L-gulono-γ-lactone Oxidase, the Key Enzyme for L-Ascorbic Acid Biosynthesis
Abstract
:1. Introduction
2. The Role of Ascorbic Acid in Biochemical Processes and Its Biosynthetic Pathways
3. Methods of Ascorbic Acid Production
3.1. Manufacturing for Commercial Purposes
3.2. Recombinant Vanillyl Alcohol Oxidase Family Members Are Usually Produced in Bacterial Cells in Apo Forms Lacking the Flavin Cofactor
3.3. Genetic Engineering of Plants and Animals to Elevate the Ascorbic Acid Level
4. Localization and Enzymatic Properties of GULO-like Enzymes from Different Species
5. Characterization of L-gulono-γ-lactone Oxidases from Different Sources
6. GULO-like Enzymes from A. thaliana and GULO from Rat
7. The Family of Aldonolactone Oxidoreductases with the Conserved HWXK Motif
8. Sequence Analysis of Rat L-gulono-γ-lactone Oxidase
9. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Figueroa-Méndez, R.; Rivas-Arancibia, S. Vitamin C in health and disease: Its role in the metabolism of cells and redox state in the brain. Front. Psychol. 2015, 6, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Hasan, L.; Vogeli, P.; Stoll, P.; Špilar Kramer, Š.; Stranzinger, G.; Neuenschwander, S. Intragenic deletion in the gene encoding L-gulonolactone oxidase causes vitamin C deficiency in pigs. Mamm. Genome 2004, 15, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Nishikimi, M. Purification and characterization of L-gulono-γ-lactone oxidase from rat and goat liver. Arch. Biochem. Biophys. 1976, 175, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, G.; Ishikawa, T.; Pornsaksit, V.; Smirnoff, N. Evolution of alternative biosynthetic pathways for vitamin C following plastid acquisition in photosynthetic eukaryotes. eLife 2015, 4, e06369. [Google Scholar] [CrossRef]
- Mansueto, A.; Good, D.J. Conservation of a chromosome 8 inversion and exon mutations confirm common gulonolactone oxidase gene evolution among primates, including H. Neanderthalensis. J. Mol. Evol. 2024, 92, 266–277. [Google Scholar] [CrossRef]
- Mandl, J.; Szarka, A.; Banhegyi, G. Vitamin C: Update on physiology and pharmacology. Br. J. Pharmacol. 2009, 157, 1097–1110. [Google Scholar] [CrossRef]
- Margittai, E.; Banhegyi, G.; Kiss, A.; Nagy, G.; Mandl, J.; Schaff, Z.; Csala, M. Scurvy leads to endoplasmic reticulum stress and apoptosis in the liver of guinea pigs. J. Nutr. 2005, 135, 2530–2534. [Google Scholar] [CrossRef]
- Dhar-Mascareno, M.; Carcamo, J.M.; Golde, D.W. Hypoxia–reoxygenation-induced mitochondrial damage and apoptosis in human endothelial cells are inhibited by vitamin C. Free Radic. Biol. Med. 2005, 38, 1311–1322. [Google Scholar] [CrossRef]
- Fischer, H.; Schwarzer, C.; Illek, B. Vitamin C controls the cystic fibrosis transmembrane conductance regulator chloride channel. Proc. Natl. Acad. Sci. USA 2004, 101, 3691–3696. [Google Scholar] [CrossRef]
- Brown, L.A.S.; Jones, D.P. Antioxidant action of vitamin C in the lung. In Vitamin C in Health and Disease; Packer, L., Fuchs, J., Eds.; CRC Press: Boca Raton, FL, USA, 1997; pp. 265–278. [Google Scholar]
- May, J.M.; Huang, J.; Qu, Z.C. Macrophage uptake and recycling of ascorbic acid: Response to activation by lipopolysaccharide. Free Radic. Biol. Med. 2005, 39, 1449–1459. [Google Scholar] [CrossRef]
- Chatterjee, I.B. Evolution and the biosynthesis of ascorbic acid. Science 1973, 182, 1271–1272. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.X.; Milne, R.I.; Ma, X.L.; Song, Y.Q.; Fang, J.Y.; Sun, H.; Zha, H.G. Characterization of a L-gulono-1, 4-lactone oxidase like protein in the floral nectar of Mucuna sempervirens, Fabaceae. Front. Plant Sci. 2018, 9, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Sivadas, S.; Mohanty, A.K.; Rajesh, S.; Muthuvel, S.K.; Vasanthi, H.R. Molecular modelling and biological evaluation of phyto-molecules as potential activators of gluconolactone oxidase (GULO). J. Biomol. Struct. Dyn. 2023, 41, 15124–15136. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Lee, S.I.; Lee, D.H.; Smith, D.; Jo, H.; Schellhorn, H.E.; Boo, Y.C. Ascorbic acid synthesis due to L-gulono-1, 4-lactone oxidase expression enhances NO production in endothelial cells. Biochem. Biophys. Res. Commun. 2006, 345, 1657–1662. [Google Scholar] [CrossRef] [PubMed]
- Grollman, A.P.; Lehninger, A.L. Enzymic synthesis of L-ascorbic acid in different animal species. Arch. Biochem. Biophys. 1957, 69, 458–467. [Google Scholar] [CrossRef]
- Jensen, P.T.; Basse, A.; Nielsen, D.H.; Larsen, H. Congenital ascorbic acid deficiency in bigs. Acta Vet. Scand. 1983, 24, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Carr, R.S.; Neff, J.M. Determination of ascorbic acid in tissues of marine animals by liquid chromatography with electrochemical detection. Anal. Chem. 1980, 52, 2428–2430. [Google Scholar] [CrossRef]
- Massie, H.R.; Shumway, M.E.; Whitney, S.J.P.; Sternick, S.M.; Aiello, V.R. Ascorbic acid in Drosophila and changes during aging. Exp. Gerontol. 1991, 26, 487–494. [Google Scholar] [CrossRef]
- Bahadorani, S.; Bahadorani, P.; Phillips, J.P.; Hilliker, A.J. The effects of vitamin supplementation on Drosophila life span under normoxia and under oxidative stress. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2008, 63, 35–42. [Google Scholar] [CrossRef]
- Barbehenn, R.V.; Bumgarner, S.L.; Roosen, E.F.; Martin, M.M. Antioxidant defenses in caterpillars: Role of the ascorbate-recycling system in the midgut lumen. J. Insect Physiol. 2001, 47, 349–357. [Google Scholar] [CrossRef]
- Kolawole, A.O.; Olajuyigbe, F.M.; Ajele, J.O.; Adedire, C.O. Activity of the antioxidant defense system in a typical bioinsecticide-and synthetic insecticide-treated cowpea storage beetle Callosobrochus maculatus F. (Coleoptera: Chrysomelidae). Int. J. Insect Sci. 2014, 6, IJIS-S19434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, H.; Xie, J.; Jiang, R.; Deng, C.; Pang, H. Transcriptome responses to heat and cold-stress in ladybirds (Cryptolaemus montrouzieri Mulasnt) analyzed by deep-sequencing. Biol. Res. 2015, 48, 66. [Google Scholar] [CrossRef] [PubMed]
- Patananan, A.N.; Budenholzer, L.M.; Pedraza, M.E.; Torres, E.R.; Adler, L.N.; Clarke, S.G. The invertebrate Caenorhabditis elegans biosynthesizes ascorbate. Arch. Biochem. Biophys. 2015, 569, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Drouin, G.; Godin, J.R.; Page, B. The genetics of vitamin C loss in vertebrates. Curr. Genom. 2011, 12, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Yamamoto, T. Purification of calcium binding substance from soluble fraction of Normal rat liver. Chem. Pharm. Bull. 1978, 26, 1915–1918. [Google Scholar] [CrossRef]
- Tsurusaki, Y.; Yamaguchi, M. Role of regucalcin in liver nuclear function: Binding of regucalcin to nuclear protein or DNA and modulation of tumor related gene expression. Int. J. Mol. Med. 2004, 14, 277–281. [Google Scholar] [CrossRef]
- Henriques, S.F.; Duque, P.; López-Fernández, H.; Vázquez, N.; Fdez-Riverola, F.; Reboiro-Jato, M.; Vieira, C.P.; Vieira, J. Multiple independent L-gulonolactone oxidase (GULO) gene losses and vitamin C synthesis reacquisition events in non-Deuterostomian animal species. BMC Evol. Biol. 2019, 19, 126. [Google Scholar] [CrossRef]
- Foyer, C.H.; Shigeoka, S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011, 155, 93–100. [Google Scholar] [CrossRef]
- Smirnof, N.; Wheeler, G.L. Ascorbic acid in plants: Biosynthesis and function. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 291–314. [Google Scholar] [CrossRef]
- Smirnoff, N. Vitamin C: The metabolism and functions of ascorbic acid in plants. In Biosynthesis of Vitamins in Plants: Vitamins B6, B8, B9, C, E, K; Rebeille, F., Ed.; Academic Press: Cambridge, MA, USA, 2011; pp. 107–177. [Google Scholar] [CrossRef]
- Liang, D.; Zhu, T.; Ni, Z.; Lin, L.; Tang, Y.; Wang, Z.; Wang, X.; Wang, J.; Lv, X.; Xia, H. Ascorbic acid metabolism during sweet cherry (Prunus avium) fruit development. PLoS ONE 2017, 12, e0172818. [Google Scholar] [CrossRef]
- Conklin, P.L.; Nprris, S.R.; Wheeler, G.L.; Williams, E.H.; Smirnof, N.; Last, R.L. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc. Natl. Acad. Sci. USA 1999, 96, 4198–4203. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zeng, W.; Xu, S.; Du, G.; Zhou, J.; Chen, J. Current challenges facing one-step production of l-ascorbic acid. Biotechnol. Adv. 2018, 36, 1882–1899. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Z.; Xia, Y.; Kai, G.; Chen, W.; Tang, K. Metabolic engineering of plant L-Ascorbic acid biosynthesis: Recent trends and applications. Crit. Rev. Biotechnol. 2007, 27, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Branduardi, P.; Fossati, T.; Sauer, M.; Pagani, R.; Porro, D. Biosynthesis of vitamin C by yeast leads to increased stress resistance. PLoS ONE 2007, 2, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.S.; Deng, Y.D.; Zhang, W.H.; Xu, J.; Gao, J.J.; Fu, X.Y.; Han, H.J.; Li, Z.J.; Wang, L.J.; Peng, R.H.; et al. Metabolic engineering of Escherichia coli for direct production of vitamin C from D-glucose. Biotechnol. Biofuels Bioprod. 2022, 15, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, G.L.; Jones, M.A.; Smirnoff, N. The biosynthetic pathway of vitamin C in higher plants. Nature 1998, 393, 365–369. [Google Scholar] [CrossRef]
- Agius, F.; Lamothe, R.G.; Caballero, J.L.; Blanco, J.M.; Botella, M.A.; Valpuesta, V. Engineering increased vitamin C levels in plants by over-expression of a D-galacturonic acid reductase. Nat. Biotechnol. 2003, 21, 177–181. [Google Scholar] [CrossRef]
- Bulley, S.; Laing, W. The regulation of ascorbate biosynthesis. Curr. Opin. Plant Biol. 2016, 33, 15–22. [Google Scholar] [CrossRef]
- Davey, M.W.; Gilot, C.; Persiau, G.; Østergaard, J.; Han, Y.; Bauw, G.C.; Van Montagu, M.C. Ascorbate biosynthesis in Arabidopsis cell suspension culture. Plant Physiol. 1999, 121, 535–543. [Google Scholar] [CrossRef]
- Wolucka, B.A.; Van Montagu, M. GDP-mannose 3′,5′-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J. Biol. Chem. 2003, 287, 47483–47490. [Google Scholar] [CrossRef]
- Aboobucker, S.I.; Suza, W.P.; Lorence, A. Characterization of two Arabidopsis L-gulono-1, 4-lactone oxidases, AtGulLO3 and AtGulLO5, involved in ascorbate biosynthesis. React. Oxyg. Species 2017, 40, 389–427. [Google Scholar] [CrossRef] [PubMed]
- Lorence, A.; Chevone, B.I.; Mendes, P.; Nessler, C.L. Myoinositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004, 134, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Wheeler, G.L. The ascorbate biosynthesis pathway in plants is known, but there is a way to go with understanding control and functions. J. Exp. Bot. 2024, 75, 2604–2630. [Google Scholar] [CrossRef] [PubMed]
- Pappenberger, G.; Hohmann, H.P. Industrial protein of L-ascorbic acid (vitamin C) and D-isoascorbic acid. Adv. Biochem. Eng. Biotechnol. 2014, 143, 143–188. [Google Scholar] [CrossRef] [PubMed]
- Reichstein, T.; Grüssner, A. Eine ergiebige synthese der 1-Ascorbinsäure (C-Vitamin). Helv. Chim. Acta. 1934, 17, 311–328. [Google Scholar] [CrossRef]
- Hancock, R.D.; Viola, R. The use of micro-organisms for l-ascorbic acid production: Current status and future perspectives. Appl. Microbiol. Biotechnol. 2001, 56, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Du, G.; Chen, J. Metabolic engineering of microorganisms for vitamin C production. In Reprogramming Microbial Metabolic Pathways Subcellular Biochemistry; Chen, J., Quinn, P., Wang, X., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 241–259. [Google Scholar]
- Yin, G.L. Production of vitamin C precursor-2-keto-l-gulonic acid from d-sorbitol by mixed culture of microorganisms. Acta Microbiol. Sin. 2001, 41, 709–715. [Google Scholar]
- Wang, E.X.; Ding, M.Z.; Ma, Q.; Dong, X.T.; Yuan, Y.J. Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation. Microb. Cell Fact. 2016, 15, 21. [Google Scholar] [CrossRef]
- Lazarus, R.A.; Seymour, J.L.; Staford, R.K.; Dennis, M.S.; Anderson, S. A biocatalytic approach to vitamin C production. In Biocatalysis Van Nostrand Reinhold Catalysis Series; Abramowicz, D.A., Ed.; Springer: Dordrecht, The Netherlands, 1990; pp. 135–155. [Google Scholar]
- Zou, Y.; Hu, M.; Lv, Y.; Wang, Y.; Song, H.; Yuan, Y.J. Enhancement of 2-ketogulonic acid yield by serial subcultivation of co-cultures of bacillus cereus and ketogulonigenium vulgare. Bioresour. Technol. 2013, 132, 370–373. [Google Scholar] [CrossRef]
- Zeng, W.; Wang, P.; Li, N.; Li, J.; Zhou, J. Production of 2-keto-l-gulonic acid by metabolically engineered Escherichia coli. Bioresour. Technol. 2020, 318, 124069–124078. [Google Scholar] [CrossRef]
- Nishikimi, M.; Kobayashi, J.; Yagi, K. Production by a baculovirus expression system of the apoprotein of L-gulono-γ-lactone oxidase, a flavoenzyme possessing a covalently-bound FAD. Mol. Biol. Int. 1994, 33, 313–320. [Google Scholar]
- Wilkinson, S.R.; Prathalingam, S.R.; Taylor, M.C.; Horn, D.; Kelly, J.M. Vitamin C biosynthesis in trypanosomes:A role for the glycosome. Proc. Natl. Acad. Sci. USA 2005, 102, 11645–11650. [Google Scholar] [CrossRef] [PubMed]
- Logan, F.J.; Taylor, M.C.; Wilkinson, S.R.; Kaur, H.; Kelly, J.M. The terminal step in vitamin C biosynthesis in Trypanosoma cruzi is mediated by a FMN-dependent galactonolactone oxidase. Biochem. J. 2007, 407, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Kudryashova, E.V.; Leferink, N.G.H.; Slot, I.G.M.; Van Berkel, W.J.H. Galactonolactone oxidoreductase from Trypanosoma cruzi employs a FAD cofactor for the synthesis of vitamin C. Biochim. Biophys. Acta 2011, 1814, 545–552. [Google Scholar] [CrossRef]
- Leferink, N.G.H.; Van den Berg, W.A.M.; van Berkel, W.J.H. L-galactono-γ-lactone dehydrogenase from Arabidopsis thaliana, a flavoprotein involved in vitamin C biosynthesis. FEBS J. 2008, 275, 713–726. [Google Scholar] [CrossRef]
- Yabuta, Y.; Yoshimura, K.; Takeda, T.; Shigeoka, S. Molecular characterization of tobacco mitochondrial L-galactono-g-dehydrogenase and its expression in Escherichia coli. Plant Cell Physiol. 2000, 41, 666–675. [Google Scholar] [CrossRef]
- Biyani, N.; Madhubala, R. Leishmania donovani encodes a functional enzyme involved in vitamin C biosynthesis: Arabino-γ-lactone oxidase. Mol. Biochem. Parasitol. 2011, 180, 76–99. [Google Scholar] [CrossRef]
- Benen, J.A.; Sánchez-Torres, P.; Wagemaker, M.J.; Fraaije, M.W.; van Berkel, W.J.; Visser, J. Molecular cloning, sequencing, and heterologous expression of the vaoA gene from Penicillium simplicissimum CBS 170.90 encoding vanillyl-alcohol oxidase. J. Biol. Chem. 1998, 273, 7865–7872. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Mazon, H.; van den Heuvel, R.H.H.; Janssen, D.B.; Fraaije, M.W. Discovery of a eugenol oxidase from Rhodococcus sp.s train RHA1. FEBS J. 2007, 274, 2311–2321. [Google Scholar] [CrossRef]
- Heuts, D.P.H.M.; van Hellemond, E.W.; Janssen, D.B.; Fraaije, M.W. Discovery, characterization, and kinetic analysis of an alditol oxidase from Streptomyces coelicolor. J. Biol. Chem. 2007, 282, 20283–20291. [Google Scholar] [CrossRef]
- Heuts, D.P.H.M.; Janssen, D.B.; Fraaije, M.W. Changing the substrate specificity of a chitooligosaccharide oxidase from Fusarium graminearum by modelinspired site-directed mutagenesis. FEBS Lett. 2007, 581, 4905–4909. [Google Scholar] [CrossRef] [PubMed]
- Ganz, P.R.; Dudani, A.K.; Tackaberry, E.S.; Sardana, R.; Sauder, C.; Cheng, X.; Altosaar, I. Expression of human blood proteins in transgenic plants: The cytokine GM-CSF as a model protein. In Transgenic Plants: A Production System for Industrial and Pharmaceutical Proteins; Owen, M.R.L., Pen, J., Eds.; Wiley: Hoboken, NJ, USA, 1996; pp. 281–297. [Google Scholar]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Emans, N. Molecular farming of pharmaceutical proteins. Transgenic Res. 2000, 9, 279–299. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, D.A.; Thomas, J.A. Biopharmaceuticals derived from genetically modified plants. QJM-Int. J. Med. 2004, 97, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Sirko, A.; Vaněk, T.; Góra-Sochacka, A.; Redkiewicz, P. Recombinant cytokines from plants. Int. J. Mol. Sci. 2011, 12, 3536–3552. [Google Scholar] [CrossRef]
- Redkiewicz, P.; Sirko, A.; Kamel, K.A.; Góra-Sochacka, A. Plant expression systems for production of hemagglutinin as a vaccine against influenza virus. Acta Biochim. Pol. 2014, 61, 551–560. [Google Scholar] [CrossRef]
- Gregory, J.A.; Topol, A.B.; Doerner, D.Z.; Mayfield, S. Alga-produced cholera toxin-Pfs25 fusion proteins as oral vaccines. Appl. Environ. Microbiol. 2013, 79, 3917–3925. [Google Scholar] [CrossRef]
- Jain, A.K.; Nessler, C.L. Metabolic engineering of an alternative pathway for ascorbic acid biosynthesis in plants. Mol. Breed. 2000, 6, 73–78. [Google Scholar] [CrossRef]
- Lim, M.Y.; Pulla, R.K.; Park, J.M.; Harn, C.H.; Jeong, B.R. Overexpression of l-gulono-γ-lactone oxidase (GLOase) gene leads to ascorbate accumulation with enhanced abiotic stress tolerance in tomato. In Vitro Cell. Dev. Biol. Plant. 2012, 48, 453–461. [Google Scholar] [CrossRef]
- Lisko, K.A.; Torres, R.; Harris, R.S.; Belisle, M.; Vaughan, M.M.; Jullian, B.; Chevone, B.I.; Mendes, P.; Nessler, C.L.; Lorence, A. Elevating vitamin C content via overexpression of myo-inositol oxygenase and L-gulono-γ-lactone oxidase in Arabidopsis leads to enhanced biomass and tolerance to abiotic stresses. In Vitro Cell. Dev. Biol. Plant. 2013, 49, 643–655. [Google Scholar] [CrossRef]
- Kwon, S.Y.; Choi, S.M.; Ahn, Y.O.; Lee, H.S.; Lee, H.B.; Park, Y.M.; Kwak, S.S. Enhanced stress-tolerance of transgenic plants expressing a human dehydroascorbate reductase gene. J. Plant Physiol. 2003, 160, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Hemavathi; Upadhyaya, C.P.; Akula, N.; Young, K.E.; Chun, S.C.; Kim, D.H.; Park, S.W. Enhanced ascorbic acid accumulation in transgenic potato confers tolerance to various abiotic stresses. Biotechnol. Lett. 2010, 32, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Todd, E.; Ling, Y.J.; Chang, S.C.; Gallie, D.R. Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc. Natl. Acad. Sci. USA 2003, 100, 3525–3530. [Google Scholar] [CrossRef] [PubMed]
- Maruta, T.; Ichikawa, Y.; Mieda, T.; Takeda, T.; Tamoi, M.; Yabuta, Y.; Ishikawa, T.; Shigeoka, S. The contribution of Arabidopsis homologs of L-gulono-1, 4-lactone oxidase to the biosynthesis of ascorbic acid, Biosci. Biotechnol. Biochem. 2010, 74, 1494–1497. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.N.; Graham, F.L.; D’ouza, C.K.; Muller, W.J.; Igdoura, S.A.; Schellhorn, H.E. Functional rescue of vitamin C synthesis deficiency in human cells using adenoviral-based expression of murine L-gulono-γ-lactone oxidase. Genomics 2004, 83, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, A.; Reinisalo, M.; Pitkänen, T.i.; Nishikimi, M.; Mölsä, H. Expression of rat gene for l-gulono-γ-lactone oxidase, the key enzyme of l-ascorbic acid biosynthesis, in guinea pig cells and in teleost fish rainbow trout (Oncorhynchus mykiss). Biochim. Biophys. Acta BBA Gen. Subj. 1998, 1381, 241–248. [Google Scholar] [CrossRef]
- Yagi, K.; Koshizaka, T.; Kito, M.; Ozawa, T.; Nishikimi, M. Expression in monkey cells of the missing enzyme in L-ascorbic acid biosynthesis, L-gulono-gamma-lactone oxidase, Biochem. Biophys. Res. Commun. 1991, 177, 659–663. [Google Scholar] [CrossRef]
- Nam, Y.K.; Cho, Y.S.; Douglas, S.E.; Gallant, J.W.; Reith, M.E.; Kim, D.S. Isolation and transient expression of a cDNA encoding l-gulono-γ-lactone oxidase, a key enzyme for l-ascorbic acid biosynthesis, from the tiger shark Scyliorhinus torazame. Aquaculture 2002, 209, 271–284. [Google Scholar] [CrossRef]
- Aboobucker, S.I.; Lorence, A. Recent progress on the characterization of aldonolactone oxidoreductases. Plant Physiol. Biochem. 2016, 98, 171–185. [Google Scholar] [CrossRef]
- Eliceiri, G.L.; Lai, E.K.; Mccay, P.B. Gulonolactone oxidase. Solubilization, properties, and partial purification. J. Biol. Chem. 1969, 244, 2641–2645. [Google Scholar] [CrossRef]
- Braun, L.; Mile, V.; Schaff, Z.; Csala, M.; Kardon, T.; Mandl, J.; Bánhegyi, G. Induction and peroxisomal appearance of gulonolactone oxidase upon clofibrate treatment in mouse liver. FEBS Lett. 1999, 458, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Kiuchi, K.; Nishikimi, M.; Yagi, K. Purification and characterization of L-gulonolactone oxidase from chicken kidney microsomes. Biochemistry 1982, 21, 5076–5082. [Google Scholar] [CrossRef] [PubMed]
- Koshizaka, T.; Nishikimi, M.; Ozawa, T.; Yagi, K. Isolation and sequence analysis of a complementary DNA encoding rat liver L -gulono-gamma-lactone oxidase, a key enzyme for L -ascorbic acid biosynthesis. J. Biol. Chem. 1988, 263, 1619–1621. [Google Scholar] [CrossRef] [PubMed]
- Puskas, F.; Braun, L.; Csala, M.; Kardon, T.; Marcolongo, P.; Benedetti, A.; Mandl, J.; Banhegyi, G. Gulonolactone oxidase activitydependent intravesicular glutathione oxidation in rat liver microsomes. FEBS Lett. 1998, 430, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Linster, C.L.; Van Schaftingen, E. Vitamin C: Biosynthesis, recycling and degradation in mammals. FEBS J. 2007, 274, 1–22. [Google Scholar] [CrossRef]
- Smirnof, N. Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free Radic Biol Med. 2018, 122, 116–129. [Google Scholar] [CrossRef]
- Douce, R. Functions of plant mitochondrial membranes. In Mitochondria in Higher Plants; American Society of Plant Physiologists Monograph Series; Academic Press: London, UK, 1985; pp. 77–153. [Google Scholar]
- Mackenzie, S.; McIntosh, L. Higher plant mitochondria. Plant Cell. 1999, 11, 571–585. [Google Scholar] [CrossRef]
- Bartoli, C.G.; Pastori, G.M.; Foyer, C.H. Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol. 2000, 123, 335–344. [Google Scholar] [CrossRef]
- Boudart, G.; Jamet, E.; Rossignol, M.; Lafitte, C.; Borderies, G.; Jauneau, A.; Esquerre-Tugaye, M.T.; Pont-Lezica, R. Cell wall proteins in apoplastic fluids of rosettes: Identification by mass spectrometry and bioinformatics. Proteomics 2005, 5, 212–221. [Google Scholar] [CrossRef]
- Mawuenyega, K.G.; Forst, C.V.; Dobos, K.M.; Belisle, J.T.; Chen, J.; Bradbury, E.M.; Bradbury, A.R.; Chen, X. Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol. Biol. Cell. 2005, 16, 396–404. [Google Scholar] [CrossRef]
- Leferink, N.G.; Fraaije, M.W.; Joosten, H.J.; Schaap, P.J.; Mattevi, A.; van Berkel, W.J. Identification of a gatekeeper residue that prevents dehydrogenases from acting as oxidases. J. Biol. Chem. 2009, 284, 4392–4397. [Google Scholar] [CrossRef] [PubMed]
- Sugisawa, T.; Setsuko, O.; Matzinger, P.K.; Hoshino, T. Isolation and characterization of a new vitamin C producing enzyme (L-gulono-g-lactone dehydrogenase) of bacterial origin. Biosci. Biotech. Biochem. 1995, 59, 190–196. [Google Scholar] [CrossRef]
- Okamura, M. Purification and properties of L-gulono-γ-lactone oxidase from Grifola frondosa. J. Nutr. Sci. Vitaminol. 2001, 47, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Kazuko, O.; Ishikawa, S.; Nishikawa, M.; Mizuno, H.; Yamamoto, T. Purification and properties of L-galactono-γ-lactone dehydrogenase, a key enzyme for ascorbic acid biosynthesis, from sweet potato roots. Biochem. J. 1995, 117, 120–124. [Google Scholar] [CrossRef]
- Leferink, N.G.H.; Van Berkel, W.J.H. Aldonolactone oxidoreductases. In Flavins and Flavoproteins: Methods and Protocols; Methods in Molecular Biology; Weber, S., Schleicher, E., Eds.; Humana Press: New York, NY, USA, 2014; Volume 1146, pp. 95–111. [Google Scholar] [CrossRef]
- Kenney, W.C.; Edmondson, D.E.; Singer, T.P. Identification of the covalently bound flavin of L-gulono-γ-lactone oxidase. Biochem. Biophys. Res. Commun. 1976, 71, 1194–1200. [Google Scholar] [CrossRef]
- Bleeg, H.S. L-ascorbic acid in yeast and isolation of L-galactono-gamma-lactone oxidase from the mitochondria. Enzymologia 1966, 31, 105–112. [Google Scholar]
- Shigeoka, S.; Nakano, Y.; Kitaoka, S. Some properties and subcellular localization of l-gulono-γ-lactone dehydrogenase in Euglena gracilis z. Agric. Biol. Chem. 1979, 43, 2187–2188. [Google Scholar] [CrossRef]
- Wolucka, B.A.; Communi, D. Mycobacterium tuberculosis possesses a functional enzyme for the synhesis of vitamin C, L-gulono-γ-lactone dehydrogenase. FEBS J. 2006, 273, 4435–4445. [Google Scholar] [CrossRef]
- Mapson, L.W.; Breslow, E. Biological synthesis of ascorbic acid: L-Galactono-γ-lactone dehydrogenase. Biochem. J. 1958, 68, 395–406. [Google Scholar] [CrossRef]
- Nakagawa, H.; Asano, A. Ascorbate synthesizing system in rat liver microsomes. I. Gulonolactone-reducible pigment as a prosthetic group of gulonolactone oxidase. J. Biochem. 1970, 68, 737e–746e. [Google Scholar] [CrossRef]
- Burns, J.J.; Peyser, P.; Moltz, A. Missing step in guinea pigs required for the biosynthesis of L-ascorbic acid. Science 1956, 124, 1148–1149. [Google Scholar] [CrossRef] [PubMed]
- Nishikimi, M.; Yagi, K. Molecular basis for the deficiency in humans of gulonolactone oxidase, a key enzyme for ascorbic acid biosynthesis. Am. J. Clin. Nutr. 1991, 4, l203S–1208S. [Google Scholar] [CrossRef] [PubMed]
- Gad, A.A.M.; Gora-Sochacka, A.; Sirko, A. Recombinant C-terminal catalytic domain of rat L-gulono lactone oxidase produced in bacterial cells is enzymatically active. Curr. Issues Mol. Biol. 2024, 46, 8958–8968. [Google Scholar] [CrossRef]
- Mapson, L.W.; Isherwood, F.A.; Chen, Y.T. Biological synthesis of L-ascorbic acid: The conversion of L-galactono-g-lactone into L-ascorbic acid by plant mitochondria. Biochem. J. 1954, 56, 21–28. [Google Scholar] [CrossRef]
- Mutsuda, M.; Ishikawa, T.; Takeda, T.; Shigeoka, S. Subcellular localization and properties of L-galactono-g-lactone dehydrogenase in spinach leaves. Biosci. Biotechnol. Biochem. 1995, 59, 1983–1984. [Google Scholar] [CrossRef]
- Oba, K.; Fukui, M.; Imai, Y.; Iriyama, S.; Nogami, K. L-Galactono-γ-lactone dehydrogenase: Partial characterization, induction of activity and role in the synthesis of ascorbic acid in wounded white potato tuber tissue. Plant Cell Physiol. 1994, 35, 473–478. [Google Scholar] [CrossRef]
- Østergaard, J.; Persiau, G.; Cavey, M.W.; Bauw, G.; Van Montagu, M. Isolation and cDNA cloning for L-galactono-γ-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. J. Biol. Chem. 1997, 272, 30009–30016. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Karita, S.; Shratori, G.; Hattori, M.; Nunome, T.; Oba, K.; Hirai, M. L-Galactono-g-lactone dehydrogenase from sweet potato: Purification and cDNA sequence analysis. Plant Cell Physiol. 1998, 39, 1350–1358. [Google Scholar] [CrossRef]
- Siendones, E.; Gonzalez-Reyes, J.A.; Santos-Ocana, C.; Navas, P.; Córdoba, F. Biosynthesis of ascorbic acid in kidney bean. L-galactono-gamma-lactone dehydrogenase is an intrinsic protein located at the mitochondrial inner membrane. Plant Physiol. 1999, 120, 907–912. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Benen, J.A.E.; Visser, J.; Mattevi, A.; van Berkel, W.J.H. A novel oxidoreductase family sharing a conserved FAD-binding domain. Trends Biochem. Sci. 1998, 23, 206–207. [Google Scholar] [CrossRef]
- Leferink, N.G.H.; Heuts, D.P.H.M.; Fraaije, M.W.; Van Berkel, W.J.H. The growing VAO flavoprotein family. Arch. Biochem. Biophys. 2008, 474, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Yuan, X.; Wang, L.; Jones, G.; Zhang, S. Recent loss of vitamin C biosynthesis ability in bats. PLoS ONE 2011, 6, e27114. [Google Scholar] [CrossRef] [PubMed]
- Fraaije, M.W.; van den Heuvel, R.H.; van Berkel, W.J.; Mattevi, A. Covalent flavinylation is essential for efficient redox catalysis in vanillylalcohol oxidase. Biol. Chem. J. 1999, 274, 35514–35520. [Google Scholar] [CrossRef]
- López-Fernández, H.; Duque, P.; Henriques, S.; Vázquez, N.; Fdez-Riverola, F.; Vieira, C.P.; Reboiro-Jato, M.; Vieira, J. A bioinformatics protocol for quickly creating large-scale phylogenetic trees. In Practical Applications of Computational Biology and Bioinformatics, 12th International Conference; Fdez-Riverola, F., Mohamad, M.S., Rocha, M., De Paz, J.F., González, P., Eds.; Springer: Cham, Switzerland, 2019; pp. 88–96. [Google Scholar] [CrossRef]
- Yang, H. Conserved or lost: Molecular evolution of the key gene GULO in vertebrate vitamin C biosynthesis. Biochem. Genet. 2013, 51, 413–425. [Google Scholar] [CrossRef] [PubMed]
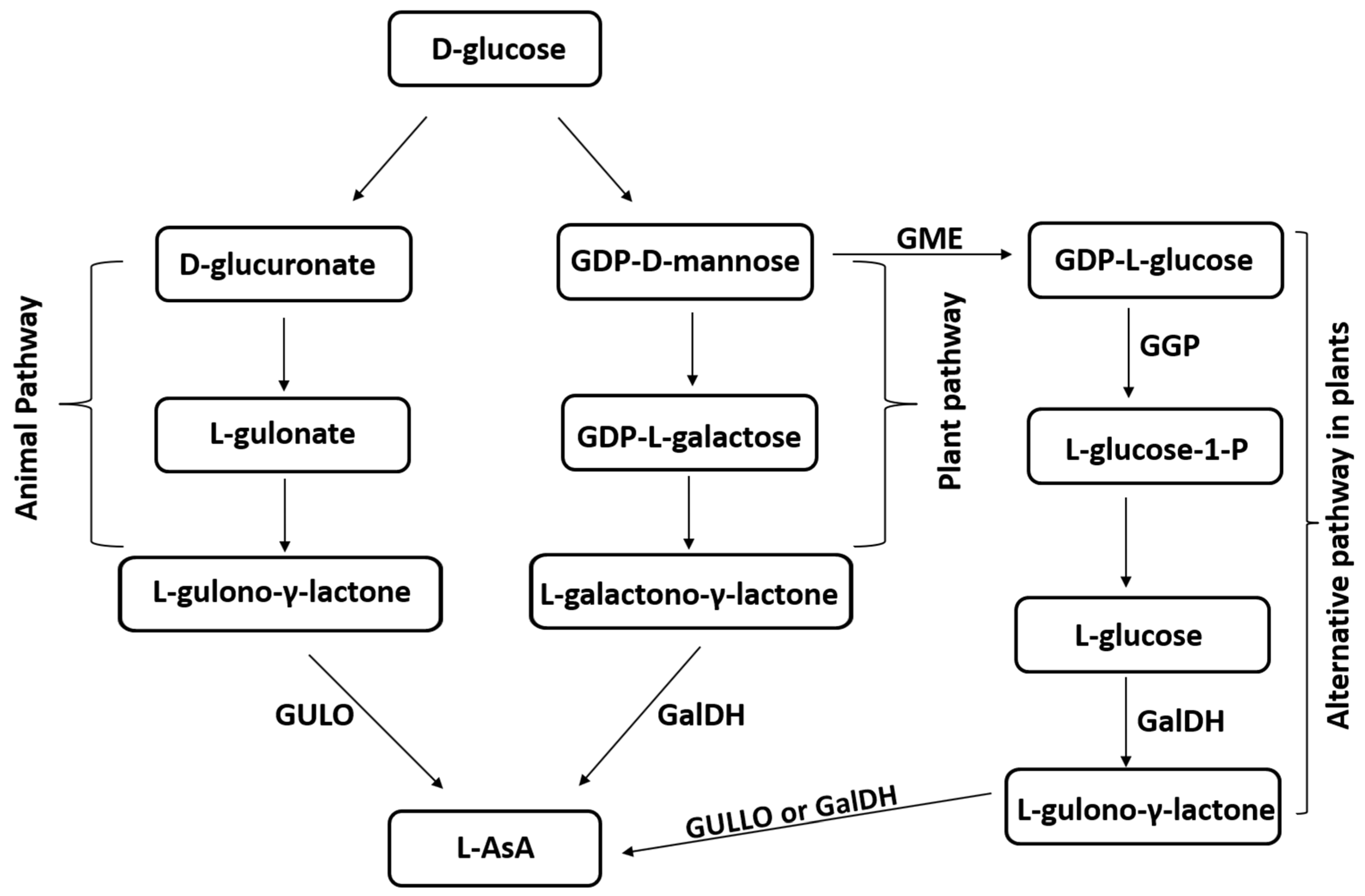
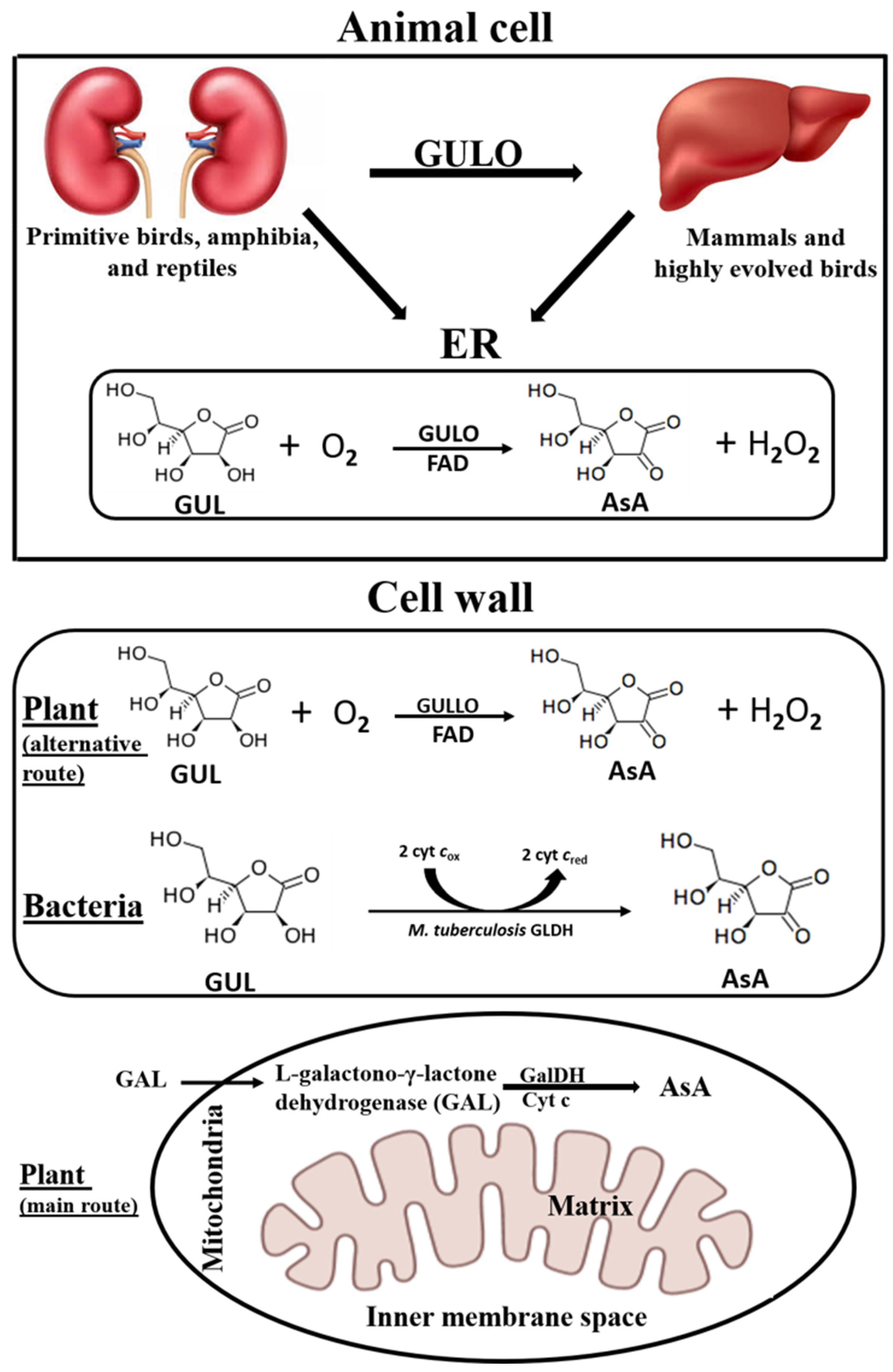
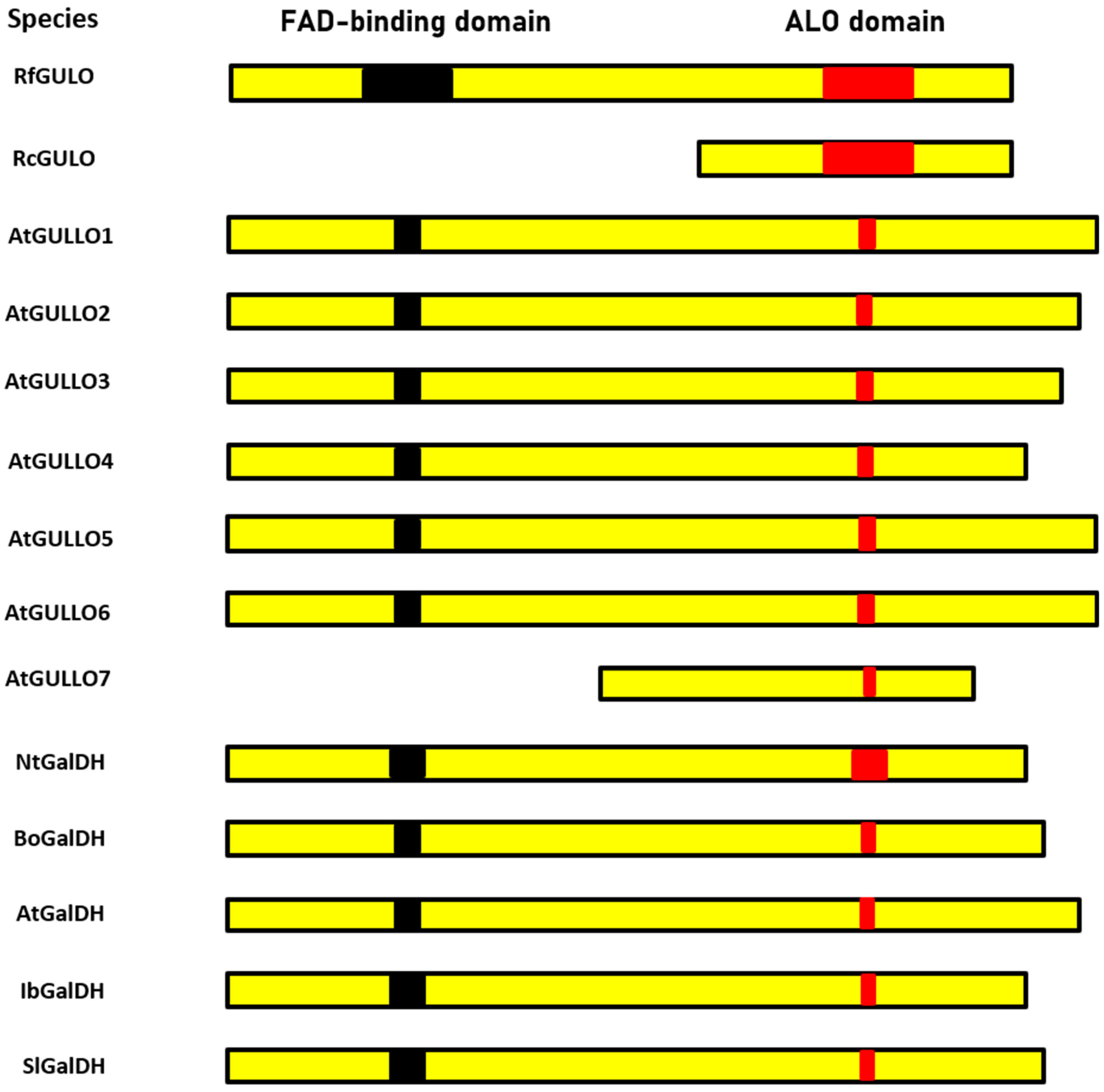

| Species | Molecular Mass (kDa) | Optimum pH | Optimum Temperature (°C) | Km (mM) | Vmax (µmol/min/mg Protein) | Reference |
|---|---|---|---|---|---|---|
| Rat liver GULO | 51 | 7.8 | 37 | 0.066 | 0.63 | [3] |
| Goat liver GULO | 51 | Nd | Nd | 0.15 | 2.7 | [3] |
| Chicken kidney GULO | 50 | 7.3 | Nd | 0.007 | 139 | [87] |
| Recombinant fGULO expressed in E. coli | 50 | 7 | 40 | 0.053 | 780 | [110] |
| Recombinant cGULO expressed in E. coli | 20 | 6.5 | 30 | 0.042 | 374 | [110] |
| GfGULLO | 69 | 7 | 45 | 24 | 2.1 | [99] |
| MtGLDH | 70 | 8 | 40 | 5.5 | 0.041 | [105] |
| AtGalDH | 60 | 8.8 | 25 | 13.1 | Nd | [59] |
| MsGULLO | 70 | Nd | Nd | Nd | Nd | [13] |
| Recombinant (AtGULLO5) expressed in N. benthamiana | 65 | 9 | 40 | 33.8 | 4.5 × 10−3 | [43] |
| Exon Number | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rat | • | • | • | • | • | • | • | • | • | • | • | • |
| Mouse | • | • | • | • | • | • | • | • | • | • | • | |
| Guinea pig | • | • | • | • | • | • | • | • | • | • | ||
| Human | • | • | • | • |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gad, A.A.M.; Sirko, A. L-gulono-γ-lactone Oxidase, the Key Enzyme for L-Ascorbic Acid Biosynthesis. Curr. Issues Mol. Biol. 2024, 46, 11057-11074. https://doi.org/10.3390/cimb46100657
Gad AAM, Sirko A. L-gulono-γ-lactone Oxidase, the Key Enzyme for L-Ascorbic Acid Biosynthesis. Current Issues in Molecular Biology. 2024; 46(10):11057-11074. https://doi.org/10.3390/cimb46100657
Chicago/Turabian StyleGad, Abdul Aziz M., and Agnieszka Sirko. 2024. "L-gulono-γ-lactone Oxidase, the Key Enzyme for L-Ascorbic Acid Biosynthesis" Current Issues in Molecular Biology 46, no. 10: 11057-11074. https://doi.org/10.3390/cimb46100657
APA StyleGad, A. A. M., & Sirko, A. (2024). L-gulono-γ-lactone Oxidase, the Key Enzyme for L-Ascorbic Acid Biosynthesis. Current Issues in Molecular Biology, 46(10), 11057-11074. https://doi.org/10.3390/cimb46100657







