salt Stress Response
Many transcription factors, such as MYB proteins, contribute to salt tolerance. MYB49 positively regulates salt tolerance by upregulating genes encoding peroxidases and late embryogenesis abundant proteins [197]. MYB44 also positively regulates salt tolerance by regulating the expression of PP2C genes [198]. RING zinc finger proteins improve salt stress tolerance by regulating ion homeostasis and ROS scavenging. SALT TOLERANCE RING FINGER1 (AtSTRF1), the rice RING-H2-type zinc finger protein OsSIRH2-14, and RING FINGER PROTEIN V6 (OsRFPv6) also positively regulate the salt tolerance of plants [199,200,201].
1.8. Implications for Crop Improvement
The mining of important functional genes facilitates molecular breeding, and the development of abiotic stress tolerance, especially saline–alkali-tolerant crop varieties, will allow utilization of the large, and increasing, area of saline–alkali land. Increasing numbers of important regulators of abiotic stress responses have been identified in crops using genome-wide association studies (GWASs) and quantitative trait loci (QTLs). In maize, ZmHAK4 (HIGH AFFINITY K+4) confers the capacity for shoot Na+ exclusion and salt tolerance [202]; ZmVPP1 (VACUOLAR-TYPE H+-PYROPHOSPHATASE) contributes to drought tolerance [203]; and ZmSRO1d (SIMILAR TO RCD-ONEs1d) helps plants to tolerate drought by regulating ROS levels in guard cells to regulate stomatal closure [204]. Alkaline tolerance 1 (AT1) in sorghum can inhibit plant alkali tolerance by negatively regulating the H2O2 transporter; knockout of AT1 through gene editing significantly improves the alkali tolerance of crops [205]. These studies suggest the importance of considering oxidative stress regulation in the development of abiotic-stress-tolerant crop cultivars. HKT1 was identified as a major regulator of salt stress tolerance in rice, wheat, and maize through QTL analysis, and marker-assisted breeding of wheat has been developed for increased yield in saline soils [206,207,208,209]. Genes encoding components of the SOS signaling pathway also contribute to phenotypic variation in salt tolerance in maize and tomato [210,211].
At present, the most important breeding methods are direct screening based on phenotypic observation combined with genetic experiments, and molecular marker genotypes [206]. With the rapid development of artificial intelligence (AI), the integration of genomics, phenomics, and environmental factors will play an important role in future breeding [212]. AI can judge data faster and more accurately than humans and precisely predict phenotypes through model training using data collected from multiple sources, including spatiotemporal omics (genomics, phenomics, and enviromics across time and space) [213]. AI technology combined with high-throughput genomics and phenomics has been applied effectively in crop breeding programs [214,215,216]. AI-assisted high-throughput phenotypic systems can effectively and accurately predict phenotypes in maize, wheat, and cassava (Manihot esculenta Crantz) [217,218,219]. AI breeding will greatly help scientists select and develop new crop cultivars that are tolerant of stress. However, AI breeding requires the collaboration of plant science, breeding science, bioinformatics, and computer engineering.
2. Conclusions and Perspectives
Plant survival under salt stress is a complex process involving crosstalk among several signaling pathways, including osmotic signaling, ROS signaling, and ABA signaling. During the past two decades, numerous studies have focused on the mechanisms regulating ionic homeostasis under salt stress. The SOS pathway has been studied extensively and found to be the most important signaling pathway regulating ionic homeostasis. Ca2+ signaling plays an essential role in the transduction of stress signals and serves as a link between different signaling pathways. Phosphorylation is the most important process regulating signaling activity, and kinases are the most important enzymes that perform these regulatory roles. However, many questions remain to be resolved.
First, the mechanism of salt stress sensing requires further study. Plant processes for sensing various ionic stresses appear to differ, prompting the need to identify a specific Na+ sensor in plants. Whether more specific sodium receptors exist and whether sodium is perceived extracellularly or intracellularly remain to be explored. Second, much research is needed in order to apply our knowledge of salt-tolerance mechanisms to the development of crops to increase their survival under high salinity. All mutants of SOS genes show a sensitive phenotype in Arabidopsis and other crops [210,211]; however, overexpressing SOS pathway genes only slightly increases plant salt tolerance [220,221]. It is likely that other factors interacting with SOS genes during the salt stress response in plants and other salt-tolerance regulators remain to be identified. Finally, for breeding salt-tolerant crop cultivars, multi-omics technologies combined with gene editing will likely serve as efficient tools and lead to wide achievements.
Halophytes are plants that can grow and reproduce under high-salinity conditions (>200 mM NaCl). In addition to sharing salt-tolerance mechanisms with glycophytic plants, halophytes also have evolved specific manners of adapting to high-salinity conditions. For instance, the capacity for osmotic adjustment of halophytes is greater than that of glycophytes [222,223]. Secretion of salt by epidermal bladder cells (EBCs) of halophytes is another specific salt-tolerance mechanism developed by some halophytic species such as Chenopodium quinoa [224]. Such specialized salt-tolerance mechanisms will be important for the cultivation of new salt-tolerant crop cultivars.
Author Contributions
H.F. and Y.Y. wrote the original draft of manuscript; H.F. and Y.Y. reviewed and corrected the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (grant No. 32070301).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This work was edited and revised by Pengpu Wang, Rong Hao, Xiao Liu, Wanjia Lv, and Qinpei Li.
Conflicts of Interest
There is no conflict of interest.
References
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Mehra, P.; Bennett, M.J. A novel Ca2+ sensor switch for elevated salt tolerance in plants. Dev. Cell 2022, 57, 2045–2047. [Google Scholar] [CrossRef]
- Ouhibi, C.; Attia, H.; Rebah, F.; Msilini, N.; Chebbi, M.; Aarrouf, J.; Urban, L.; Lachaal, M. Salt stress mitigation by seed priming with UV-C in lettuce plants: Growth, antioxidant activity and phenolic compounds. Plant Physiol. Biochem. 2014, 83, 126–133. [Google Scholar] [CrossRef]
- Hoque, M.N.; Imran, S.; Hannan, A.; Paul, N.C.; Mahamud, M.A.; Chakrobortty, J.; Sarker, P.; Irin, I.J.; Brestic, M.; Rhaman, M.S. Organic Amendments for Mitigation of Salinity Stress in Plants: A Review. Life 2022, 12, 1632. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Pandey, G.K.; Tuteja, N. Calcium- and salt-stress signaling in plants: Shedding light on SOS pathway. Arch. Biochem. Biophys. 2008, 471, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.; El-Sharkawy, I.; Sherif, S. Salt stress signals on demand: Cellular events in the right context. Int. J. Mol. Sci. 2020, 21, 3918. [Google Scholar] [CrossRef]
- Ismail, A.; Takeda, S.; Nick, P. Life and death under salt stress: Same players, different timing? J. Exp. Bot. 2014, 65, 2963–2979. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.H.; Luan, S. AtKuP1: A dual-affinity K+ transporter from Arabidopsis. Plant Cell 1998, 10, 63–73. [Google Scholar] [PubMed]
- Lazof, D.B.; Bernstein, N. The NaCl induced inhibition of shoot growth: The case for distributed nutrition with special consideration of calcium. Adv. Bot. Res. 1999, 29, 113–189. [Google Scholar]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of plant responses to salt stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salt. Annu. Rev. Plant Phys. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Arya, A.; Nyamathulla, S.; Noordin, M.I.; Mohd, M.A. Antioxidant and hypoglycemic activities of leaf extracts of three Popular Terminalia species. J. Chem. 2012, 9, 883–892. [Google Scholar]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151–160. [Google Scholar] [CrossRef]
- Genisel, M.; Erdal, S.; Kizilkaya, M. The mitigating effect of cysteine on growth inhibition in salt-stressed barley seeds is related to its own reducing capacity rather than its effects on antioxidant system. Plant Growth Regul. 2015, 75, 187–197. [Google Scholar] [CrossRef]
- Shabala, S.; Wu, H.; Bose, J. Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci. 2015, 241, 109–119. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigomoreno, A.; Lai, D.; Xie, Y.; Shen, W.; Shabala, S. Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann. Bot. 2015, 115, 481–494. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nat. Cell Biol. 2019, 572, 341–346. [Google Scholar] [CrossRef]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 2019, 48, 697–709. [Google Scholar] [CrossRef]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef]
- Li, W.; Guan, Q.; Wang, Z.Y.; Wang, Y.; Zhu, J. A bi-functional xyloglucan galactosyltransferase is an indispensable salt stress tolerance determinant in Arabidopsis. Mol. Plant 2013, 6, 1344–1354. [Google Scholar] [CrossRef]
- Van der Does, D.; Boutrot, F.; Engelsdorf, T.; Rhodes, J.; McKenna, J.F.; Vernhettes, S.; Koevoets, I.; Tintor, N.; Veerabagu, M.; Miedes, E.; et al. The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet. 2017, 13, e1006832. [Google Scholar] [CrossRef]
- Dinneny, J.R.; Long, T.A.; Wang, J.Y.; Jung, J.W.; Mace, D.; Pointer, S.; Barron, C.; Brady, S.M.; Schiefelbein, J.; Benfey, P.N. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 2008, 320, 942–945. [Google Scholar] [CrossRef]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I.; et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 2018, 28, 666–675. [Google Scholar] [CrossRef]
- Xu, S.L.; Rahman, A.; Baskin, T.I.; Kieber, J.J. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and acc synthase in Arabidopsis. Plant Cell 2008, 20, 3065–3079. [Google Scholar] [CrossRef]
- Moura, J.C.; Bonine, C.A.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Sewelam, N.; Oshima, Y.; Mitsuda, N.; Ohme-Takagi, M. A step towards understanding plant responses to multiple environmental stresses: A genome-wide study. Plant Cell Environ. 2014, 37, 2024–2035. [Google Scholar] [CrossRef]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, Q.-R.; Liu, L.-L.; Zhang, H.-M.; Gao, J.-W.; Pei, Z.-M. Osmotic stress alters circadian cytosolic Ca2+ oscillations and OSCA1 is required in circadian gated stress adaptation. Plant Signal. Behav. 2020, 15, 1836883. [Google Scholar] [CrossRef]
- Pei, S.; Liu, Y.; Li, W.; Krichilsky, B.; Dai, S.; Wang, Y.; Wang, X.; Johnson, D.M.; Crawford, B.M.; Swift, G.B.; et al. OSCA1 is an osmotic specific sensor: A method to distinguish Ca2+-mediated osmotic and ionic perception. New Phytol. 2022, 235, 1665–1678. [Google Scholar] [CrossRef]
- Han, Y.; Wang, Y.; Zhai, Y.; Wen, Z.; Liu, J.; Xi, C.; Zhao, H.; Wang, Y.; Han, S. OsOSCA1.1 mediates hyperosmolality and salt stress sensing in Oryza sativa. Biology 2022, 11, 678. [Google Scholar] [CrossRef]
- Niu, X.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Ion homeostasis in NaCI stress environments. Plant Physiol. 1995, 109, 735–742. [Google Scholar] [CrossRef]
- Beilby, M.J. Salt tolerance at single cell level in giant-celled Characeae. Front. Plant Sci. 2015, 6, 226. [Google Scholar] [CrossRef]
- Blumwald, E.; Aharon, G.S.; Apse, M.P. Sodium transport in plant cells. Biochim. Biophys. Acta Biomembr. 2000, 1465, 140–151. [Google Scholar] [CrossRef]
- Tuteja, N. Mechanisms of high salinity tolerance in plants. Methods Enzymol. 2007, 428, 419–438. [Google Scholar] [PubMed]
- Amtmann, A.; Jelitto, T.C.; Sanders, D. K+-selective inward-rectifying channels and apoplastic pH in barley roots. Plant Physiol. 1999, 120, 331–338. [Google Scholar] [CrossRef]
- Wada, M.; Satoh, S.; Kasamo, K.; Fujii, T. Presence of a Na+-activated ATPase in the plasma membrane of the marine raphidophycean Heterosigma akashiwo. Plant Cell Physiol. 1989, 30, 923–928. [Google Scholar] [CrossRef]
- Wada, M.; Urayama, O.; Satoh, S.; Hara, Y.; Ikawa, Y.; Fujii, T. A marine algal Na+-activated ATPase possesses an immunologcally identical epitope to Na+, K+-ATPase. FEBS Lett. 1992, 309, 272–274. [Google Scholar] [CrossRef]
- Sussman, M.R. Molecular analysis of proteins in the plant plasma membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1994, 45, 211–234. [Google Scholar] [CrossRef]
- Li, J.; Guo, Y.; Yang, Y. The molecular mechanism of plasma membrane H+-ATPases in plant responses to abiotic stress. J. Genet. Genom. 2022, 49, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Apse, M.P.; Aharon, G.S.; Snedden, W.A.; Blumwald, E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 1999, 285, 1256–1258. [Google Scholar] [CrossRef] [PubMed]
- Uozumi, N.; Kim, E.J.; Rubio, F.; Yamaguchi, T.; Muto, S.; Tsuboi, A.; Bakker, E.P.; Nakamura, T.; Schroeder, J.I. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000, 122, 1249–1259. [Google Scholar] [CrossRef]
- Ali, A.; Raddatz, N.; Pardo, J.M.; Yun, D.J. HKT sodium and potassium transporters in Arabidopsis thaliana and related halophyte species. Physiol. Plant. 2021, 171, 546–558. [Google Scholar] [CrossRef]
- Davenport, R.J.; Muñoz-Mayor, A.; Jha, D.; Essah, P.A.; Rus, A.; Tester, M. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 2007, 30, 497–507. [Google Scholar] [CrossRef]
- Hamamoto, S.; Horie, T.; Hauser, F.; Deinlein, U.; Schroeder, J.I.; Uozumi, N. HKT transporters mediate salt stress resistance in plants: From structure and function to the field. Curr. Opin. Biotechnol. 2015, 32, 113–120. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, J.K. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943–1945. [Google Scholar] [CrossRef]
- Ishitani, M.; Liu, J.; Halfter, U.; Kim, C.S.; Shi, W.; Zhu, J.K. SOS3 function in plant salt tolerance requires n-myristoylation and calcium binding. Plant Cell 2000, 12, 1667–1677. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Halfter, U.; Ishitani, M.; Zhu, J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Yang, Y.; Quan, R.; Mendoza, I.; Wu, Y.; Du, W.; Zhao, S.; Schumaker, K.S.; Pardo, J.M.; Guo, Y. Phosphorylation of SOS3 LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 2009, 21, 1607–1619. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [PubMed]
- Schachtman, D.P.; Liu, W. Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci. 1999, 4, 281–287. [Google Scholar] [CrossRef]
- Nunez-Ramırez, R.; Sanchez-Barrena, M.J.; Villalta, I.; Vega, J.F.; Pardo, J.M.; Quintero, F.J.; Martinez-Salazar, J.; Albert, A. Structural insights on the plant salt-overly-sensitive 1 (SOS1) Na+/H+ antiporter. J. Mol. Biol. 2012, 424, 283–294. [Google Scholar] [CrossRef]
- Quintero, F.J.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.; Kim, W.Y.; Ali, Z.; Fujii, H.; Mendoza, I.; Yun, D.J.; Zhu, J.K.; et al. Activation of the plasma membrane Na+/H+ antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Quintero, F.J.; Pardo, J.M.; Schumaker, K.S.; Zhu, J.K. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 2004, 279, 207–215. [Google Scholar] [CrossRef]
- Guo, Y.; Halfter, U.; Ishitani, M.; Zhu, J.K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 2001, 13, 1383–1400. [Google Scholar] [CrossRef]
- Chaves-Sanjuan, A.; Sanchez-Barrena, M.J.; Gonzalez-Rubio, J.M.; Moreno, M.; Ragel, P.; Jimenez, M.; Pardo, J.M.; Martinez-Ripoll, M.; Quintero, F.J.; Albert, A. Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress. Proc. Natl. Acad. Sci. USA 2014, 111, 4532–4541. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019, 10, 1199. [Google Scholar] [CrossRef]
- Zhou, H.; Lin, H.; Chen, S.; Becker, K.; Yang, Y.; Zhao, J.; Kudla, J.; Schumaker, K.S.; Guo, Y. Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 2014, 26, 1166–1182. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 273–275. [Google Scholar] [CrossRef]
- Li, J.; Zhou, H.; Zhang, Y.; Li, Z.; Yang, Y.; Guo, Y. The GSK3-like ki-nase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev. Cell 2020, 55, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Ohta, M.; Guo, Y.; Halfter, U.; Zhu, J.K. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 2003, 100, 11771–11776. [Google Scholar] [CrossRef]
- Barajas-Lopez, J.; Moreno, J.; Gamez-Arjona, F.; Pardo, J.; Punkkinen; Zhu, J.; Quintero, F.; Fujii, H. Upstream kinases of plant SnRKs are involved in salt stress tolerance. Plant J. 2018, 93, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.S.; Zhu, J.K.; Bressan, R.A.; Hasegawa, P.M.; Shi, H. Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 2008, 53, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhao, J.; Yang, Y.; Chen, C.; Liu, Y.; Jin, X.; Chen, L.; Li, X.; Deng, X.; Schumaker, K.S.; et al. Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na+/H+ antiport activity and serine hydroxy-methyltransferase stability. Plant Cell 2012, 24, 5106–5122. [Google Scholar] [CrossRef]
- Verslues, P.E.; Batelli, G.; Grillo, S.; Agius, F.; Kim, Y.S.; Zhu, J.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.K. Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Mol. Cell Biol. 2007, 27, 7771–7780. [Google Scholar] [CrossRef]
- Quan, R.; Wang, J.; Yang, D.; Zhang, H.; Zhang, Z.; Huang, R. EIN3 and SOS2 synergistically modulate plant salt tolerance. Sci. Rep. 2017, 7, 44637–44647. [Google Scholar] [CrossRef]
- Yu, L.; Nie, J.; Cao, C.; Jin, Y.; Yan, M.; Wang, F.; Liu, J.; Xiao, Y.; Liang, Y.; Zhang, W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010, 188, 762–773. [Google Scholar] [CrossRef]
- Fu, H.; Yu, X.; Jiang, Y.; Wang, Y.; Yang, Y.; Chen, S.; Chen, Q.; Guo, Y. SALT OVERLY SENSITIVE 1 is inhibited by clade D Protein phosphatase 2C D6 and D7 in Arabidopsis thaliana. Plant Cell 2023, 35, 279–297. [Google Scholar] [CrossRef] [PubMed]
- Nieves-Cordones, M.; Alemán, F.; Martínez, V.; Rubio, F. K+ uptake in plant roots. The systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 2014, 171, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.L.; Qi, G.N.; Feng, H.Q.; Zhao, S.; Zhao, S.S.; Wang, Y.; Wu, W.H. Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 2013, 74, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guan, C.; Wang, P.; Ma, Q.; Bao, A.K.; Zhang, J.L.; Wang, S.M. The Effect of AtHKT1;1 or AtSOS1 Mutation on the Expressions of Na+ or K+ Transporter Genes and Ion Homeostasis in Arabidopsis thaliana under Salt Stress. Int. J. Mol. Sci. 2019, 20, 1085. [Google Scholar] [CrossRef]
- Latz, A.; Becker, D.; Hekman, M.; Müller, T.; Beyhl, D.; Marten, I.; Eing, C.; Fischer, A.; Dunkel, M.; Bertl, A.; et al. TPK1, a Ca2+-regulated Arabidopsis vacuole two-pore K+ channel is activated by 14-3-3 proteins. Plant J. 2007, 52, 449–459. [Google Scholar] [CrossRef]
- Latz, A.; Mehlmer, N.; Zapf, S.; Mueller, T.D.; Wurzinger, B.; Pfister, B.; Csaszar, E.; Hedrich, R.; Teige, M.; Becker, D. Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol. Plant 2013, 6, 1274–1289. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, W.Y.; Yun, A.D.J. A new insight of salt stress signaling in plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef]
- Stephan, A.B.; Kunz, H.H.; Yang, E.; Schroeder, J. Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and involvement of plastidial KEA transporters. Proc. Natl. Acad. Sci. USA 2016, 113, E5242–E5249. [Google Scholar] [CrossRef]
- Hamilton, E.S.; Jensen, G.S.; Maksaev, G.; Katims, A.; Sherp, A.M.; Haswell, E.S. Mechanosensitive channel MSL8 regulates osmotic forces during pollen hydration and germination. Science 2015, 350, 438–441. [Google Scholar] [CrossRef]
- Chen, K.; Gao, J.; Sun, S.; Zhang, Z.; Yu, B.; Li, J.; Xie, C.; Li, G.; Wang, P.; Bressan, R.A.; et al. The calcium-responsive phospholipid-binding BONZAI proteins control global osmotic stress responses in plants through repression of immune signaling. SSRN Electron. J. 2020, 30, 4815. [Google Scholar] [CrossRef]
- Gasulla, F.; Barreno, E.; Parages, M.L.; Camara, J.; Jimenez, C.; Doermann, P.; Bartels, D. The role of phospholipase D and MAPK signaling cascades in the adaption of Lichen microalgae to desiccation: Changes in membrane lipids and phosphoproteome. Plant Cell Physiol. 2016, 57, 1908–1920. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Kim, S.H.; Woo, D.H.; Lee, S.Y.; Park, H.Y.; Seok, H.Y.; Chung, W.S.; Moon, Y.H. Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell Rep. 2012, 31, 217–224. [Google Scholar] [CrossRef]
- Kim, S.H.; Woo, D.H.; Kim, J.M.; Lee, S.Y.; Chung, W.S.; Moon, Y.H. Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem. Biophys. Res. Commun. 2011, 412, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Naguro, I.; Ichijo, H.; Watanabe, K. Mitogen-activated protein kinases as key players in osmotic stress signaling. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 2037–2052. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, K.; Abu-Qamar, S.; Jarrar, M.; Al-Rajab, A.J.; Trémouillaux-Guiller, J. MAPK cascades and major abiotic stresses. Plant Cell Rep. 2014, 33, 1217–1225. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Soma, F.; Mogami, J.; Yoshida, T.; Abekura, M.; Takahashi, F.; Kidokoro, S.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants 2017, 3, 16204. [Google Scholar] [CrossRef]
- Thalmann, M.; Pazmino, D.; Seung, D.; Horrer, D.; Nigro, A.; Meier, T.; Ko€lling, K.; Pfeifhofer, H.W.; Zeeman, S.C.; Santelia, D. Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 2016, 28, 1860–1878. [Google Scholar] [CrossRef]
- Chen, J.; Yu, F.; Liu, Y.; Du, C.; Li, X.; Zhu, S.; Wang, X.; Lan, W.; Rodriguez, P.L.; Liu, X.; et al. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E5519–E5527. [Google Scholar] [CrossRef]
- Marusig, D.; Tombesi, S. Abscisic acid mediates drought and salt stress responses in Vitis vinifera—A review. Int. J. Mol. Sci. 2020, 21, 8648. [Google Scholar] [CrossRef] [PubMed]
- Apse, M.P.; Blumwald, E. Engineering salt tolerance. Curr. Opin. Biotechnol. 2002, 13, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Yancey, P.H.; Clark, M.E.; Hand, S.C.; Bowlus, R.D.; Somero, G.N. Living with water stress: Evolution of osmolyte systems. Science 1982, 217, 1214–1222. [Google Scholar] [CrossRef]
- Blumwald, E. Engineering salt tolerance in plants. Curr. Opin. Biotechnol. 2003, 13, 261–275. [Google Scholar] [CrossRef]
- Parvanova, D.; Ivanov, S.; Konstantinova, T.; Karanov, E.; Atanassov, A.; Tsvetkov, T.; Alexieva, V.; Djilianov, D. Transgenic tobacco plants accumulating osmolytes show reduced oxidative damage under freezing stress. Plant Physiol. Biochem. 2004, 42, 57–63. [Google Scholar] [CrossRef]
- Yancey, P.H. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef]
- Pirzad, A.; Shakiba, M.R.; Zehtab-Salmasi, S.; Mohammadi, S.A.; Darvishzadeh, R.; Samadi, A. Effect of water stress on leaf relative water content, chlorophyll, proline and soluble carbohydrates in Matricaria chamomilla L. J. Med. Plants Res. 2011, 5, 2483–2488. [Google Scholar]
- Sailaja, B.; Mangrauthia, S.; Sarla, N.; Voleti, S.R. Transcriptomics of heat stress in plants. In Improvement of Crops in the Era of Climatic Changes; Ahmad, P., Wani, M.R., Azooz, M.M., Tran, L.P., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 2, pp. 49–89. [Google Scholar]
- Hanson, A.D.; Rathinasabapathi, B.; Rivoal, J.; Burnet, M.; Dillon, M.O.; Gage, D.A. Osmoprotective compounds in the Plumbaginaceae-a natural experiment in metabolic engineering of stress tolerance. Proc. Natl. Acad. Sci. USA 1994, 91, 306–310. [Google Scholar] [CrossRef]
- Summers, P.S.; Nolte, K.D.; Cooper, A.J.L.; Borgeas, H.; Leustek, T. Identification and stereospecificity of the first three enzymes of 3-dimethylsulfoniopropionate biosynthesis in a chlorophyte alga. Plant Physiol. 1998, 116, 369–378. [Google Scholar] [CrossRef]
- Bouchereau, A.; Aziz, A.; Larher, F.; Martin-Tanguy, J. Polyamines and environmental challenges: Recent development. Plant Sci. 1999, 140, 103–125. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, P.; Jiang, Y.; Fu, J. Metabolomic analysis revealed differential adaptation to salinity and alkalinity stress in kentucky bluegrass (Poa pratensis). Plant Mol. Biol. Rep. 2015, 33, 56–68. [Google Scholar] [CrossRef]
- Nelson, D.E.; Koukoumanos, M.; Bohnert, H.J. Myo-inositol-dependent sodium uptake in ice plant. Plant Physiol. 1999, 119, 165–172. [Google Scholar] [CrossRef]
- Ferjani, A.; Mustardy, L.; Sulpice, R.; Marin, K.; Suzuki, I.; Hagemann, M.; Murata, N. Glucosylglycerol, a compatible solute, sustains cell division under salt stress. Plant Physiol. 2003, 131, 1628–1637. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A. Osmoregulation in Dunaliella, Part II: Photosynthesis and starch contribute carbon for glycerol synthesis during a salt stress in Dunaliella tertiolecta. Plant Physiol. Biochem. 2007, 45, 705–710. [Google Scholar] [CrossRef]
- Lee, G.; Carrow, R.N.; Duncan, R.R.; Eiteman, M.A.; Rieger, M.W. Synthesis of organic osmolytes and salt tolerance mechanisms in Paspalum vaginatum. Environ. Exp. Bot. 2008, 63, 19–27. [Google Scholar] [CrossRef]
- Conde, A.; Silva, P.; Agasse, A.; Conde, C.; Geros, H. Mannitol transport and mannitol dehydrogenase activities are coordinated in Olea europaea under salt and osmotic stresses. Plant Cell Physiol. 2011, 52, 1766–1775. [Google Scholar] [CrossRef] [PubMed]
- Gil, R.; Lull, C.; Boscaiu, M.; Bautista, I.; Vicente, O. Soluble carbohydrates as osmolytes in several halophytes from a mediterranean salt marsh. Not. Bot. Horti. Agrobot. 2011, 39, 9–17. [Google Scholar] [CrossRef]
- Bertrand, A.; Dhont, C.; Bipfubusa, M.; Chalifour, F.P.; Drouin, P.; Beauchamp, C.J. Improving salt stress responses of the symbiosis in alfalfa using salt-tolerant cultivar and rhizobial strain. Appl. Soil Ecol. 2015, 87, 108–117. [Google Scholar] [CrossRef]
- Rodriguez, H.G.; Drew, M.C. Growth, water relations, and accumulation of organic and inorganic solutes in roots of maize seedlings during salt stress. Plant Physiol. 1997, 113, 881–893. [Google Scholar] [CrossRef]
- Page-Sharp, M.; Behm, C.A.; Smith, G.D. Involvement of the compatible solutes trehalose and sucrose in the response to salt stress of a cyanobacterial Scytonema, species isolated from desert soils. Biochim. Biophys. Acta 1999, 1472, 519–528. [Google Scholar] [CrossRef]
- Kerepesi, I.; Galiba, G. Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop Sci. 2000, 40, 482–487. [Google Scholar] [CrossRef]
- Liu, T.; Staden, J.V. Partitioning of carbohydrates in salt-sensitive and salt-tolerant soybean callus cultures under salinity stress and its subsequent relief. Plant Growth Regul. 2001, 33, 13–17. [Google Scholar] [CrossRef]
- Pommerrenig, B.; Papini-Terzi, F.S.; Sauer, N. Differential regulation of sorbitol and sucrose loading into the phloem of Plantago major in response to salt stress. Plant Physiol. 2007, 144, 1029–1038. [Google Scholar] [CrossRef]
- Nedjimi, B. Is salinity tolerance related to osmolytes accumulation in Lygeum spartum L. seedlings? J. Saudi Soc. Agric. Sci. 2011, 10, 81–87. [Google Scholar] [CrossRef]
- Redillas, M.R.; Park, S.H.; Lee, J.W.; Kim, J.W.; Jeong, J.S.; Jung, H.; Bang, S.W.; Hahn, T.; Kim, J. Accumulation of trehalose increases soluble sugar contents in rice plants conferring tolerance to drought and salt stress. Plant Biotechnol. Rep. 2012, 6, 89–96. [Google Scholar] [CrossRef]
- Singh, P.; Choudhary, K.; Chaudhary, N.; Gupta, S.; Sahu, M.; Tejaswini, B.; Sarkar, S. Salt stress resilience in plants mediated through osmolyte accumulation and its crosstalk mechanism with phytohormones. Front Plant Sci. 2022, 13, 1006617. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive oxygen species in plant signaling. Ann. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef]
- Huihui, Z.; Xin, L.; Zisong, X.; Yue, W.; Zhiyuan, T.; Meijun, A.; Yuehui, Z.; Wenxu, Z.; Nan, X.; Guangyu, S. Toxic effects of heavy metals Pb and Cd on mulberry (Morus alba L.) seedling leaves: Photosynthetic function and reactive oxygen species (ROS) metabolism responses. Ecotoxicol. Environ. Saf. 2020, 195, 110469. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef]
- Li, C.; Wang, G.; Zhao, J.; Zhang, L.; Ai, L.; Han, Y.; Sun, D.; Zhang, S.; Sun, Y. The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef]
- Pan, J.; Zhang, M.; Kong, X.; Xing, X.; Liu, Y.; Zhou, Y.; Liu, Y.; Sun, L.; Li, D. ZmMPK17, a novel maize group D MAP kinase gene, is involved in multiple stress responses. Planta 2011, 235, 661–676. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, S.; Pan, J.; Kong, X.; Zhou, Y.; Liu, Y.; Li, D. The overexpression of a maize mitogen-activated protein kinase gene (ZmMPK5) confers salt stress tolerance and induces defence responses in tobacco. Plant Biol. 2013, 16, 558–570. [Google Scholar] [CrossRef]
- Perez-Salamo, I.; Papdi, C.; Rigo, G.; Zsigmond, L.; Vilela, B.; Lumbreras, V.; Nagy, I.; Horvath, B.; Domoki, M.; Darula, Z.; et al. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 2014, 165, 319–334. [Google Scholar] [CrossRef]
- Nadarajah, K.K. ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef]
- Pitzschke, A.; Djamei, A.; Bitton, F.; Hirt, H. A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant 2009, 2, 120–137. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Wang, G.; Cha, J.Y.; Li, G.; Chen, S.; Li, Z.; Guo, J.; Zhang, C.; Yang, Y.; et al. A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 2015, 27, 908–925. [Google Scholar] [CrossRef]
- Del Rio, L.A.; Lopez-Huertas, E. ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol. 2016, 57, 1364–1376. [Google Scholar] [CrossRef]
- Li, G.; Li, J.; Hao, R.; Guo, Y. Activation of catalase activity by a peroxisome-localized small heat shock protein Hsp17.6CII. J. Genet. Genom. 2017, 44, 395–404. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Grimm, B.; Wobus, U.; Weschke, W. Differential response of antioxidant compounds to salinity stress in salt-tolerant and salt- sensitive seedlings of foxtail millet (Setaria italica). Physiol. Plant 2000, 109, 435–442. [Google Scholar] [CrossRef]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Amako, K.; Ushimaru, T. Dehydroascorbate reductase and salt stress. CABI Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2009, 4, 1–7. [Google Scholar] [CrossRef]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Begaramorales, J.C.; Sanchezcalvo, B.; Chaki, M.; Mataperez, C.; Valderrama, R.; Padilla, M.N.; Lopez-Jaramillo, J.; Luque, F.; Corpas, F.J.; Barroso, J.B. Differential molecular response of monodehydroascorbate reductase and glutathione reductase by nitration and s-nitrosylation. J. Exp. Bot. 2015, 66, 5983–5996. [Google Scholar] [CrossRef]
- Wang, X.; Fang, G.; Yang, J.; Li, Y. A thioredoxin-dependent glutathione peroxidase (OsGPX5) is required for rice normal development and salt stress tolerance. Plant Mol. Biol. Rep. 2017, 35, 333–342. [Google Scholar] [CrossRef]
- Hernandez, J.A.; Jimenez, A.; Mullineaux, P.; Sevilia, F. Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ. 2000, 23, 853–862. [Google Scholar] [CrossRef]
- De Pascale, S.; Maggio, A.; Angelino, G.; Graziani, G. Effect of salt stress on water relations and antioxidant activity in tomato. Acta Hortic. 2003, 613, 39–46. [Google Scholar] [CrossRef]
- Huang, C.; He, W.; Guo, J.; Chang, X.; Su, P.; Zhang, L. Increased sensitivity to salt stress in an ascorbate-deficient Arabidopsis mutant. J. Exp. Bot. 2005, 56, 3041–3049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Li, C.J.; Nan, Z.B. Effects of salt and drought stress on alkaloid production in endophyte-infected drunken horse grass (Achnatherum inebrians). Biochem. Syst. Ecol. 2011, 39, 471–476. [Google Scholar] [CrossRef]
- Borghesi, E.; Gonzalezmiret, M.L.; Escuderogilete, M.L.; Malorgio, F.; Heredia, F.J.; Melendezmartinez, A.J. Effects of salinity stress on carotenoids, anthocyanins, and color of diverse tomato genotypes. J. Agric. Food Chem. 2011, 59, 11676–11682. [Google Scholar] [CrossRef]
- Petridis, A.; Therios, I.; Samouris, G.; Tananaki, C. Salinity-induced changes in phenolic compounds in leaves and roots of four olive cultivars (Olea europaea L.) and their relationship to antioxidant activity. Environ. Exp. Bot. 2012, 79, 37–43. [Google Scholar] [CrossRef]
- Abdallah, S.B.; Aung, B.; Amyot, L.; Lalin, I.; Lachaal, M.; Karray-Bouraoui, N.; Hannoufa, A. Salt stress (NaCl) affects plant growth and branch pathways of carotenoid and flavonoid biosyntheses in Solanum nigrum. Acta Physiol. Plant. 2016, 38, 72. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, C.; Tang, D.; Yan, L.; Wang, D.; Yang, Y.; Gui, J.; Zhao, X.; Li, L.; Tang, X.; et al. The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. New phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef]
- Tamaki, S.; Mochida, K.; Suzuki, K. Diverse biosynthetic pathways and protective functions against environmental stress of antioxidants in microalgae. Plants 2021, 10, 1250. [Google Scholar] [CrossRef]
- Tiong, S.H.; Looi, C.Y.; Hazni, H.; Arya, A.; Paydar, M.; Wong, W.F.; Cheah, S.-C.; Mustafa, M.R.; Awang, K. Antidiabetic and antioxidant properties of alkaloids from Catharanthus roseus (L.) G. Don. Molecules 2013, 18, 9770–9784. [Google Scholar] [CrossRef]
- Kumar, S.; Thakur, P.; Kaushal, N.; Malik, J.A.; Gaur, P.; Nayyar, H. Effect of varying high temperatures during reproductive growth on reproductive function, oxidative stress and seed yield in chickpea genotypes differing in heat sensitivity. Arch. Agron. Soil Sci. 2013, 59, 823–843. [Google Scholar] [CrossRef]
- Hussain, H.A.; Men, S.; Hussain, S.; Chen, Y.; Ali, S.; Zhang, S.; Zhang, K.; Li, Y.; Xu, Q.; Liao, C. Interactive effects of drought and heat stresses on morpho-physiological attributes, yield, nutrient uptake and oxidative status in maize hybrids. Sci. Rep. 2019, 9, 3890. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ju, J.; Xia, G. Identification of the flavonoid3-hydroxylase and flavonoid 3,5-hydroxylase genes from Antarctic moss and their regulation during abiotic stress. Gene 2014, 543, 145–152. [Google Scholar] [CrossRef]
- Di Ferdinando, M.; Brunetti, C.; Fini, A.; Tattini, M. Flavonoids as antioxidants in plants under abiotic stresses. In Abiotic Stress Responses in Plants; Ahmad, P., Prasad, M., Eds.; Springer: New York, NY, USA, 2012; pp. 159–179. [Google Scholar]
- Park, J.E.; Park, J.Y.; Kim, Y.S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.Y.; Kim, J.; Lee, Y.H.; Park, C.M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef]
- Lee, H.S.; Li, H.; Xu, B.; Deng, X.; Kwak, S.S. Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Biochem. 2015, 94, 19–27. [Google Scholar]
- Ribba, T.; Garrido-Vargas, F.; O’Brien, J.A. Auxin-mediated responses under salt stress: From developmental regulation to biotechnological applications. J. Exp. Bot. 2020, 71, 3843–3853. [Google Scholar] [CrossRef]
- Yan, S.; Che, G.; Ding, L.; Chen, Z.; Liu, X.; Wang, H.; Zhao, W.; Ning, K.; Zhao, J.; Tesfamichael, K.; et al. Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 2016, 6, 20760. [Google Scholar] [CrossRef]
- Ke, Q.; Wang, Z.; Ji, C.Y.; Jeong, J.C.; Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2015, 25, 1117–1130. [Google Scholar] [CrossRef]
- Naser, V.; Shani, E. Auxin response under osmotic stress. Plant Mol. Biol. 2016, 91, 661–672. [Google Scholar] [CrossRef]
- Jiang, K.; Moe-Lange, J.; Hennet, L.; Feldman, L.J. Salt stress affects the redox status of Arabidopsis root meristems. Front. Plant Sci. 2016, 7, 81. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef]
- Su, Q.; Zheng, X.; Tian, Y.; Wang, C. Exogenous brassinolide alleviates salt stress in Malus hupehensis Rehd. by regulating the transcription of NHX-type Na+(K+)/H+ antiporters. Front. Plant Sci. 2020, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Guan, L.; Sun, Y.; Zhu, Y.; Liu, L.; Lu, R.; Jiang, M.; Tan, M.; Zhang, A. Calcium and ZmCCaMK are involved in brassinosteroid-induced antioxidant defense in maize leaves. Plant Cell Physiol. 2015, 56, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.; Ding, J.; Lee, D.; Lu, X.; Feng, Y.; Song, W. Overexpression of SoCYP85A1, a spinach cytochrome p450 gene in transgenic tobacco enhances root development and drought stress tolerance. Front. Plant Sci. 2017, 8, 1909. [Google Scholar] [CrossRef]
- Zeng, H.T.; Tang, Q.; Hua, X.J. Arabidopsis brassinosteroid mutants det2-1 and bin2-1 display altered salt tolerance. J. Plant Growth Regul. 2010, 29, 44–52. [Google Scholar] [CrossRef]
- Li, Z.Y.; Xu, Z.S.; He, G.Y.; Yang, G.X.; Chen, M.; Li, L.C.; Ma, Y.Z. A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Commun. 2012, 426, 522–527. [Google Scholar] [CrossRef]
- Geng, Y.; Wu, R.; Wee, C.W.; Xie, F.; Wei, X.; Chan, P.M.; Tham, C.; Duan, L.; Dinneny, J.R.A. spatiotemporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 2013, 25, 2132–2154. [Google Scholar] [CrossRef]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H.; Guo, H. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 2014, 10, e1004664. [Google Scholar] [CrossRef]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Aziz, U.; Alsahli, A.A.; Alyemeni, M.N.; Ahmad, P. Influence of exogenous salicylic acid and nitric oxide on growth, photosynthesis, and ascorbate-glutathione cycle in salt stressed Vigna angularis. Biomolecules 2019, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, S.G.; Park, C.M. Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 2010, 188, 626–637. [Google Scholar] [CrossRef]
- Wang, J.; Qin, H.; Zhou, S.; Wei, P.; Zhang, H.; Zhou, Y.; Miao, Y.; Huang, R. The ubiquitin-binding protein OsDSK2a mediates seedling growth and salt responses by regulating gibberellin metabolism in rice. Plant Cell 2020, 32, 414–428. [Google Scholar] [CrossRef]
- Zhou, J.; Li, Z.; Xiao, G.; Zhai, M.; Pan, X.; Huang, R.; Zhang, H. CYP71D8L is a key regulator involved in growth and stress responses by mediating gibberellin homeostasis in rice. J. Exp. Bot. 2020, 71, 1160–1170. [Google Scholar]
- Wang, C.; Yang, Y.; Wang, H.; Ran, X.; Li, B.; Zhang, J.; Zhang, H. Ectopic expression of a cytochrome P450 monooxygenase gene PtCYP714A3 from Populus trichocarpa reduces shoot growth and improves tolerance to salt stress in transgenic rice. Plant Biotechnol. J. 2016, 14, 1838–1851. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Salinity stress enhances color parameters, bioactive leaf pigments, vitamins, polyphenols, flavonoids and antioxidant activity in selected amaranthus leafy vegetables. J. Sci. Food Agric. 2019, 99, 2275–2284. [Google Scholar] [CrossRef]
- Kaya, C.; Tuna, A.L.; Yokas, I. The role of plant hormones in plants under salinity stress. Salin. Water Stress 2009, 44, 45–50. [Google Scholar]
- Abdel Latef, A.A.H.; Chaoxing, H. Effect of arbuscular mycorrhizal fungi on growth, mineral nutrition, antioxidant enzymes activity and fruit yield of tomato grown under salinity stress. Sci. Hortic. 2011, 127, 228–233. [Google Scholar] [CrossRef]
- Ashraf, M.; Akram, N.A. Improving salinity tolerance of plants through conventional breeding and genetic engineering: An analytical comparison. Biotechnol. Adv. 2009, 27, 744–752. [Google Scholar] [CrossRef]
- Charfeddine, M.; Charfeddine, S.; Ghazala, I.; Bouaziz, D.; Bouzid, R.G. Investigation of the response to salinity of transgenic potato plants overexpressing the transcription factor StERF94. J. Biosci. 2019, 44, 141. [Google Scholar] [CrossRef]
- Levy, D.; Coleman, W.K.; Veilleux, R.E. Adaptation of potato to water shortage: Irrigation management and enhancement of tolerance to drought and salinity. Am. J. Potato Res. 2013, 90, 186–206. [Google Scholar] [CrossRef]
- Kolomeichuk, L.V.; Efimova, M.V.; Zlobin, I.E.; Kreslavski, V.D.; Murgan, O.K.; Kovtun, I.S.; Khripach, V.A.; Kuznetsov, V.V.; Allakhverdiev, S.I. 24-epibrassinolide alleviates the toxic effects of NaCl on photosynthetic processes in potato plants. Photosynth. Res. 2020, 146, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Oi, T.; Enomoto, S.; Nakao, T.; Arai, S.; Yamane, K.; Taniguchi, M. Three-dimensional ultrastructural change of chloroplasts in rice mesophyll cells responding to salt stress. Ann. Bot. 2020, 125, 833–840. [Google Scholar] [CrossRef]
- Zhu, D.; Luo, F.; Zou, R.; Liu, J.; Yan, Y. Integrated physiological and chloroplast proteome analysis of wheat seedling leaves under salt and osmotic stresses. J. Proteom. 2021, 234, 104097. [Google Scholar] [CrossRef]
- Goussi, R.; Manaa, A.; Derbali, W.; Cantamessa, S.; Abdelly, C.; Barbato, R. Comparative analysis of salt stress, duration and intensity, on the chloroplast ultrastructure and photosynthetic apparatus in Thellungiella salsuginea. J. Photochem. Photobiol. B Biol. 2018, 183, 275–287. [Google Scholar] [CrossRef]
- Bose, J.; Munns, R.; Shabala, S.; Gilliham, M.; Pogson, B.; Tyerman, S.D. Chloroplast function and ion regulation in plants growing on saline soils: Lessons from halophytes. J. Exp. Bot. 2017, 68, 3129–3143. [Google Scholar] [CrossRef]
- Rahman, M.M.; Rahman, M.A.; Miah, M.G.; Saha, S.R.; Karim, M.; Mostofa, M.G. Mechanistic insight into salt tolerance of Acacia auriculiformis: The importance of ion selectivity, osmoprotection, tissue tolerance, and Na+ exclusion. Front. Plant Sci. 2017, 8, 155. [Google Scholar] [CrossRef]
- Qin, C.; Ahanger, M.; Zhou, J.; Ahmed, N.; Wei, C.; Yuan, S.; Ashraf, M.; Zhang, L. Beneficial role of acetylcholine in chlorophyll metabolism and photosynthetic gas exchange in Nicotiana benthamiana seedlings under salinity stress. Plant Biol. 2020, 22, 357–365. [Google Scholar] [CrossRef]
- Zahra, N.; Al Hinai, M.S.; Hafeez, M.B.; Rehman, A.; Wahid, A.; Siddique, K.H.M.; Farooq, M. Regulation of photosynthesis under salt stress and associated tolerance mechanisms. Plant Physiol. Biochem. 2022, 178, 55–69. [Google Scholar] [CrossRef]
- Goussi, R.; Manfredi, M.; Marengo, E.; Derbali, W.; Cantamessa, S.; Barbato, R.; Manaa, A. Thylakoid proteome variation of Eutrema salsugineum in response to drought and salinity combined stress. Biochim. Biophy. Acta-Bioener. 2021, 1862, 148482. [Google Scholar] [CrossRef]
- Pompelli, M.F.; Ferreira, P.P.; Chaves, A.R.; Figueiredo, R.C.; Martins, A.O.; Jarma-Orozco, A.; Bhatt, A.; Batista-Silva, W.; Endres, L.; Araújo, W.L. Physiological, metabolic, and stomatal adjustments in response to salt stress in Jatropha curcas. Plant Physiol. Biochem. 2021, 168, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Killi, D.; Haworth, M. Diffusive and metabolic constraints to photosynthesis in quinoa during drought and salt stress. Plants 2017, 6, 49. [Google Scholar] [CrossRef]
- Wanichthanarak, K.; Boonchai, C.; Kojonna, T.; Chadchawan, S.; Sangwongchai, W.; Thitisaksakul, M. Deciphering rice metabolic flux reprograming under salinity stress via in silico metabolic modeling. Comput. Struct. Biotechnol. J. 2020, 18, 3555–3566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, R.; Yang, X.; Ju, Q.; Li, W.; Lü, S.; Tran, L.P.; Xu, J. The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 2020, 43, 1925–1943. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef]
- Park, Y.; Lim, S.; Moon, J.; Jang, C. A rice really interesting new gene H2-type E3 ligase, OsSIRH2-14, enhances salinity tolerance via ubiquitin/26 S proteasome-mediated degradation of salt-related proteins. Plant Cell Environ. 2019, 42, 3061–3076. [Google Scholar] [CrossRef]
- Tian, M.; Lou, L.; Liu, L.; Yu, F.; Zhao, Q.; Zhang, H.; Wu, Y.; Tang, S.; Xia, R.; Zhu, B.; et al. The RING finger E3 ligase STRF1 is involved in membrane trafficking and modulates salt-stress response in Arabidopsis thaliana. Plant J. 2015, 82, 81–92. [Google Scholar] [CrossRef]
- Kim, J.; Lim, S.; Jang, C. Oryza sativa, C4HC3-type really interesting new gene (RING), OsRFPv6, is a positive regulator in response to salt stress by regulating Na+ absorption. Physiol. Plant 2021, 173, 883–895. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, X.; Wang, L.; Cao, Y.; Song, W.; Shi, J.; Lai, J.; Jiang, C. A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat. Plants 2019, 5, 1297–1308. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, X.; Qin, F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef]
- Gao, H.; Cui, J.; Liu, S.; Wang, S.; Lian, Y.; Bai, Y.; Zhu, T.; Wu, H.; Wang, Y.; Yang, S.; et al. Natural variations of ZmSRO1d modulate the trade-off between drought resistance and yield by affecting ZmRBOHC-mediated stomatal ROS production in maize. Mol. Plant 2022, 15, 1558–1574. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, F.; Xie, P.; Sun, S.; Qiao, X.; Tang, S.; Chen, C.; Yang, S.; Mei, C.; Yang, D.; et al. A Gg protein regulates alkaline sensitivity in crops. Science 2023, 379, eade8416. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, S.J.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M.; et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012, 30, 360-U173. [Google Scholar] [CrossRef]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef]
- Zhang, M.; Li, Y.; Liang, X.; Lu, M.; Lai, J.; Song, W.; Jiang, C. A teosinte-derived allele of an HKT1 family sodium transporter improves salt tolerance in maize. Plant Biotechnol. J. 2023, 21, 97–108. [Google Scholar] [CrossRef]
- Zhou, X.; Li, J.; Wang, Y.; Liang, X.; Zhang, M.; Lu, M.; Guo, Y.; Qin, F.; Jiang, C. The classical SOS pathway confers natural variation of salt tolerance in maize. New Phytol. 2022, 236, 479–494. [Google Scholar] [CrossRef]
- Wang, Z.; Hong, Y.; Li, Y.; Shi, H.; Yao, J.; Liu, X.; Wang, F.; Huang, S.; Zhu, G.; Zhu, J.K. Natural variations in SlSOS1 contribute to the loss of salt tolerance during tomato domestication. Plant Biotechnol. J. 2021, 19, 20–22. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, X.; Li, H.; Zheng, H.; Zhang, J.; Olsen, M.; Varshney, R.; Prasanna, B.; Qian, Q. Smart breeding driven by big data, artificial intelligence, and integrated genomic-enviromic prediction. Mol. Plant 2022, 15, 1664–1695. [Google Scholar] [CrossRef]
- Khan, M.-U.; Wang, S.; Wang, J.; Ahmar, S.; Saeed, S.; Khan, S.; Xu, X.; Chen, H.; Bhat, J.; Feng, X. Applications of Artificial Intelligence in Climate-Resilient Smart-Crop Breeding. Int. J. Mol. Sci. 2022, 23, 11156. [Google Scholar] [CrossRef]
- Esposito, S.; Carputo, D.; Cardi, T.; Tripodi, P. Applications and trends of machine learning in genomics and phenomics for next-generation breeding. Plants 2020, 9, 34. [Google Scholar] [CrossRef]
- Reinoso-Peláez, E.L.; Gianola, D.; González-Recio, O. Genome-enabled prediction methods based on machine learning. In Genomic Prediction of Complex Traits; Methods in Molecular Biology; Ahmadi, N., Bartholomé, J., Eds.; Humana: New York, NY, USA, 2022; Volume 2467. [Google Scholar]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; De Los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Selvaraj, M.G.; Valderrama, M.; Guzman, D.; Valencia, M.; Ruiz, H.; Acharjee, A. Machine learning for high-throughput field phenotyping and image processing provides insight into the association of above and below-ground traits in cassava (Manihot esculenta Crantz). Plant Methods 2020, 16, 87. [Google Scholar] [CrossRef]
- Sadeghi-Tehran, P.; Sabermanesh, K.; Virlet, N.; Hawkesford, M.J. Automated method to determine two critical growth stages of wheat: Heading and flowering. Front. Plant Sci. 2017, 8, 252. [Google Scholar] [CrossRef]
- Brichet, N.; Fournier, C.; Turc, O.; Strauss, O.; Artzet, S.; Pradal, C.; Welcker, C.; Tardieu, F.; Cabrera-Bosquet, L. A robot-assisted imaging pipeline for tracking the growths of maize ear and silks in a high-throughput phenotyping platform. Plant Methods 2017, 13, 96. [Google Scholar] [CrossRef]
- Yang, Q.; Chen, Z.Z.; Zhou, X.F.; Yin, H.B.; Li, X.; Xin, X.F.; Hong, X.H.; Zhu, J.K.; Gong, Z. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2009, 2, 22–31. [Google Scholar] [CrossRef]
- Ma, D.M.; Xu, W.R.; Li, H.W.; Jin, F.X.; Guo, L.N.; Wang, J.; Da, H.J.; Xu, X. Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.). Protoplasma 2014, 251, 219–231. [Google Scholar] [CrossRef]
- Gong, Q.; Li, P.; Ma, S.; Indu Rupassara, S.; Bohnert, H. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005, 44, 826–839. [Google Scholar] [CrossRef]
- Wang, X.; Chang, L.; Wang, B.; Wang, D.; Li, P. Comparative proteomics of Thellungiella halophila leaves from plants subjected to salinity reveals the importance of chloroplastic starch and soluble sugars in halophyte salt tolerance. Mol. Cell. Proteom. 2013, 12, 2174–2195. [Google Scholar] [CrossRef]
- Kiani-Pouya, A.; Roessner, U.; Jayasinghe, N.; Lutz, A.; Rupasinghe, T. Epidermal bladder cells confer salinity stress tolerance in the halophyte quinoa and Atriplex species. Plant Cell Environ. 2017, 40, 1900–1915. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).