Abstract
To explore the mitogenome characteristics of Tetratomidae and the phylogenetic position of this family in Tenebrionoidea, the mitogenome of Penthe kochi Mařan, 1940 was sequenced, annotated, and analyzed. The P. kochi mitogenome is consistent with Tenebrionoidea species in gene length, genomic organization, codon usage, and secondary structures of transfer genes (tRNAs). Most protein-coding genes (PCGs) originate with a typical ATN start codon, except nad1 and nad3, which start with TTG. In total, 10 PCGs are terminated with complete stop codon TAA and TAG, while cox1, cox2, and nad 4 contain an incomplete stop codon T-. Among the 13 PCGs, nad2 (Pi = 0.282) has the most diverse nucleotide composition, and cox2 is the most conserved gene with the lowest value (Pi = 0.154). The Ka/Ks ratio of cox1 (0.076) and cox2 (0.124) has a lower value. All the tRNAs can be folded in a typical clover-leaf secondary structure, except trnS1, which lacked a dihydrouridine arm. And phylogenetic analyses were performed based on 13 PCGs using the Bayesian inference (BI) method. The results showed that the clade of Tenebrionoidea was well separated from the outgroups, and Tetratomidae and Mycetophagidae were not well resolved. Phylogenetic analyses with more mitogenome samplings are needed to resolve the phylogeny of Tenebrionoidea.
1. Introduction
The family Tetratomidae, including 13 genera over 150 species in five subfamilies [1], is a small group in the superfamily Tenebrionoidea. The tetratomid species are widely distributed in the Palearctic, Oriental, Sino-Japanese, Australian, Oceanian, Afrotropical, and Nearctic regions. To date, eight genera and twenty-two species in five subfamilies have been recorded from China, primarily distributed in the southwest and southeast [2,3,4,5,6,7,8,9,10]. Adults are frequently found on the surface of Polyporaceae or Tricholomataceae at night, and their larvae feed internally on hyphal tissue [1].
Although many taxonomists have made important contributions to the Tenebrionoidea classification during phylogenetic analyses based on morphological characteristics and molecular data, the problems of the overall classification remain unsettled. Based on the morphological characteristics, Tenebrionoidea (26 families) was divided into five lineages, one of which was composed of Tetratomidae, Melandryidae, Mordellidae, and Ripiphoridae [11]. Based on the molecular data, Hunt et al. performed a comprehensive phylogenetic analysis of beetles inferred from three genes (16S, 18S, cox1); the results suggested that the 23 sampled families of Tenebrionoidea formed a clade [12]. Mckenna and Farrell constructed a Timetree of life of Coleoptera based on morphological characteristics, fossils, and molecular data, and the results suggested that the 21 sampled families of Tenebrionoidea formed a clade with the origin of Tenebrionoidea being ca. 236 Ma [13]. Gunter et al. reconstructed the phylogenetic tree of 24 tenebrionoid families based on four genes (ssu, lsu, rrnL, cox1), and the results showed that topology included four lineages [14]. Then, McKenna et al. reconstructed the phylogenetic tree of Coleoptera based on eight nuclear genes; the results exhibited that Tenebrionoidea included the 28 sampled families and Lymexyloidea was recovered within Tenebrionoidea [15]. And then, McKenna et al. inferred the phylogeny of beetles using 4818 genes for 146 species [16]; the results suggested that the phylogenetic position of Lymexylidae in Tenebrionoidea is suitable. However, Cai et al. used 68 single-copy nuclear protein-coding genes of 129 extant families to explore beetle evolution, and the results suggested that Lymexyloidea is a sister group of Tenebrionoidea [17]. However, Tetratomidae is not included in these studies. In Tenebrionoidea, the phylogenetic position of Tetratomidae is rather unclear.
Recently, the mitogenome emerged as a valuable source for higher-level phylogenetic analysis [18,19,20,21]. In this study, the first complete mitochondrial genome for Tetratomidae, the mitogenome of Penthe kochi Mařan, 1940, was sequenced, annotated, and comparatively analyzed. Moreover, phylogenetic analyses based on the Bayesian inference (BI) method was carried out to assess the phylogenetic position of Tetratomidae in Tenebrionoidea and to contribute to further understanding the phylogenetic relationships of each group of Tenebrionoidea.
2. Materials and Methods
2.1. Sampling, Identification, and DNA Extraction
The specimens of Penthe kochi Mařan, 1940 were collected from Dayaoshan Mountains, Jinxiu County, Guangxi Zhuang Autonomous Region, China, on 11 February 2021. Specimens were immediately preserved in 95% ethanol in the field after they were collected and then stored at −24 °C in laboratory. The specimens were examined using Olympus SZX10 and were identified based on the morphological characteristics described by Mařan [22]. Genomic DNA was extracted from the legs and thoracic muscle tissue. Next-generation sequencing and the assembly of mitogenome were performed by Beijing Aoweisen Gene Technology Co., Ltd. (Beijing, China).
2.2. Mitogenome Assembly, Annotation and Analysis
A whole genome shotgun strategy was used based on the Illumina HiSeq platform when the total genome DNA was quantified. Then, sequencing was performed with a strategy of 150 bp paired-end reads. The assembler MITO-bim was used for mitogenome assembly [23]. To check the correctness of the assembly, clean data were manually mapped on the mitochondrial scaffold using Geneious 11.0.2 software [24]. The 13 protein-coding genes (PCGs) and two ribosomal RNA genes (rRNAs) were identified using Geneious 11.0.2. The 22 transfer RNA genes (tRNAs) were re-identified using a tRNAScan-SE server v 1.21 and MITOS WebSever [25,26]. The mitogenome map was illustrated using the online tool Organellar Genome DRAW (OGDRAW; https://chlorobox.mpimp-golm.mpg.de/OGDraw.html, 11 September 2024) [27]. In an A + T-rich region, the tandem repeat elements were identified using an online tool, Tandem Repeats Finder [28]. The relative synonymous codon usage (RSCU) was calculated by Mega X [29]. The nucleotide diversity (Pi) and the ratio of nonsynonymous/synonymous (Ka/Ks) PCGS were calculated with DnaSP v 5 [30].
2.3. Phylogenetic Analysis
The mitochondrial genome of 33 species from 15 families of Tenebrionoidea were selected as ingroups (Table 1), and mitochondrial genomes of Lymexylidae species were chosen as the outgroups, as they are phylogenetically distant from Tenebrionoidea in the Coleoptera [17,31]. The nucleotide sequences (13 PCGs) of 33 mitogenomes were aligned using ClustalW and trimmed using trimAl v 1.2 [32,33]. The best-fit model was determined by ModelFinder based on Bayesian information criterion. The phylogenetic trees were constructed using PhyloSuite v 1.2.2 [34], based on the Bayesian inference (BI) method. Bayesian analyses were run with two independent chains spanning 2,000,000 generations, four Markov chains, with sampling at every 100 generations and a burn-in period of 0.25 for each chain. The phylogenetic trees were edited and visualized by Figtree v 1.4.3.

Table 1.
The mitogenomes of Tenebrionoidea and outgroups were used for phylogenetic analyses.
3. Results and Discussion
3.1. Genome Organization and Base Composition
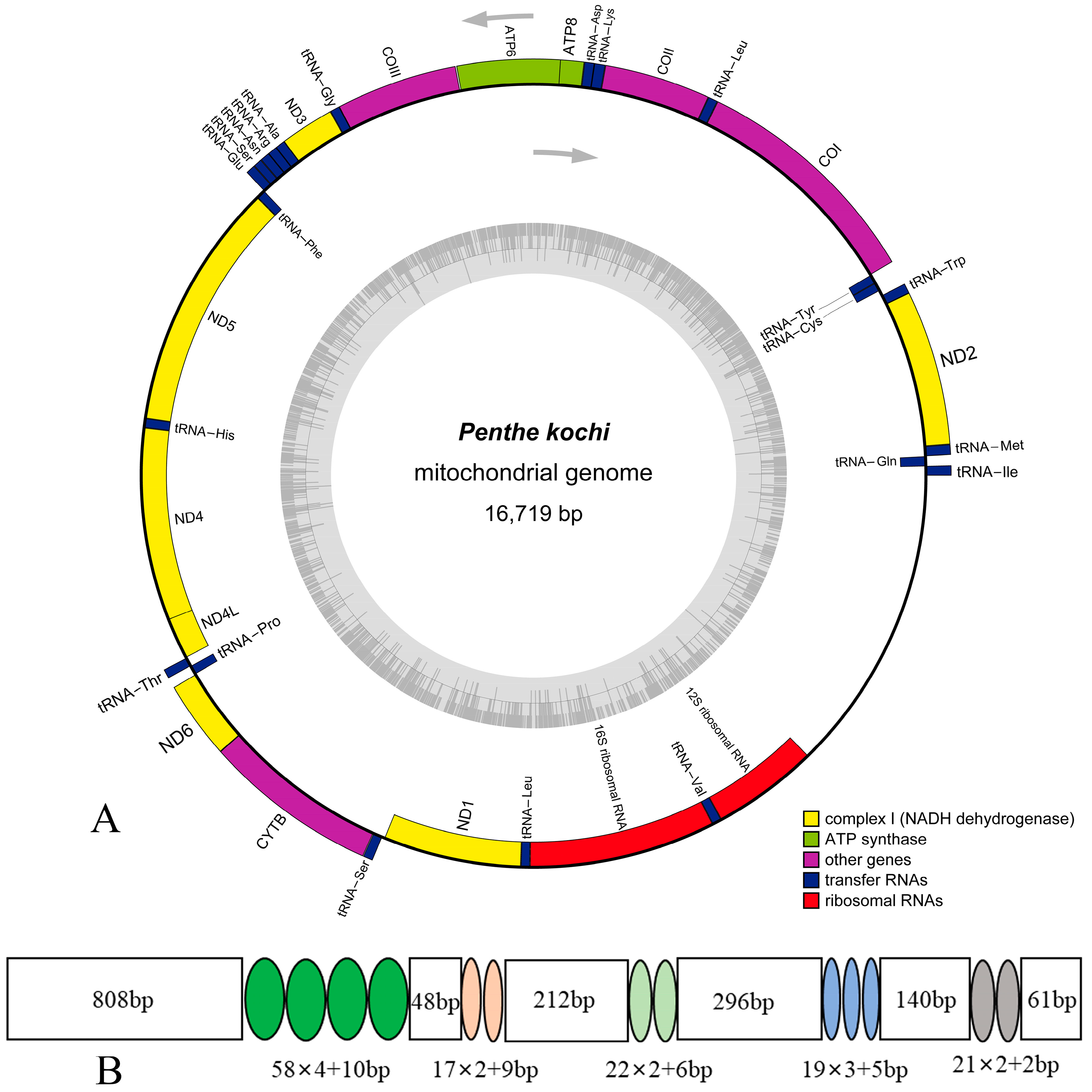
The mitogenome of P. kochi is deposited in GenBank with accession number (ON113044). The mitogenome sequence is 16,719 bp in length and encodes 37 typical mitochondrial genes: 13 protein-coding genes (13 PCGs), two ribosomal RNA genes (2 rRNAs), 22 transfer RNA genes (22 tRNAs), and an A + T-rich region (control region, CR) (Figure 1). The gene arrangement of the P. kochi mitogenome is identical to the hypothetical ancestral insect pattern [43]: 14 genes (4 PCGs, 8 tRNAs, and 2 RNAs) are N-strand, and the other genes (9 PCGs and 14 tRNAs) are J-strand (Table 2). This mitogenome contains 11 overlapping genes, and the longest overlap is 7 bp between nad4 and nad4l. In tRNAs, the gene overlap probably represents the lower evolutionary constraints [39,44]. In the P. kochi mitogenome, intergenic nucleotides range from 1 to 34 bp, with the longest region located between trnW and trnC.
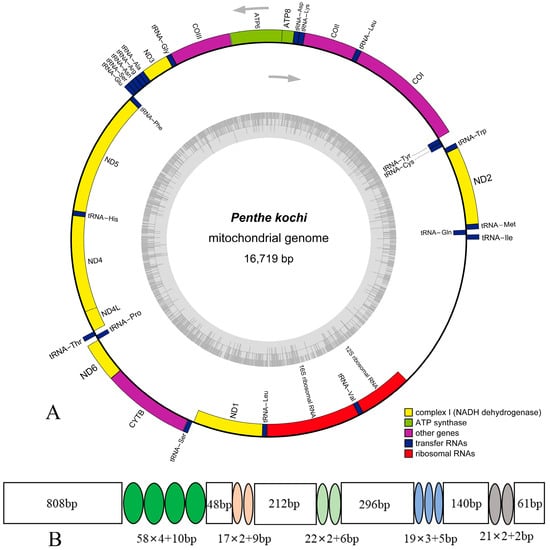
Figure 1.
(A) The mitogenome map of Penthe kochi; (B) the organization of the A+T-rich region in the P. kochi mitogenome.

Table 2.
Summary of the characteristics of the mitogenome of Penthe kochi.
The base composition of the mitogenome of P. kochi is A (40.97%), C (11.69%), G (7.70%), and T (39.63%) (Table 2 and Table S2). The nucleotide composition has a high A/T bias (80.60%), the same as other beetles [42,45,46,47,48], and exhibits a positive AT-skew (0.02) and a negative GC-skew (−0.21), indicating that A and C are more abundant than T and G.
3.2. Protein-Coding Genes and Codon Usage
The total length of 13 protein-coding genes (PCGs) is 11,130 bp, approximately accounting for 66.57% of the entire mitochondrial genome. In total, 4 of the 13 PCGs (nad1, nad4, and4L, and nad5) are located on the minority strand (N-strand), and the other nine (nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad6, and cytb) are located on the majority strand (J-strand) (Table 2). The A/T nucleotide composition of PCGs is 78.99%, exhibiting a high A/T bias (Table S1). The AT-skew of the entire PCGs is −0.14. Among the 13 PCGs, nad5 (1713 bp) and atp8 (156 bp) were found to be the largest and smallest genes, respectively. Most PCGs originated with a typical ATN (ATA, ATT, ATG) start codon, except nad1 and and3, which started with TTG. Moreover, with the exception of cox1, cox2, and nad 4 that contain an incomplete stop codon T-, the other 10 PCGs were terminated with complete stop codon TAA (nad6, cytb, nad4l, nad5, atp8, atp6, cox3, and nad2) and TAG (nad1, nad3). In insects, lots of mitochondrial genes have incomplete stop codons, which is consistent with previous research [49].
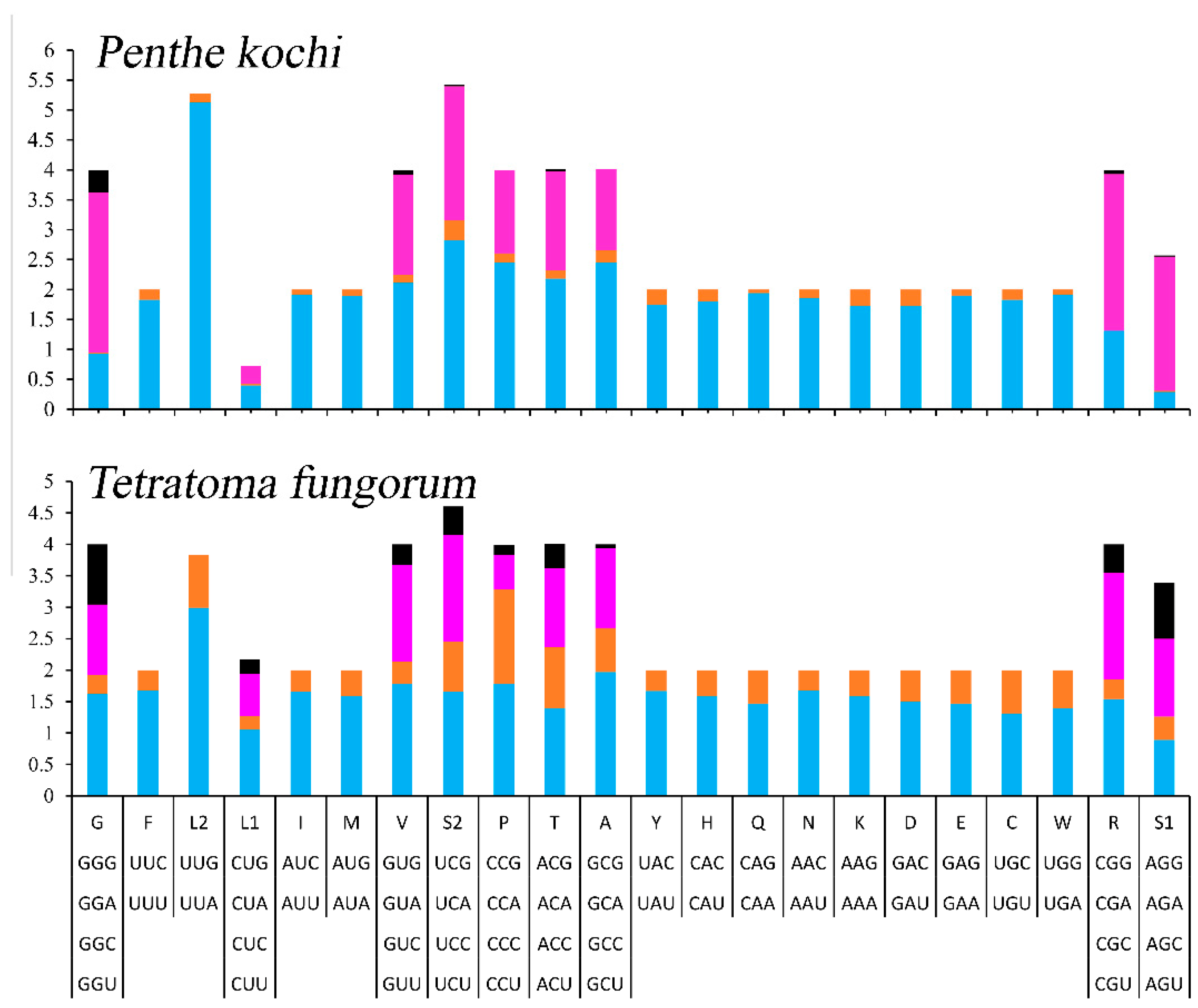
The relative synonymous codon usage (RSCU) of the mitogenomes was calculated and is exhibited (Figure 2). The 13 PCGs contain 3699 codons, excluding stop codons (Table S2), and the most frequently used codons are UUA (508), AUU (429), UUU (311), and AAU (214). Accordingly, L2, I, F, and N are the most frequently used amino acids, accounting for 13.73%, 11.60%, 8.41%, and 5.78% of the total amino acids, respectively.
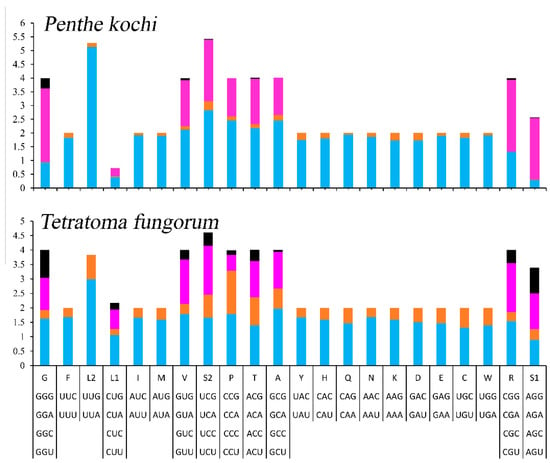
Figure 2.
The relative synonymous codon usage of mitogenomes of Tetratomidae.
To determine the differences between Penthinae (P. kochi) and Tetratominae (T. fungorum), the authors compared the length, acid composition, and RSCU of PCGs. The whole mitogenome of P. kochi (16,719) is longer than the T. fungorum (15,089). In the 13 PCGs, the size of nad2, cox2, nad5, nad4l, nad6, cytb, and nad1 is longer in P. kochi than in T. fungorum, but cox1 is the opposite (Table S3). And the length of the rrnL and rrnS of P. kochi (1285 bp and 735 bp) is shorter than that of T. fungorum (1364 bp and 768 bp). In the P. kochi mitogenome, the CUG codon used for L1, the CCG codon used for P, the CGC codon used for R, and the GCG codon used for A are absent (Figure 2); however, their stability requires further investigation. In Penthinae (P. kochi) and Tetratominae (T. fungorum), the amino acid composition and proportion are similar.
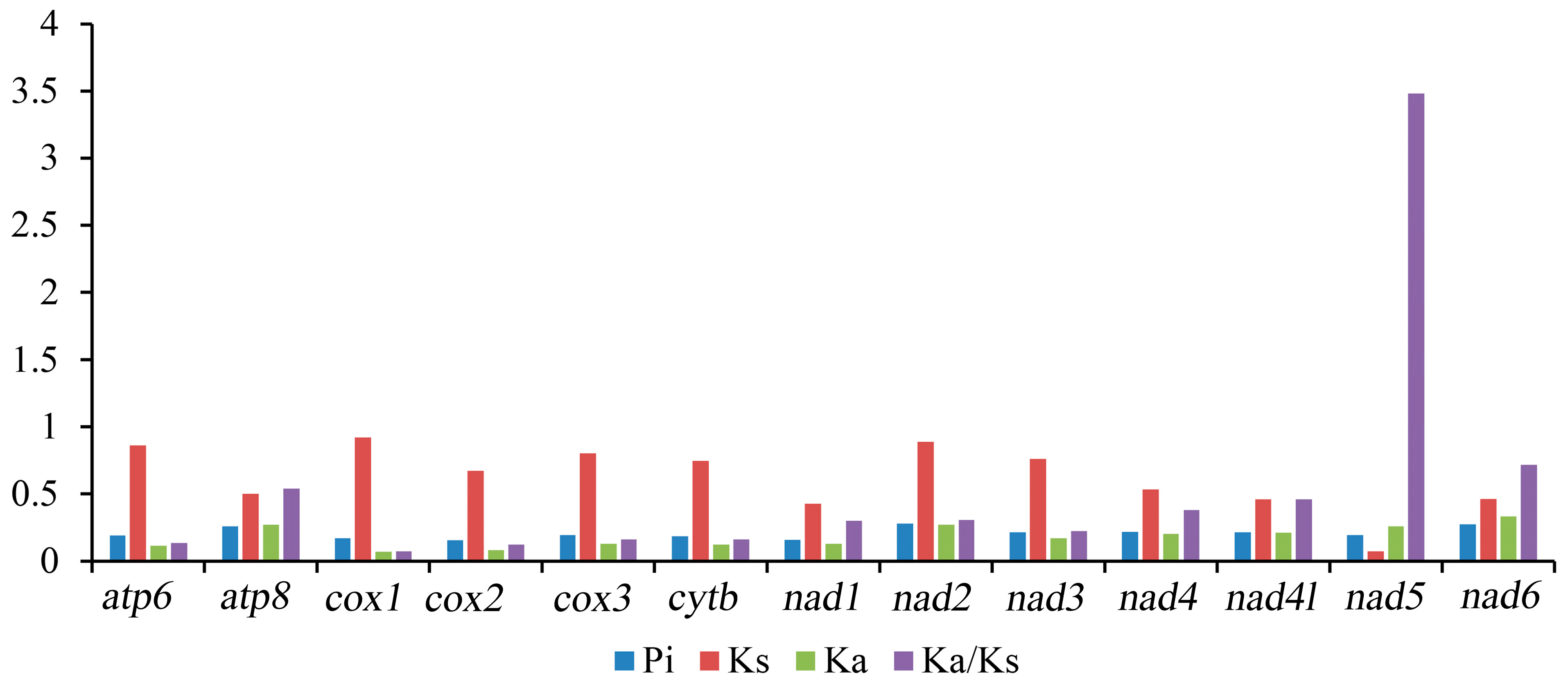
In Tetratomidae, the nucleotide diversity (Pi) of 13 PCGs ranged from 0.154 (cox2) to 0.282 (nad2) (Figure 3, Table S1). Among the 13 PCGs, the gene nad2 (Pi = 0.282) was the most diverse nucleotide, followed by nad6 (Pi = 0.277) and atp8 (Pi = 0.260). In contrast, the gene cox2 was the most conserved gene with the lowest value (Pi = 0.154).
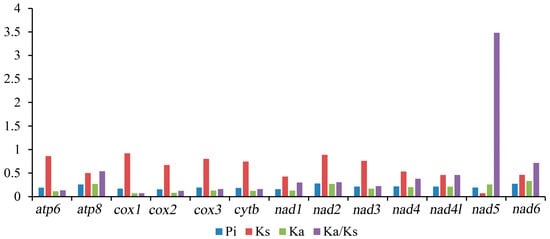
Figure 3.
The Pi, Ka, Ks, and Ka/Ks of the 13 PCGs of mitogenomes in Tetratomidae. Pi. Nucleotide diversity, Ka. Nonsynoymous, Ks. Synoymous, Ka/Ks. Nonsynoymous/Synoymous.
The ratio of nonsynonymous/synonymous (Ka/Ks) of PCGs was also calculated, which is representative of the evolutionary rate [50]. The Ka/Ks ratio of nad5 (3.484) is distinctly larger than one, indicating that this gene is under positive selection [51], while the other 12 PCGs are under purifying selection (Figure 3; Table S4). Among them, cox1 (0.076) and cox2 (0.124) have a lower value. These data suggest that the genes cox1 and cox2 can be used as barcodes for deducing the phylogenetic relationships of Tetratomidae.
3.3. Transfer and Ribosomal RNA Genes
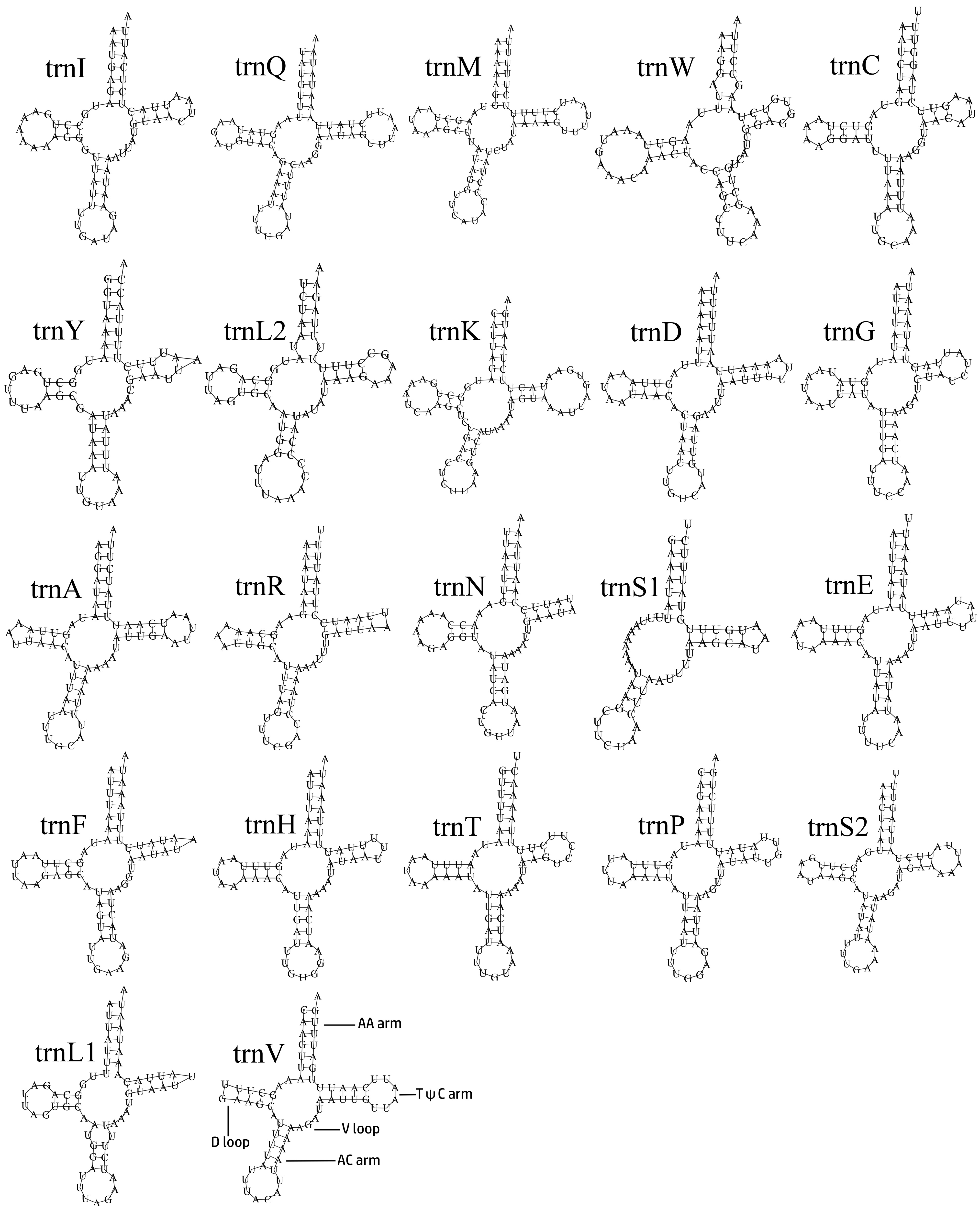
The transfer RNA genes (tRNAs) are 1416 bp in total length, with a high A/T bias (A + T content is 80.08%). The AT-skew of whole tRNAs is 0.03 (Table S1). The tRNAs range from 58 to 71 bp in length. All the tRNAs can be folded in the typical clover-leaf secondary structure, except trnS1, which lacked a dihydrouridine arm (Figure 4), which is a common phenomenon for insects [52,53,54,55,56]. In the whole mitogenome, the longest intergenic nucleotide (34 bp) is located between trnC and trnW.
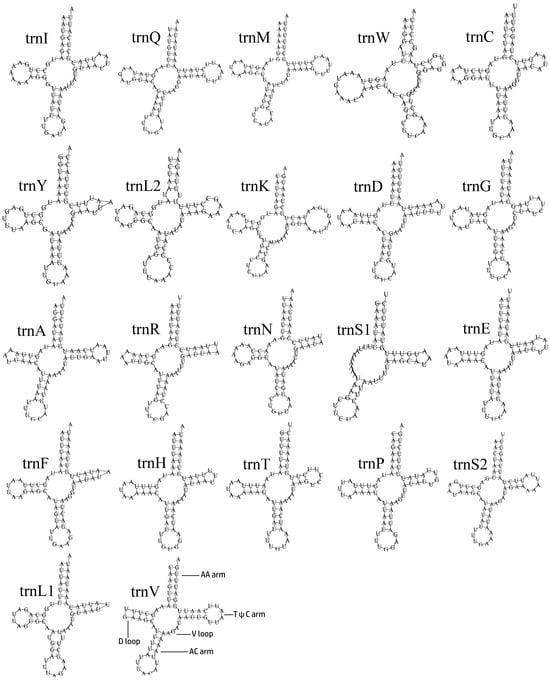
Figure 4.
The predicted secondary structures of tRNAs in the mitogenomes of Penthe kochi.
The lengths of rrnL and rrnS are 1285 bp and 735 bp, respectively. The A + T content is 82.52%, with a negative AT-skew (−0.04) and positive GC-skew (0.33). The rrnL subunit is located between trnV and trnL1, and the rrnS is located at the trnV and A + T-rich region.
3.4. A + T-Rich Region
It is well known that the A + T-rich region (control region, CR) acts in transcription initiation, transcription elongation, and DNA replication [57,58]. This non-coding region is 2119 bp in total length, located between rrnS and trnI. The A + T content is 87.54% (Table S5), with positive AT-Skew (0.04) and negative GC-skew (−0.39). There are five tandem repeat sequence units in this region, of which the length and position are provided (Figure 1).
4. Phylogenetic Analysis
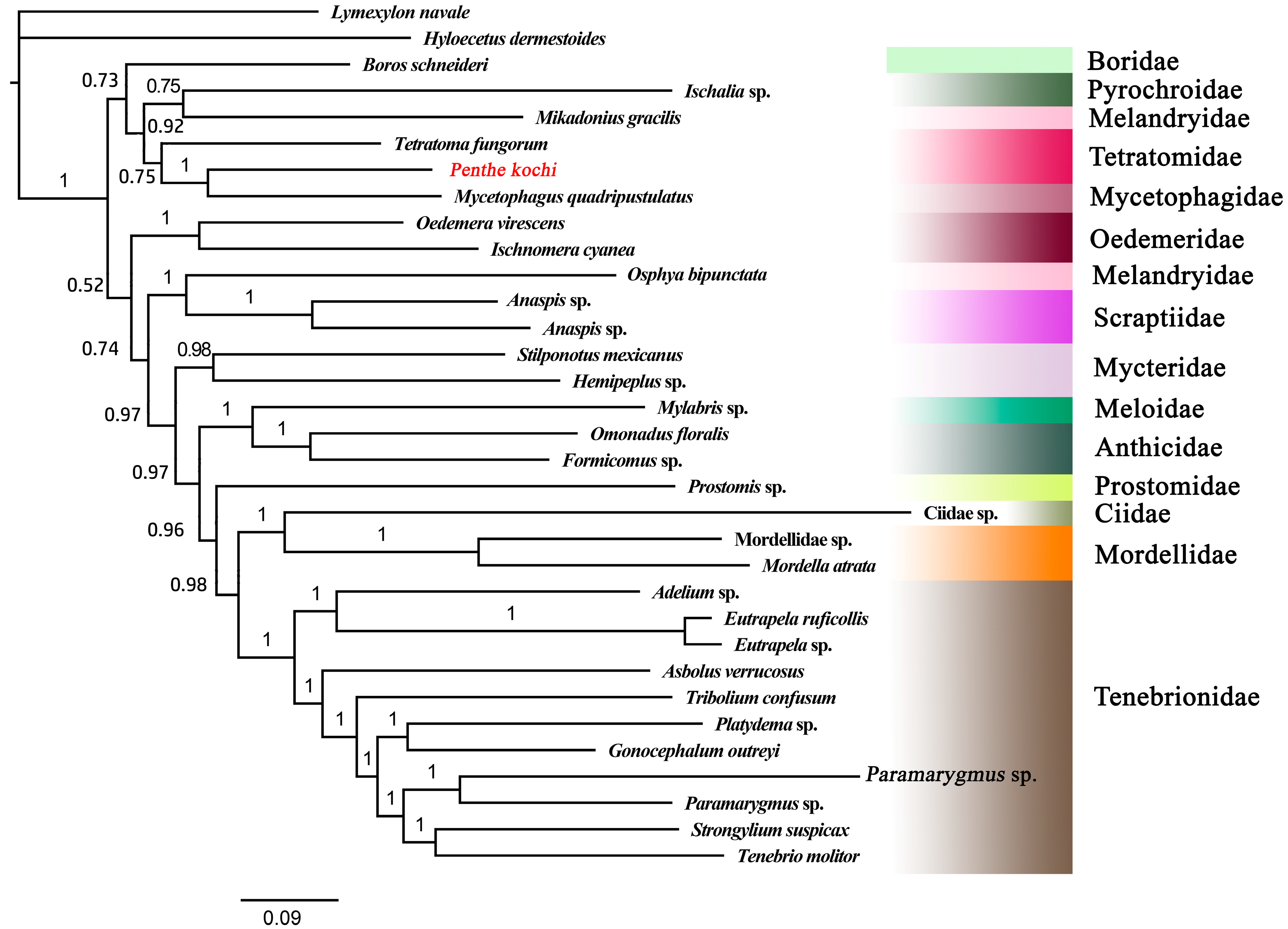
The BI tree was constructed based on the best-fit model GTR + F+ I + G4. Based on the BI tree topology, the monophyly of Tenebrionoidea was again confirmed, as all the tenebrionoid species converged together as an independent clade with high-value support (PP = 1) (Figure 5). The Tetratomidae, Boridae, Mycetophagidae, Melandryidae, and Pyrochroidae converged together as an independent clade with high support (PP = 0.98), which formed a sister group of other tenebrionid families. All the Tenebrionidae species formed a clade. The Meloidae was close to Anthicidae and formed a clade. And Ciidae and Mordellidae were clustered together and formed a sister group of Tenebrionidae with high support (PP = 1). The results also showed that the Melandryidae is not monophyletic. So, the interrelationships among the families of Tenebrionoidea still require more data to be determined completely. These questions will be well addressed in the future when sufficient numbers of complete mitogenomes of Tenebrionoidea species are accumulated.
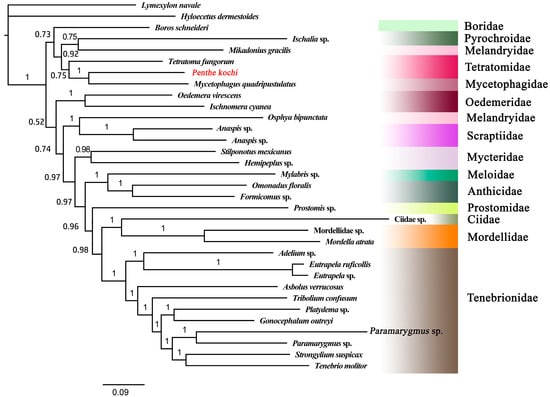
Figure 5.
The BI tree based on 13 PCGs of Tenebrionoidea. The value at each branch shows posterior probability. The target species is displayed in red font.
5. Conclusions
In the present study, the mitochondrial genome of Penthe kochi, which is the first complete mitogenome of Penthinae, was sequenced, annotated, and analyzed. It was found to be consistent with the known mitogenome of Tenebrionoidea in genomic organization, the codon usage of 13 PCGs, A + T biased base composition, and the secondary structures of tRNAs. Among the 13 PCGs, the gene nad2 (Pi = 0.282) is the most diverse nucleotide, and cox2 (Pi = 0.154) is the most conserved gene. The A + T-rich region has five tandem repeat sequence units with different lengths. Phylogenetic analyses based on 13 PCGs from 33 species show that Tetratomidae is close to Mycetophagidae.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cimb46100641/s1, Table S1. Nucleotide characteristics of the mitogenome of Penthe kochi; Table S2. The codon count and RSCU of the mitogenome of Penthe kochi; Table S3. The length of 13 PCGs in Tetratomidae species; Table S4. mitogenomeThe nonsynonymous (Ka) and synonymous (Ks) of Penthe kochi mitogenome; Table S5. The A + T content of the Tetratomidae; Figure S1. The nucleotide diversity of 13 PCGs of Penthe kochi.
Author Contributions
Conceptualization, Z.W. and A.S.; Methodology, Z.W.; Validation, Z.W. and A.S.; Investigation, A.S.; Resources, Z.W.; Data curation: B.O., J.W. and Y.L.; Formal analysis: B.O. and Z.W. Funding acquisition: Z.W. Software: J.W. and Y.L. Writing—original draft: B.O. and Z.W. Writing—review and editing: Z.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Natural Science Foundation of Sichuan Province (2024NSFSC0076, 2022NSFSC1707) and the Doctoral Scientific Research Foundation of China West Normal University (20E054).
Institutional Review Board Statement
This study was approved by the Ethics Committee of China West Normal University (ID: 2024LLSC0014).
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete mitogenome is available at NCBI (ON113044).
Acknowledgments
We wish to thank Qiaoqiao Ji (Beijing, China) who identified the specimens.
Conflicts of Interest
The authors have declared that no competing interests exist.
References
- Lawrence, J.F.; Leschen, R.A.B. Tetratomidae Billberg. In Handbook of Zoology, Arthropoda: Insecta, Coleoptera, Beetles. Volume 2: Morphology and Systematics (Elateroidea, Bostrichiformia, Cucujiformia partim), 2nd ed.; Leschen, R.A.B., Beutel, R.G., Lawrence, J.F., Eds.; Walter de Gruyter: Berlin, Germany; Boston, MA, USA, 2016; pp. 514–520. [Google Scholar]
- Nikitsky, N.B. Generic Classification of the Beetle Family Tetratomidae (Coleoptera, Tenebrionoidea) of the World, with Description of New Taxa; Pensoft Series Faunistica No. 9; BioInform Services, Ltd.: Moscow, Russia, 1998; pp. 1–80. [Google Scholar]
- Nikitsky, N.B. The beetles of the subfamily Tetratominae Billberg, 1820 (Coleoptera, Tetratomidae) of the world fauna. Byulleten’ Mosk. Obs. Ispyt. Prir. Otd. Biol. 2004, 109, 25–36, (In Russian with English Summary). [Google Scholar]
- Nikitsky, N.B. The beetles of the subfamily Penthinae Lacordaire, 1859 (Coleoptera, Tenebrionoidea, Tetratomidae) of the world fauna. Byulleten’ Mosk. Obs. Ispyt. Prir. Otd. Biol. 2005, 110, 16–26, (In Russian with English Summary). [Google Scholar]
- Johnson, P.J.; Löbl, I.; Smetana, A. Catalogue of Palaearctic Coleoptera. Volume 3: Scarabaeoidea–Scirtoidea–Dascilloidea–Buprestoidea–Byrrhoidea; and Volume 4: Elateroidea–Derodontoidea–Bostrichoidea–Lymexyloidea–Cleroidea–Cucujoidea. Ann. Entomol. Soc. Am. 2009, 102, 735–736. [Google Scholar] [CrossRef][Green Version]
- Nikitsky, N. A new species of the genus Tetratoma Fabricius (Coleoptera, Tetratomidae) from China. Zootaxa 2016, 4154, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Pollock, D. Review of the Eustrophinae (Coleoptera, Tetratomidae) of America north of Mexico. ZooKeys 2012, 188, 1–153. [Google Scholar] [CrossRef]
- Hsiao, Y.; Pollock, D.A.; Barclay, M.V.L. Two new species of Cyanopenthe Nikitsky from Taiwan (Coleoptera, Tetratomidae, Penthinae). Zootaxa 2015, 4058, 578–588. [Google Scholar] [CrossRef]
- Saitô, M.; Konvička, O. A new species of Holostrophus (Paraholostrophus) (Coleoptera: Tetratomidae) from central Honshu Island, Japan. Acta Musei Silesiae Sci. Nat. 2017, 66, 1–5. [Google Scholar] [CrossRef][Green Version]
- Ji, Q.; Ren, G. Two new species of the genus Cyanopenthe Nikitsky, 1998 (Coleoptera, Tetratomidae) from southwest China. ZooKeys 2019, 874, 19–30. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Newton, A.F. Evolution and classification of beetles. Annu. Rev. Ecol. Syst. 1982, 13, 261–290. [Google Scholar] [CrossRef]
- Hunt, T.; Bergsten, J.; Levkanicova, Z.; Papadopoulou, A.; John, O.S.; Wild, R.; Hammond, P.M.; Ahrens, D.; Balke, M.; Caterino, M.S.; et al. A Comprehensive Phylogeny of Beetles Reveals the Evolutionary Origins of a Superradiation. Science 2007, 318, 1913–1916. [Google Scholar] [CrossRef]
- McKenna, D.D.; Farrell, B.D. Beetles (Coleoptera). In The Timetree of Life; Hedges, S.B., Kumar, S., Eds.; Oxford University Press: Oxford, UK, 2009; pp. 278–289. [Google Scholar]
- Gunter, N.L.; Levkaničová, Z.; Weir, T.H.; Ślipiński, A.; Cameron, S.L.; Bocak, L. Towards a phylogeny of the Tenebrionoidea (Coleoptera). Mol. Phylogenetics Evol. 2014, 79, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Mckenna, D.D.; Wild, A.L.; Kanda, K.; Bellamy, C.L.; Beutel, R.G.; Caterino, M.S.; Farnum, C.W.; Hawks, D.C.; Ivie, M.A.; Jameson, M.L.; et al. The beetle tree of life reveals that Coleoptera survived end-permian mass extinction to diversify during the Cretaceous terrestrial revolution. Syst. Entomol. 2015, 40, 835–880. [Google Scholar] [CrossRef]
- McKenna, D.D.; Shin, S.; Ahrens, D.; Balke, M.; Beza-Beza, C.; Clarke, D.J.; Donath, A.; Escalona, H.E.; Friedrich, F.; Letsch, H.; et al. The evolution and genomic basis of beetle diversity. Proc. Natl. Acad. Sci. USA 2019, 116, 24729–24737. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Tihelka, E.; Giacomelli, M.; Lawrence, J.F.; Ślipiński, A.; Kundrata, R.; Yamamoto, S.; Thayer, M.K.; Newton, A.F.; Leschen, R.A.B.; et al. Integrated phylogenomics and fossil data illuminate the evolution of beetles. R. Soc. Open Sci. 2022, 9, 211771. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Qin, J.; Zhang, Y.; Zhou, X.; Kong, X.; Wei, S.; Ward, R.D.; Zhang, A.-B. Mitochondrial phylogenomics and genetic relationships of closely related pine moth (Lasiocampidae: Dendrolimus) species in China, using whole mitochondrial genomes. BMC Genom. 2015, 16, 428. [Google Scholar] [CrossRef]
- Wang, W.; Huang, Y.; Bartlett, C.R.; Zhou, F.; Meng, R.; Qin, D. Characterization of the complete mitochondrial genomes of two species of the genus Aphaena Guérin-Méneville (Hemiptera: Fulgoridae) and its phylogenetic implications. Int. J. Biol. Macromol. 2019, 141, 29–40. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Liu, G.-H.; Wang, W.; James, P.; Colwell, D.D.; Tran, A.; Gong, S.; Cai, W.; Shao, R. Mitochondrial genome fragmentation unites the parasitic lice of Eutherian mammals. Syst. Biol. 2018, 68, 430–440. [Google Scholar] [CrossRef]
- Mařan, J. Norý druh rodu Penthe newm. z činy. Novae speciei generis Penthe newm. Descr. (Coleoptera: Melandryidae). Časopis Ceskoslov. Společnosti Entomol. 1940, 37, 87–88. [Google Scholar]
- Hahn, C.; Bachmann, L.; Chevreux, B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—A baiting and iterative mapping approach. Nucleic Acids Res. 2013, 41, e129. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2012, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Timmermans, M.J.T.N.; Barton, C.; Haran, J.; Ahrens, D.; Culverwell, C.L.; Ollikainen, A.; Dodsworth, S.; Foster, P.G.; Bocak, L.; Vogler, A.P. Family-level sampling of mitochondrial genomes in Coleoptera: Compositional heterogeneity and phylogenetics. Genome Biol. Evol. 2016, 8, 161–175. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2019, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Linard, B.; Arribas, P.; Andújar, C.; Crampton-Platt, A.; Vogler, A.P. Lessons from genome skimming of arthropod-preserving ethanol. Mol. Ecol. Resour. 2016, 16, 1365–1377. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L.; Sullivan, J.; Song, H.; Miller, K.B.; Whiting, M.F. A mitochondrial genome phylogeny of the Neuropterida (lace-wings, alderflies and snakeflies) and their relationship to the other holometabolous insect orders. Zool. Scr. 2009, 38, 575–590. [Google Scholar] [CrossRef]
- Timmermans, M.J.T.N.; Dodsworth, S.; Culverwell, C.L.; Bocak, L.; Ahrens, D.; Littlewood, D.T.J.; Pons, J.; Vogler, A.P. Why barcode? High-throughput multiplex sequencing of mitochondrial genomes for molecular systematics. Nucleic Acids Res. 2010, 38, e197. [Google Scholar] [CrossRef] [PubMed]
- Rider, S.D. The complete mitochondrial genome of the desert darkling beetle Asbolus verrucosus (Coleoptera, Tenebrionidae). Mitochondrial DNA Part A 2015, 27, 2447–2449. [Google Scholar] [CrossRef] [PubMed]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst. Biol. 2009, 58, 381–394. [Google Scholar] [CrossRef]
- Liu, L.-N.; Wang, C.-Y. Complete mitochondrial genome of yellow meal worm (Tenebrio molitor). Zool. Res. 2014, 35, 537–545. [Google Scholar] [CrossRef]
- Ou, J.; Yao, F.-J.; Li, Y.-X.; Yang, Y.; Jin, C.; Wei, Z.-M. The complete mitochondrial genome of the confused flour beetle Tribolium confusum (Coleoptera: Tenebrionidae). Mitochondrial DNA Part A 2015, 27, 3297–3298. [Google Scholar] [CrossRef]
- Song, N.; Liu, H.-Y.; Yang, X.-J.; Zhao, X.-C.; Lin, A.-L. Complete mitochondrial genome of the darkling beetle Gonocephalum outreyi (Coleoptera: Tenebrionidae) with phylogenetic implications. J. Asia-Pac. Entomol. 2018, 21, 721–730. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol. Biol. Evol. 2008, 25, 2499–2509. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wei, Z.; Shi, A. The complete mitochondrial genome of the jewel beetle, Anthaxia chinensis (Coleoptera: Buprestidae). Mitochondrial DNA Part B 2021, 6, 2962–2963. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Kamiński, M.J.; Kanda, K.; Sweet, A.D.; Betancourt, J.L.; Holmgren, C.A.; Hempel, E.; Alberti, F.; Hofreiter, M. Recovery and analysis of ancient beetle DNA from subfossil packrat middens using high-throughput sequencing. Sci. Rep. 2021, 11, 12635. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Ge, X.; Xie, G.; Liu, H.; Yang, Y. First complete mitochondrial genome of Melyridae (Coleoptera, Cleroidea): Genome description and phylogenetic implications. Insects 2021, 12, 87. [Google Scholar] [CrossRef]
- Wei, Z. The complete mitochondrial genomes of five Agrilinae (Coleoptera, Buprestidae) species and phylogenetic implications. ZooKeys 2022, 1092, 195–212. [Google Scholar] [CrossRef]
- Miya, M.; Kawaguchi, A.; Nishida, M. Mitogenomic exploration of higher Teleostean phylogenies: A case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol. Biol. Evol. 2001, 18, 1993–2009. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Mori, S.; Matsunami, M. Signature of positive selection in mitochondrial DNA in Cetartiodactyla. Genes Genet. Syst. 2018, 93, 65–73. [Google Scholar] [CrossRef]
- Park, J.S.; Cho, Y.; Kim, M.J.; Nam, S.-H.; Kim, I. Description of complete mitochondrial genome of the black-veined white, Aporia crataegi (Lepidoptera: Papilionoidea), and comparison to papilionoid species. J. Asia-Pac. Entomol. 2012, 15, 331–341. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, M.; Gao, Y.; Pape, T.; Zhang, D. First mitogenome for the subfamily Miltogramminae (Diptera: Sarcophagidae) and its phylogenetic implications. Eur. J. Entomol. 2017, 114, 422–429. [Google Scholar] [CrossRef]
- Yu, P.; Cheng, X.; Ma, Y.; Yu, D.; Zhang, J. The complete mitochondrial genome of Brachythemis contaminata (Odonata: Libellulidae). Mitochondrial DNA Part A 2014, 27, 2272–2273. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Shu, X.; Li, X.; Meng, L.; Li, B. Comparative mitogenome analysis of three species and monophyletic inference of Catantopinae (Orthoptera: Acridoidea). Genomics 2018, 111, 1728–1735. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.-H.; Jia, J.-G.; Murphy, R.W.; Huang, D.-W. Rapid evolution of the mitochondrial genome in chalcidoid wasps (Hymenoptera: Chalcidoidea) driven by parasitic lifestyles. PLoS ONE 2011, 6, e26645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-X.; Szymura, J.M.; Hewitt, G.M. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 1995, 40, 382–391. [Google Scholar] [CrossRef]
- Zhang, D.-X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).