Genome-Wide Analysis of TCP Transcription Factors and Their Expression Pattern Analysis of Rose Plants (Rosa chinensis)
Abstract
:1. Introduction
2. Methods
2.1. Sequence Identification and Annotation of TCP Genes in Rosa chinensis
2.2. Physicochemical Analysis and Prediction of TCP Gene Family Members
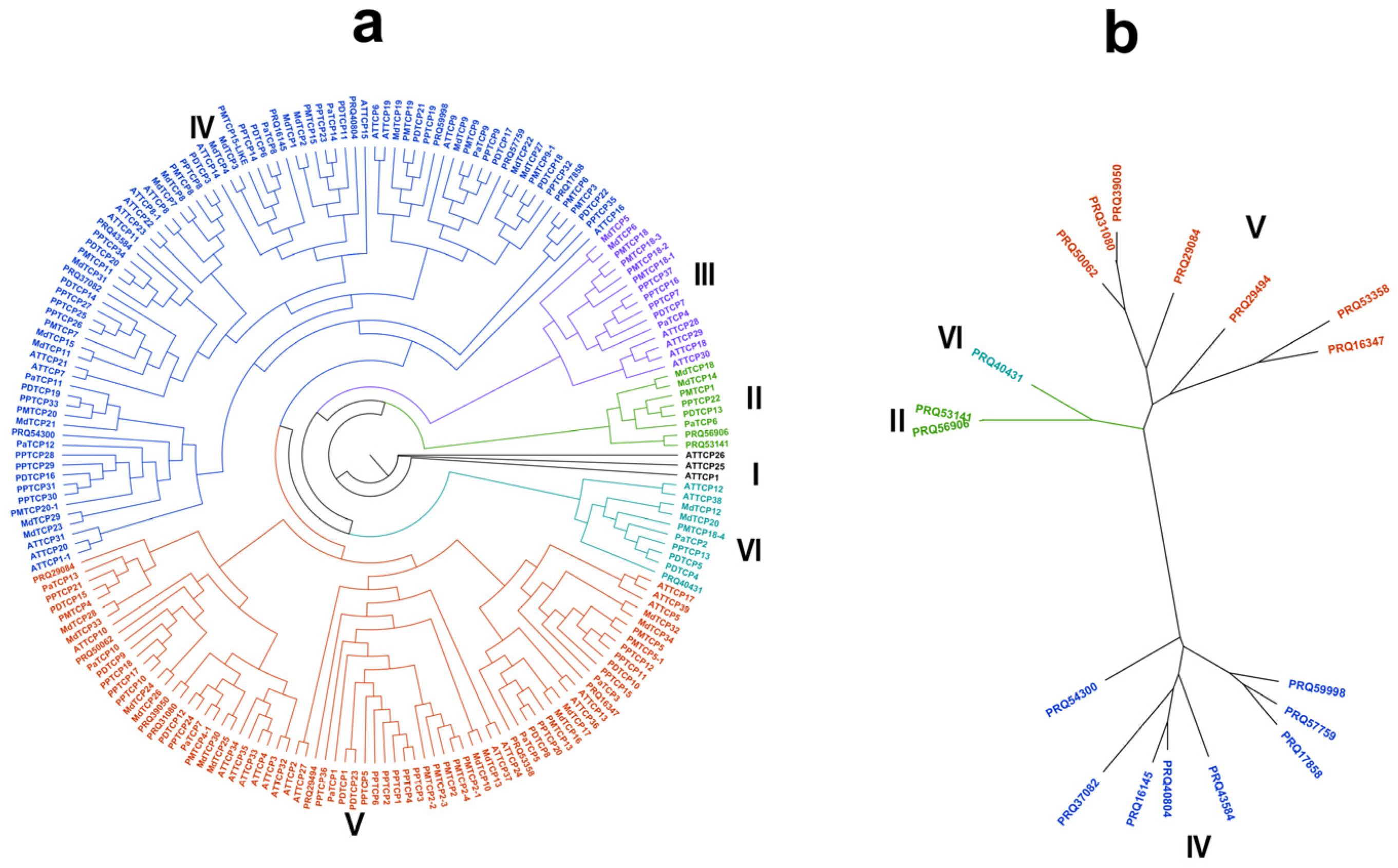
2.3. Phylogenetic Analysis of the TCP Genes
2.4. Chromosomal Distribution and Gene Structure Analysis for TCP Genes
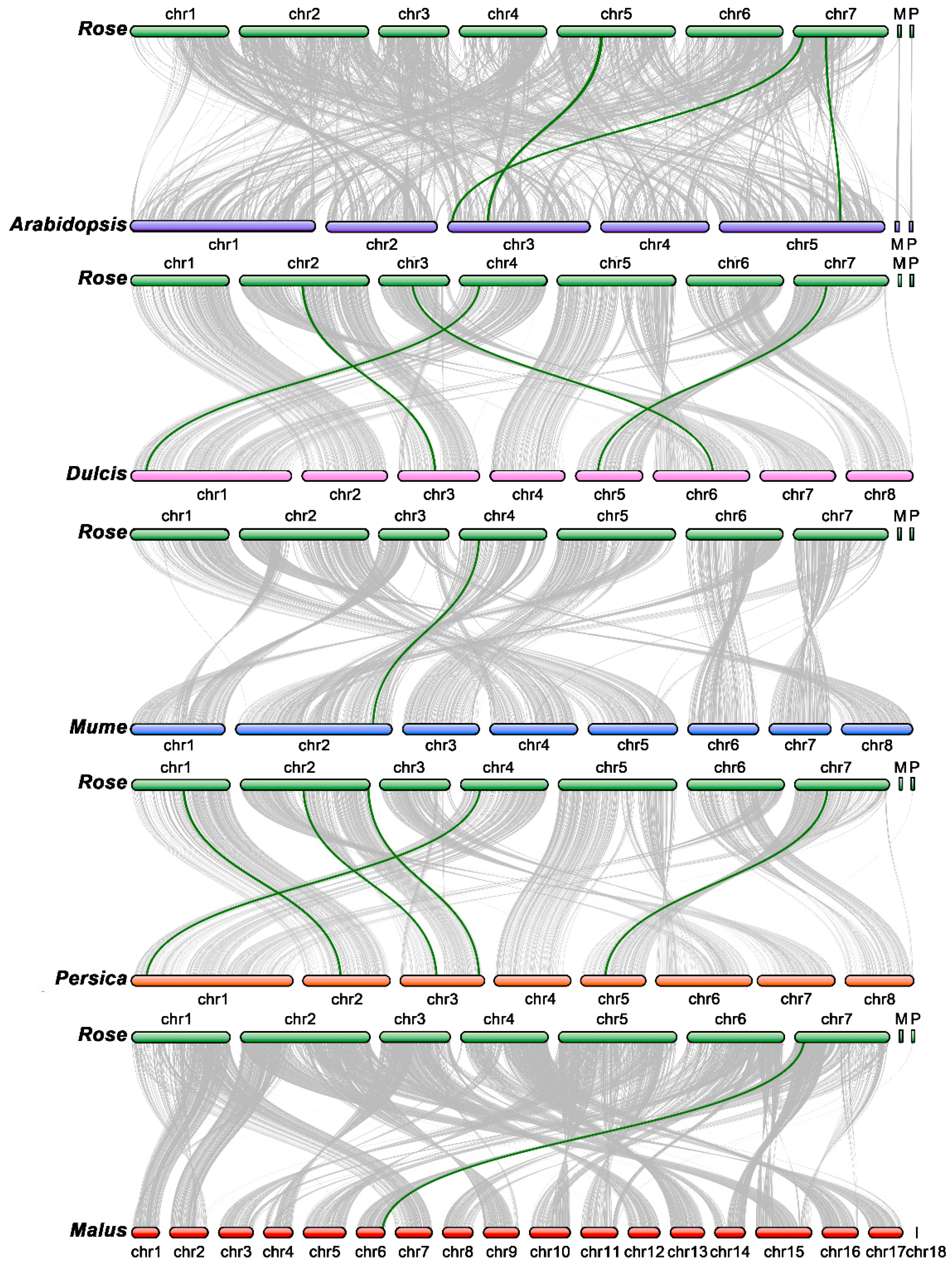
2.5. Synteny Analysis of TCP Genes
2.6. Gene Expression Patterns Based on Transcriptome Data
2.7. qPCR for Certification
3. Results
3.1. Identification of TCP Proteins in Roses
3.2. Chromosomal Distribution of Rose TCP Genes
3.3. Phylogenetic Analysis of TCP Genes
3.4. Intron-Exon Distribution and Motif Analyses of TCP Genes
3.5. Synteny Analysis for TCP Genes in Rose and Other Species
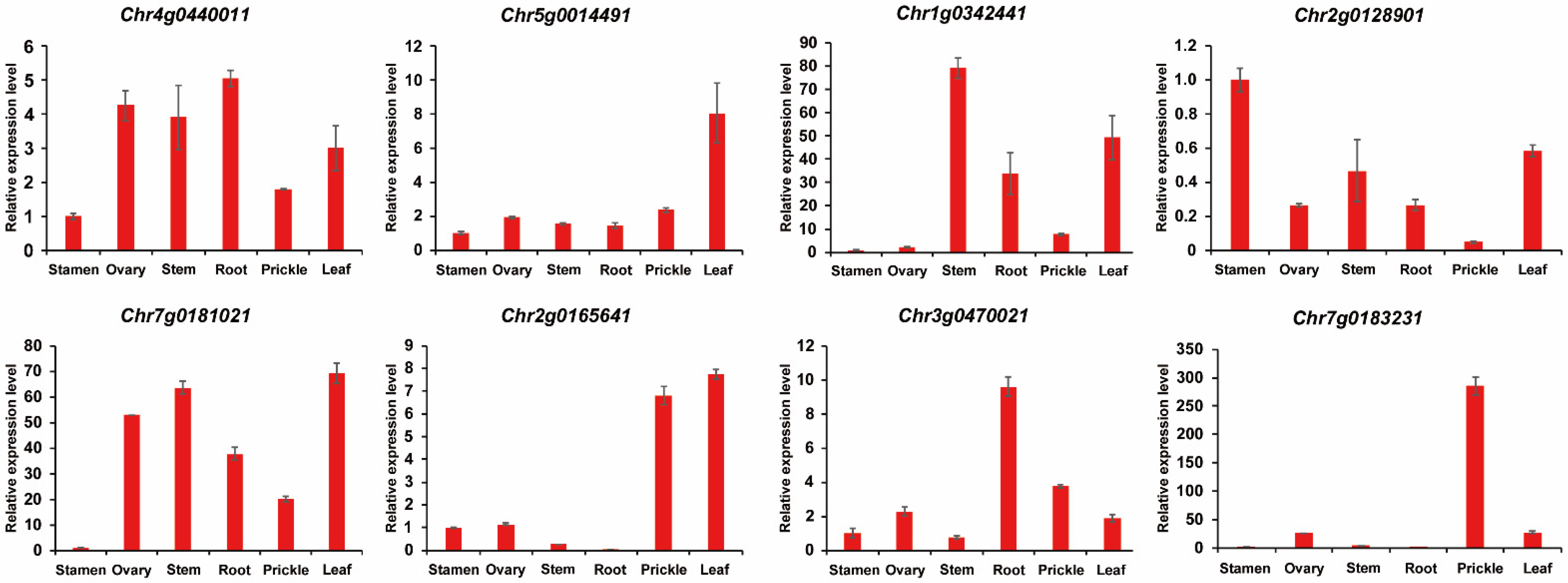
3.6. Analyses of Gene Expression of TCPs in Roses
3.7. Expression Characteristics of Rose TCP Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhan, W.; Cui, L.; Guo, G.; Zhang, Y. Genome-wide identification and functional analysis of the TCP gene family in rye (Secale cereale L.). Gene 2023, 854, 147104. [Google Scholar] [CrossRef]
- Wang, F.; Cai, X.X.; Wei, H.Z.; Zhang, L.H.; Dong, A.W.; Su, W. Histone methylation readers MRG1/MRG2 interact with the transcription factor TCP14 to positively modulate cytokinin sensitivity in Arabidopsis. J. Genet. Genomics 2023, in press. [Google Scholar] [CrossRef]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.; Deckena, M.; Almeida-Trapp, M.; Kopischke, S.; Kock, C.; Schüssler, E.; Tsiantis, M.; Mithöfer, A.; Zachgo, S. Mp TCP 1 controls cell proliferation and redox processes in Marchantia polymorpha. New Phytol. 2019, 224, 1627–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef]
- Lukens, L.; Doebley, J. Molecular evolution of the teosinte branched gene among maize and related grasses. Mol. Biol. Evol. 2001, 18, 627–638. [Google Scholar] [CrossRef] [Green Version]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 1997, 9, 1607–1619. [Google Scholar]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [Green Version]
- Leng, X.P.; Wei, H.R.; Xu, X.Z.; Ghuge, S.A.; Jia, D.J.; Liu, G.S.; Wang, Y.Z.; Yuan, Y.B. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genom. 2019, 20, 786. [Google Scholar] [CrossRef] [Green Version]
- Nie, Y.M.; Han, F.X.; Ma, J.J.; Chen, X.; Song, Y.T.; Niu, S.H.; Wu, H.X. Genome-wide TCP transcription factors analysis provides insight into their new functions in seasonal and diurnal growth rhythm in Pinus tabuliformis. BMC Plant Biol. 2022, 22, 167. [Google Scholar] [CrossRef]
- Li, D.L.; Tang, X.; Dong, Y.X.; Wang, Y.Y.; Shi, S.L.; Li, S.H.; Liu, Y.; Ge, H.Y.; Chen, H.Y. Comparative genomic investigation of TCP gene family in eggplant (Solanum melongena L.) and expression analysis under divergent treatments. Plant Cell Rep. 2022, 41, 2213–2228. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Tatematsu, K.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 2008, 53, 42–52. [Google Scholar] [CrossRef]
- Aguilar-Martınez, J.A.; Poza-Carrion, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef]
- Sun, X.D.; Wang, C.D.; Xiang, N.; Li, X.; Yang, S.H.; Du, J.C.; Yang, Y.P.; Yang, Y.Q. Activation of secondary cell wall biosynthesis by miR319-targeted. Plant Biotechnol. J. 2017, 15, 1284–1294. [Google Scholar] [CrossRef] [Green Version]
- Rubio-somoza, I.; Weigel, D. Coordination of flower maturation by a regulatory circuit of three MicroRNAs. PLoS Genet. 2013, 9, e1003374. [Google Scholar] [CrossRef] [Green Version]
- Hammani, K.; Gobert, A.; Hleibieh, K.; Choulier, L.; Small, I.; Giege, P. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 2011, 23, 730–740. [Google Scholar] [CrossRef] [Green Version]
- Pagnussat, G.C.; Yu, H.J.; Ngo, Q.A.; Rajani, S.; Mayalagu, S.; Johnson, C.S.; Capron, A.; Xie, L.-F.; Ye, D.; Sundaresan, V. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 2005, 132, 603–614. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.R.; Li, D.Y.; Yan, J.P.; Wang, K.X.; Luo, H.; Zhang, W.J. MiR319-mediated ethylene biosynthesis, signalling and salt stress response in switchgrass. Plant Biotechnol. J. 2019, 17, 2370–2383. [Google Scholar] [CrossRef] [Green Version]
- Viola, I.L.; Camoirano, A.; Gonzalez, D.H. Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol. 2016, 170, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Giraud, E.; Ng, S.; Carrie, C.; Duncan, O.; Low, J.; Lee, C.P.; Aken, O.V.; Millar, H.A.; Murcha, M.; Whelan, J. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 2010, 22, 3921–3934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.L.; Gao, Y.M.; Wu, M.; Shi, Y.N.; Wang, H.; Wu, L.; Xiang, Y. TCP10, a TCP transcription factor in moso bamboo (Phyllostachys edulis), confers drought tolerance to transgenic plants. Environ. Exp. Bot. 2020, 172, 104002. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002, 30, 337–348. [Google Scholar] [CrossRef] [Green Version]
- Danisman, S.; Wal, F.; Dhondt, S.; Waites, R.; Folter, S.; Bimbo, A.; Dijk, A.D.J.; Muino, J.M.; Cutri, L.; Dornelas, M.C.; et al. Arabidopsis Class I and Class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar] [CrossRef] [Green Version]
- Parapunova, V.; Busscher, M.; Busscher-lange, J.; Lammers, M.; Karlova, R.; Bovy, A.G.; Angenent, G.C.; de Maagd, R.A. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014, 14, 157. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Hu, Y.; Cui, M.Y.; Han, Y.T.; Gao, K.; Feng, J.Y. Identification and Transcript Analysis of the TCP Transcription Factors in the Diploid Woodland Strawberry Fragaria vesca. Front. Plant Sci. 2016, 7, 1937. [Google Scholar] [CrossRef] [Green Version]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [Green Version]
- Martín-Trillo, M.; Grandío, E.G.; Serra, F.; Marcel, F.; Rodríguez-Buey, M.L.; Schmitz, G.; Theres, K.; Bendahmane, A.; Dopazo, H.; Cubas, P. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 2011, 67, 701–714. [Google Scholar] [CrossRef]
- Yao, X.; Ma, H.; Wang, J.; Zhang, D. Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 2007, 49, 885–897. [Google Scholar] [CrossRef]
- Xu, R.R.; Sun, P.; Jia, F.J.; Lu, L.T.; Li, Y.Y.; Zhang, S.Z.; Huang, J.G. Genome wide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 2014, 93, 733–746. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Q.L.; Sun, R.R.; Xie, F.L.; Jones, D.C.; Zhang, B.H. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 2014, 4, 6645. [Google Scholar] [CrossRef] [Green Version]
- Bao, S.; Zhang, Z.X.; Lian, Q.; Sun, Q.H.; Zhang, R.F. Evolution and expression of genes encoding TCP transcription factors in Solanum tuberosum reveal the involvement of StTCP23 in plant defence. BMC Genet. 2019, 20, 91. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.B.; Guy, K.M.; Wu, W.F.; Fang, B.S.; Yang, J.H.; Zhang, M.F.; Hu, Z.Y. Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 2016, 16, 85. [Google Scholar] [CrossRef] [Green Version]
- Shang, X.; Han, Z.; Zhang, D.; Wang, Y.; Qin, H.; Zou, Z.; Zhou, L.; Zhu, X.; Fang, W.; Ma, Y. Genome-Wide Analysis of the TCP Gene Family and Their Expression Pattern Analysis in Tea Plant (Camellia sinensis). Front. Plant Sci. 2022, 13, 840350. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.K.; Zhang, C.; Zhao, X.; Ke, S.; Li, Y.; Zhang, D.; Zheng, Q.; Li, M.H.; Lan, S.; Liu, Z.J. Genome-wide analysis of the TCP gene family and their expression pattern in Cymbidium goeringii. Front. Plant Sci. 2022, 13, 1068969. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Geng, Z.W.; Zhang, C.P.; Wang, K.L.; Jiang, X.Q. Whole-genome characterization of Rosa chinensis AP2/ERF transcription factors and analysis of negative regulator RcDREB2B in Arabidopsis. BMC Genom. 2021, 22, 90. [Google Scholar] [CrossRef]
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.; Wong, T.; Haeseler, A.; Jermiin, L. Model Finder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Chao, J.T.; Kong, Y.Z.; Wang, Q.; Sun, Y.H.; Gong, D.P.; Lv, J.; Liu, G.S. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Hereditas 2015, 37, 91–97. [Google Scholar]
- Bailey, T.L.; Bodén, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. Meme Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; De Barry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Xun, Q.; Zhang, D.; Lv, M.; Ou, Y.; Li, J. TCP Transcription Factors Associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to Regulate Thermomorphogenesis in Arabidopsis thaliana. iScience 2019, 15, 600–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarvepalli, K.; Nath, U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 2011, 67, 595–607. [Google Scholar] [CrossRef] [Green Version]
- Hao, J.; Lou, P.; Han, Y.; Chen, Z.; Chen, J.; Ni, J.; Yang, Y.; Jiang, Z.; Xu, M. GrTCP11, a Cotton TCP Transcription Factor, Inhibits Root Hair Elongation by Down-Regulating Jasmonic Acid Pathway in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 769675. [Google Scholar] [CrossRef]
- Xiao, X.; Lin, W.Q.; Feng, E.Y.; Wu, C.Y.; Ou, X.C. Genome-Wide Identification of Binding Sites for SmTCP7a Transcription Factors of Eggplant during Bacterial Wilt Resistance by ChIP-Seq. Int. J. Mol. Sci. 2022, 23, 6844. [Google Scholar] [CrossRef]
- Liu, Y.; Guan, X.Y.; Liu, S.N.; Yang, M.; Ren, J.H.; Guo, M.; Huang, Z.H.; Zhang, Y.W. Genome-Wide Identification and Analysis of TCP Transcription Factors Involved in the Formation of Leafy Head in Chinese Cabbage. Int. J. Mol. Sci. 2018, 19, 847. [Google Scholar] [CrossRef] [Green Version]
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Wang, H.W.; Cao, Y.N.; Kan, S.L.; Liu, Y.Y. Comprehensive evolutionary analysis of the TCP gene family: Further insights for its origin, expansion, and diversification. Front. Plant Sci. 2022, 13, 994567. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snap-shot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Xu, H.; Lantzouni, O.; Bruggink, T.; Benjamins, R.; Lanfermeijer, F.; Denby, K.; Schwechheimer, C.; Bassel, G.W. A Molecular Signal Integration Network Underpinning Arabidopsis Seed Germination. Curr. Biol. 2020, 30, 3703–3712. [Google Scholar] [CrossRef]
- Zhang, W.; Cochet, F.; Ponnaiah, M.; Lebreton, S.; Matheron, L.; Pionneau, C.; Boudsocq, M.; Resentini, F.; Huguet, S.; Blázquez, M.Á.; et al. The MPK8-TCP14 pathway promotes seed germination in Arabidopsis. Plant J. 2019, 100, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, L.V.; Gastaldi, V.; Ariel, F.D.; Viola, I.L.; Gonzalez, D.H. Class I TCP proteins TCP14 and TCP15 are required for elongation and gene expression responses to auxin. Plant Mol. Biol. 2021, 105, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Rath, M.; Challa, K.R.; Sarvepalli, K.; Nath, U. CINCINNATA-Like TCP Transcription Factors in Cell Growth—An Expanding Portfolio. Front. Plant Sci. 2022, 13, 825341. [Google Scholar] [CrossRef] [PubMed]
- Saini, K.; Dwivedi, A.; Ranjan, A. High temperature restricts cell division and leaf size by coordination of PIF4 and TCP4 transcription factors. Plant Physiol. 2022, 190, 2380–2397. [Google Scholar] [CrossRef]
- Koyama, T.; Sato, F.; Ohme-Takagi, M. Roles of miR319 and TCP Transcription Factors in Leaf Development. Plant Physiol. 2017, 175, 874–885. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.Y.; Zhang, L.; Wang, W.Y.; Tian, P.; Wang, W.; Wang, K.Y.; Gao, Z.; Liu, S.; Zhang, Y.X.; Irish, V.F.; et al. TCP5 controls leaf margin development by regulating KNOX and BEL-like transcription factors in Arabidopsis. J. Exp. Bot. 2021, 72, 1809–1821. [Google Scholar] [CrossRef]
- Hur, Y.S.; Kim, J.; Kim, S.; Son, O.; Kim, W.Y.; Kim, G.T.; Ohme-Takagi, M.; Cheon, C.I. Identification of TCP13 as an Upstream Regulator of ATHB12 during Leaf Development. Genes 2019, 10, 644. [Google Scholar] [CrossRef] [Green Version]
- Challa, K.R.; Aggarwal, P.; Nath, U. Activation of YUCCA5 by the Transcription Factor TCP4 Integrates Developmental and Environmental Signals to Promote Hypocotyl Elongation in Arabidopsis. Plant Cell 2016, 28, 2117–2130. [Google Scholar] [CrossRef] [Green Version]




| Protein ID | Gene ID | Length of CDS (bp) | Number of Amino Acids | Molecular Weight (Da) | Theoretical pI | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|
| PRQ40804 | Chr4g0440011 | 1278 | 425 | 44,862.52 | 6.57 | Nucleus |
| PRQ16145 | Chr7g0181021 | 1281 | 426 | 45,529.8 | 6.81 | Nucleus |
| PRQ31080 | Chr5g0031541 | 1317 | 438 | 48,250.47 | 6.68 | Nucleus |
| PRQ29494 | Chr5g0014491 | 1326 | 441 | 47,923.63 | 8.42 | Nucleus |
| PRQ40431 | Chr4g0435921 | 1347 | 448 | 50,039.06 | 9.34 | Nucleus |
| PRQ39050 | Chr4g0420791 | 873 | 290 | 32,732.35 | 9.08 | Nucleus |
| PRQ54300 | Chr2g0175971 | 1008 | 335 | 35,745.35 | 9.02 | Nucleus |
| PRQ56906 | Chr1g0342441 | 1083 | 360 | 39,668.36 | 8.4 | Nucleus |
| PRQ53141 | Chr2g0163221 | 942 | 313 | 35,026.29 | 9.26 | Nucleus |
| PRQ50062 | Chr2g0128901 | 1095 | 364 | 39,858.69 | 6.45 | Nucleus |
| PRQ59998 | Chr1g0376331 | 981 | 326 | 33,999.1 | 6.32 | Nucleus |
| PRQ57759 | Chr1g0351841 | 924 | 307 | 32,831.23 | 8.57 | Nucleus |
| PRQ17858 | Chr7g0199561 | 1182 | 393 | 40,894.44 | 6.4 | Nucleus |
| PRQ43584 | Chr3g0470021 | 750 | 249 | 26,700.61 | 7.29 | Nucleus |
| PRQ37082 | Chr4g0398661 | 831 | 276 | 29,026.25 | 9.2 | Nucleus |
| PRQ29084 | Chr5g0010031 | 741 | 246 | 28,087.84 | 5.09 | Nucleus |
| PRQ16347 | Chr7g0183231 | 1155 | 384 | 42,654.47 | 8.93 | Nucleus |
| PRQ53358 | Chr2g0165641 | 1104 | 367 | 40,483.52 | 6.48 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Q.; Dong, Q.; Tian, D.; Mao, L.; Cao, X.; Zhu, K. Genome-Wide Analysis of TCP Transcription Factors and Their Expression Pattern Analysis of Rose Plants (Rosa chinensis). Curr. Issues Mol. Biol. 2023, 45, 6352-6364. https://doi.org/10.3390/cimb45080401
Zou Q, Dong Q, Tian D, Mao L, Cao X, Zhu K. Genome-Wide Analysis of TCP Transcription Factors and Their Expression Pattern Analysis of Rose Plants (Rosa chinensis). Current Issues in Molecular Biology. 2023; 45(8):6352-6364. https://doi.org/10.3390/cimb45080401
Chicago/Turabian StyleZou, Qingcheng, Qing Dong, Danqing Tian, Lihui Mao, Xuerui Cao, and Kaiyuan Zhu. 2023. "Genome-Wide Analysis of TCP Transcription Factors and Their Expression Pattern Analysis of Rose Plants (Rosa chinensis)" Current Issues in Molecular Biology 45, no. 8: 6352-6364. https://doi.org/10.3390/cimb45080401
APA StyleZou, Q., Dong, Q., Tian, D., Mao, L., Cao, X., & Zhu, K. (2023). Genome-Wide Analysis of TCP Transcription Factors and Their Expression Pattern Analysis of Rose Plants (Rosa chinensis). Current Issues in Molecular Biology, 45(8), 6352-6364. https://doi.org/10.3390/cimb45080401





