DNA Gene’s Basic Structure as a Nonperturbative Circuit Quantum Electrodynamics: Is RNA Polymerase II the Quantum Bus of Transcription?
Abstract
:1. Introduction
2. DNA Gene’s Basic Structure as a Nonperturbative Circuit Quantum Electrodynamics: Quantum Description of Transcription
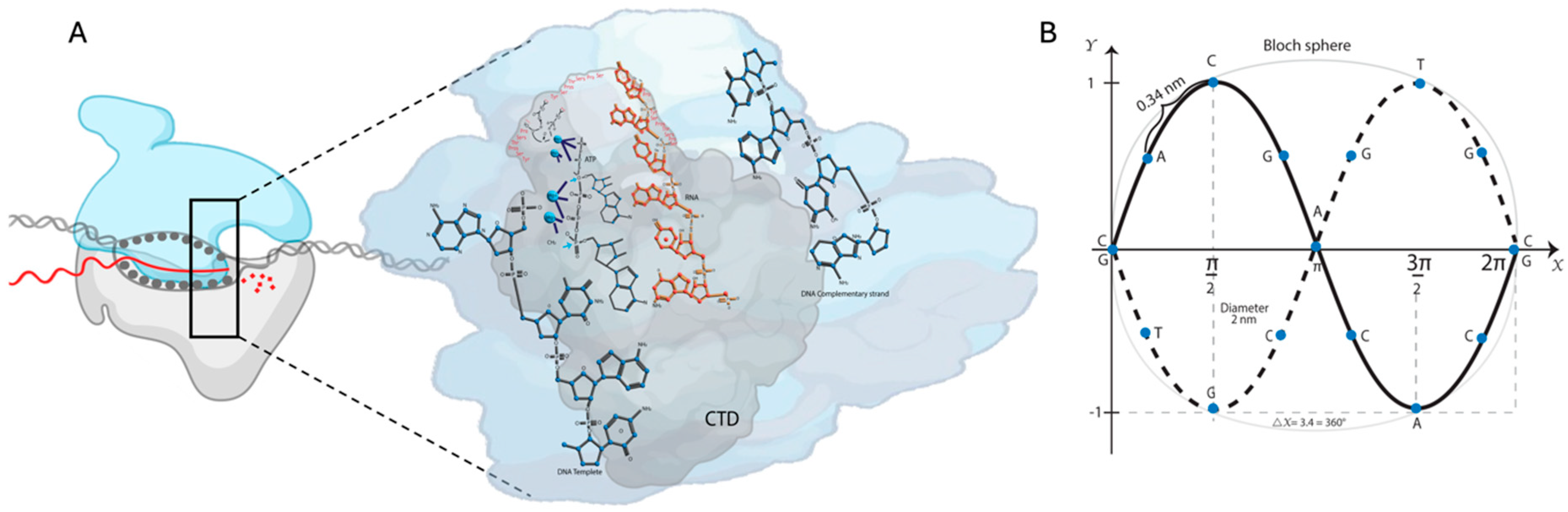
2.1. DNA and Transcription: Structural and Biochemical Studies
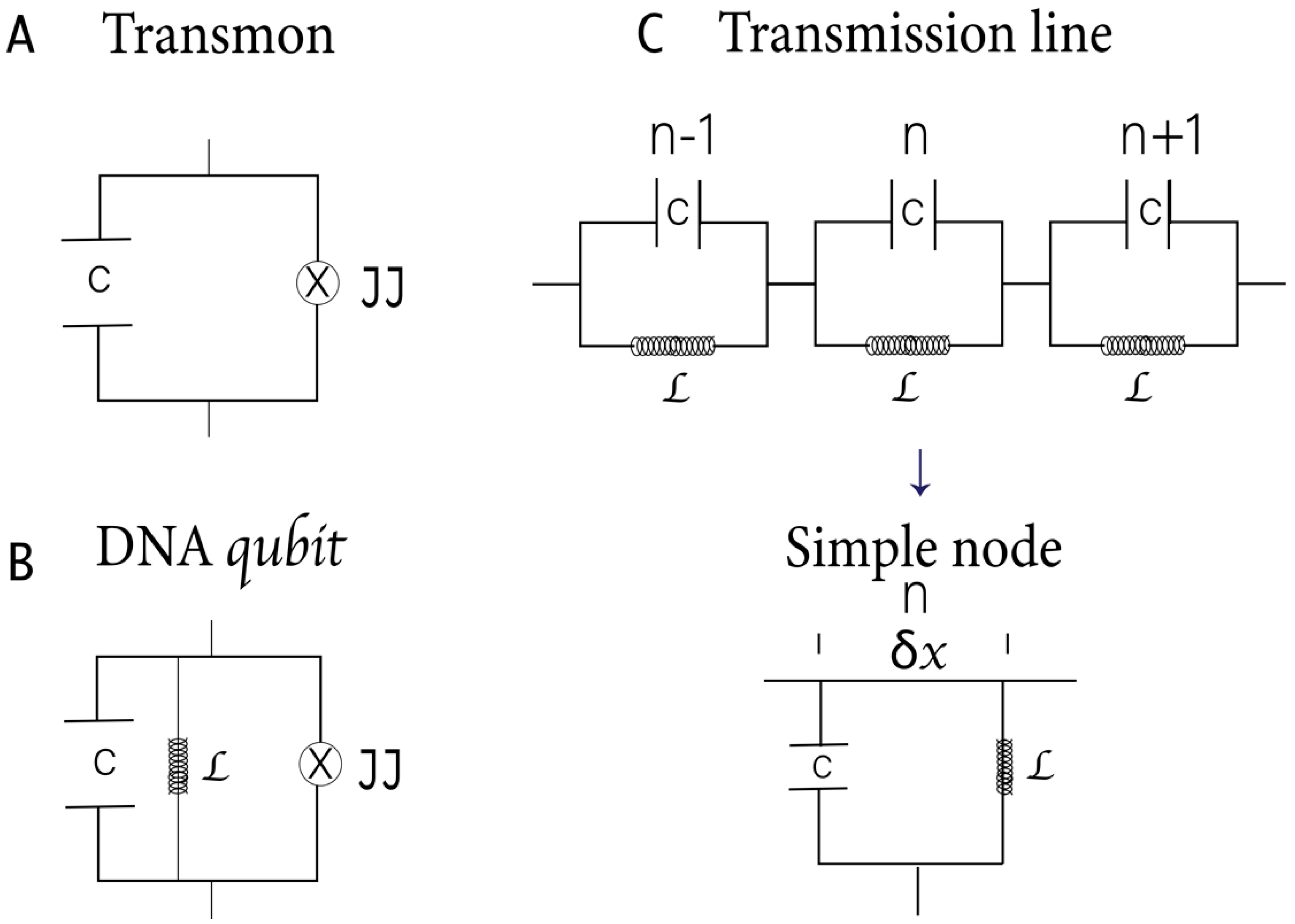
2.2. A–T, T–A, G–C, and C–G Base Pairs as RF Squids: The Hamiltonian Scheme
- is a single-electron and hole-pair momentum.
- is the probability density of finding an electron and hole pair in a given volume.
- of the phase associated with each electron and hole pair in space.
- : flow in the JJ;
- : external magnetic flux; ;
- : capacitance;
- : inductance;
- : Josephson energy;
- : magnetic flux quantum;
- : JJ energy.
2.3. DNA Backbone as a Finite Transmission Line with Distributed Parameters: The Hamiltonian Scheme
2.4. DNA as a Quantum Electrodynamic Circuit: The Hamiltonian Scheme
- , the cavity losses and the decay rate.
- , decoherence or decay of the two-level system.
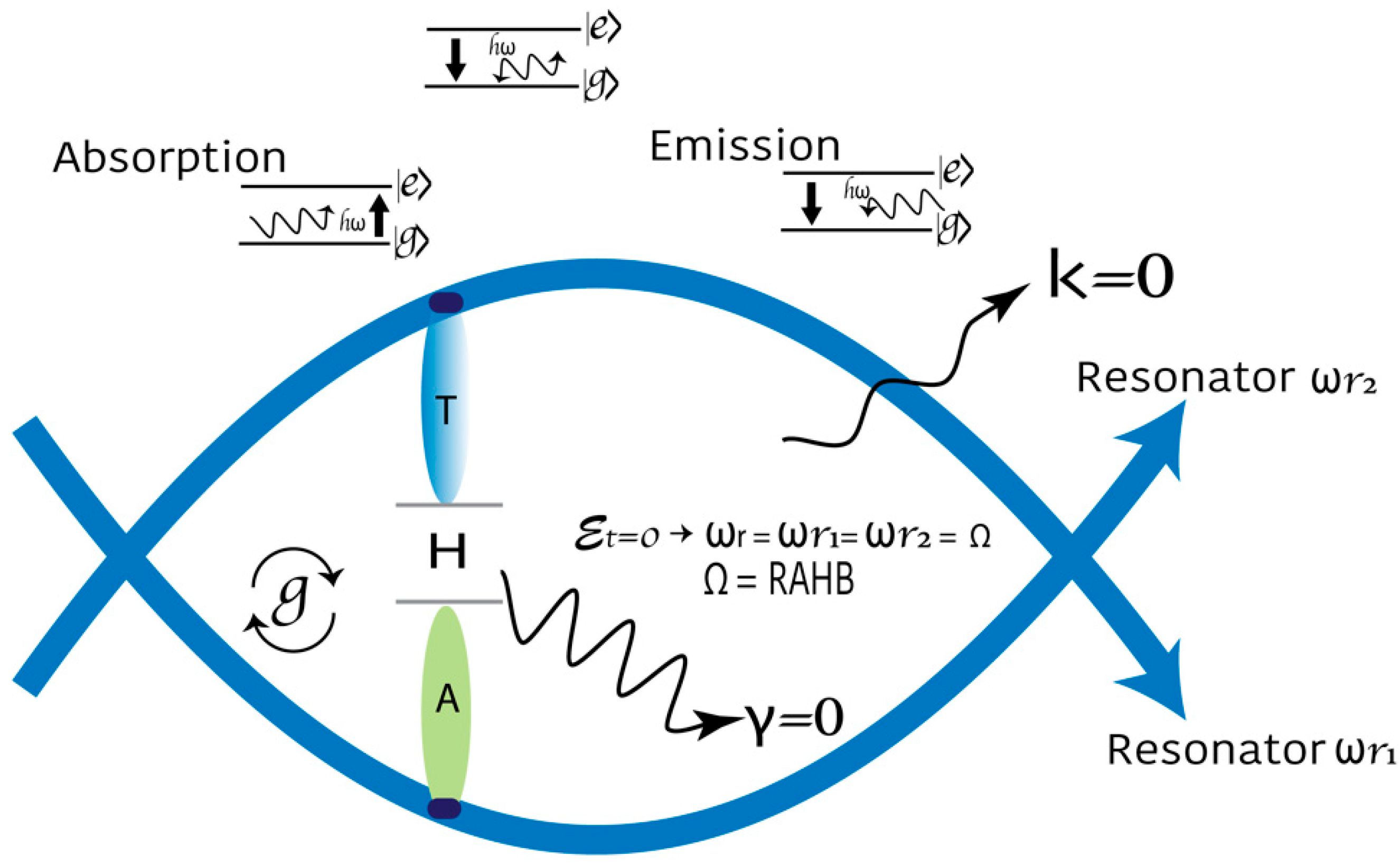

2.5. Quantum Description of Transcription
- Due to the Josephson effect, when the pairs coherently pass through a JJ, the supercurrent related to the phase difference between electrodes can be represented by . is the maximum critical current the junction can maintain. In our model of DNA, the maximum crucial current of the H-bond is ;
- Due to the supercurrent, an intrinsic phase shift appears when computing the line integral of momentum pairs moving along the curve;
- Due to external magnetic flux, classical electromagnetism and Maxwell’s laws show that a magnetic field that changes with time induces an electric field. This, in turn, modifies the momentum of the charged particles by exerting a force on them. The moment of the electron pairs is given by equation . The first component corresponds to the kinetic part, and the second to the field contribution. The potential vector is .
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nudler, E. RNA Polymerase Active Center: The Molecular Engine of Transcription. Annu. Rev. Biochem. 2009, 78, 335–361. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rucobo, F.W.; Cramer, P. Structural basis of transcription elongation. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2013, 1829, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Werner, F.; Grohmann, D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 2011, 9, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Vannini, A.; Cramer, P. Conservation between the RNA Polymerase I, II, and III Transcription Initiation Machineries. Mol. Cell 2012, 45, 439–446. [Google Scholar] [CrossRef]
- Lu, F. Several Ways to Implement Qubits in Physics. J. Phys. Conf. Ser. 2021, 1865, 022007. [Google Scholar] [CrossRef]
- Basilewitsch, D.; Schmidt, R.; Sugny, D.; Maniscalco, S.; Koch, C.P. Beating the limits with initial correlations. New J. Phys. 2017, 19, 113042. [Google Scholar] [CrossRef]
- Fursina, A.A.; Sinitskii, A. Toward Molecular Spin Qubit Devices: Integration of Magnetic Molecules into Solid-State Devices. ACS Appl. Electron. Mater. 2023, 5, 3531–3545. [Google Scholar] [CrossRef]
- Riera Aroche, R.; Ortiz García, Y.M.; Martínez Arellano, M.A.; Riera Leal, A. DNA as a perfect quantum computer based on the quantum physics principles. Sci. Rep. 2024, 14, 11636. [Google Scholar] [CrossRef]
- Dos Santos, C.S.; Filho, E.D.; Ricotta, R.M. Quantum confinement in hydrogen bond of DNA and RNA. J. Phys. Conf. Ser. 2015, 597, 012033. [Google Scholar] [CrossRef]
- Majer, J.; Chow, J.M.; Gambetta, J.M.; Koch, J.; Johnson, B.R.; Schreier, J.A.; Frunzio, L.; Schuster, D.I.; Houck, A.A.; Wallraff, A.; et al. Coupling superconducting qubits via a cavity bus. Nature 2007, 449, 443–447. [Google Scholar] [CrossRef]
- Clarke, J.; Wilhelm, F.K. Superconducting quantum bits. Nature 2008, 453, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Karamitros, D.; McKelvey, T.; Pilaftsis, A. Quantum coherence of critical unstable two-level systems. Phys. Rev. D 2023, 108, 016006. [Google Scholar] [CrossRef]
- Linke, N.M.; Maslov, D.; Roetteler, M.; Debnath, S.; Figgatt, C.; Landsman, K.A.; Wright, K.; Monroe, C. Experimental comparison of two quantum computing architectures. Proc. Natl. Acad. Sci. USA 2017, 114, 3305–3310. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.P. Coplanar Waveguide: A Surface Strip Transmission Line Suitable for Nonreciprocal Gyromagnetic Device Applications. IEEE Trans. Microw. Theory Tech. 1969, 17, 1087–1090. [Google Scholar] [CrossRef]
- Bennett, R.; Barlow, T.M.; Beige, A. A physically motivated quantization of the electromagnetic field. Eur. J. Phys. 2016, 37, 014001. [Google Scholar] [CrossRef]
- Sarabi, B.; Ramanayaka, A.N.; Burin, A.L.; Wellstood, F.C.; Osborn, K.D. Cavity quantum electrodynamics using a near-resonance two-level system: Emergence of the Glauber state. Appl. Phys. Lett. 2015, 106, 172601. [Google Scholar] [CrossRef]
- Blais, A.; Grimsmo, A.L.; Girvin, S.M.; Wallraff, A. Circuit quantum electrodynamics. Rev. Mod. Phys. 2021, 93, 025005. [Google Scholar] [CrossRef]
- Hwang, M.-J.; Choi, M.-S. Variational study of a two-level system coupled to a harmonic oscillator in an ultrastrong-coupling regime. Phys. Rev. A 2010, 82, 025802. [Google Scholar] [CrossRef]
- Joo, J.; Lee, C.-W.; Kono, S.; Kim, J. Logical measurement-based quantum computation in circuit-QED. Sci. Rep. 2019, 9, 16592. [Google Scholar] [CrossRef]
- Han, J.-X.; Wu, J.-L.; Wang, Y.; Jiang, Y.-Y.; Xia, Y.; Song, J. Multi-qubit phase gate on multiple resonators mediated by a superconducting bus. Opt. Express 2020, 28, 1954. [Google Scholar] [CrossRef]
- Blais, A.; Girvin, S.M.; Oliver, W.D. Quantum information processing and quantum optics with circuit quantum electrodynamics. Nat. Phys. 2020, 16, 247–256. [Google Scholar] [CrossRef]
- Travers, A.; Muskhelishvili, G. DNA structure and function. FEBS J. 2015, 282, 2279–2295. [Google Scholar] [CrossRef]
- Wing, R.; Drew, H.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Crystal structure analysis of a complete turn of B-DNA. Nature 1980, 287, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Kohestani, H.; Wereszczynski, J. The effects of RNA.DNA-DNA triple helices on nucleosome structures and dynamics. Biophys. J. 2023, 122, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.H.; Jin, Y.; Struhl, K. TFIIH Phosphorylation of the Pol II CTD Stimulates Mediator Dissociation from the Preinitiation Complex and Promoter Escape. Mol. Cell 2014, 54, 601–612. [Google Scholar] [CrossRef]
- Tripathi, S.; Brahmachari, S.; Onuchic, J.N.; Levine, H. DNA supercoiling-mediated collective behavior of co-transcribing RNA polymerases. Nucleic Acids Res. 2022, 50, 1269–1279. [Google Scholar] [CrossRef]
- Mayfield, J.E.; Irani, S.; Escobar, E.; Zhang, Z.; Burkholder, N.T.; Robinson, M.R.; Mehaffey, M.R.; Sipe, S.N.; Yang, W.; Prescott, N.; et al. Tyr1 phosphorylation promotes phosphorylation of Ser2 on the C-terminal domain of eukaryotic RNA polymerase II by P-TEFb. Elife 2019, 8, e48725. [Google Scholar] [CrossRef]
- Boehning, M.; Dugast-Darzacq, C.; Rankovic, M.; Hansen, A.S.; Yu, T.; Marie-Nelly, H.; McSwiggen, D.T.; Kokic, G.; Dailey, G.M.; Cramer, P.; et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018, 25, 833–840. [Google Scholar] [CrossRef]
- Lu, H.; Yu, D.; Hansen, A.S.; Ganguly, S.; Liu, R.; Heckert, A.; Darzacq, X.; Zhou, Q. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018, 558, 318–323. [Google Scholar] [CrossRef]
- McNamara, R.P.; Reeder, J.E.; McMillan, E.A.; Bacon, C.W.; McCann, J.L.; D’orso, I. KAP1 Recruitment of the 7SK snRNP Complex to Promoters Enables Transcription Elongation by RNA Polymerase II. Mol. Cell 2016, 61, 39–53. [Google Scholar] [CrossRef]
- Du, X.; Qin, W.; Yang, C.; Dai, L.; San, M.; Xia, Y.; Zhou, S.; Wang, M.; Wu, S.; Zhang, S.; et al. RBM22 regulates RNA polymerase II 5′ pausing, elongation rate, and termination by coordinating 7SK-P-TEFb complex and SPT5. Genome Biol. 2024, 25, 102. [Google Scholar] [CrossRef] [PubMed]
- Turecka, K.; Firczuk, M.; Werel, W. Alteration of the −35 and −10 sequences and deletion the upstream sequence of the −35 region of the promoter A1 of the phage T7 in dsDNA confirm the contribution of non-specific interactions with E. coli RNA polymerase to the transcription initiation process. Front. Mol. Biosci. 2024, 10, 1335409. [Google Scholar]
- Klein, C.A.; Teufel, M.; Weile, C.J.; Sobetzko, P. The bacterial promoter spacer modulates promoter strength and timing by length, TG-motifs and DNA supercoiling sensitivity. Sci. Rep. 2021, 11, 24399. [Google Scholar] [CrossRef] [PubMed]
- Paget, M.S.; Helmann, J.D. The σ70family of sigma factors. Genome Biol. 2003, 4, 203. [Google Scholar] [CrossRef]
- Borukhov, S.; Nudler, E. RNA polymerase: The vehicle of transcription. Trends Microbiol. 2008, 16, 126–134. [Google Scholar] [CrossRef]
- Martinez-Rucobo, F.W.; Sainsbury, S.; Cheung, A.C.; Cramer, P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011, 30, 1302–1310. [Google Scholar] [CrossRef]
- Gnatt, A.L.; Cramer, P.; Fu, J.; Bushnell, D.A.; Kornberg, R.D. Structural Basis of Transcription: An RNA Polymerase II Elongation Complex at 3.3 Å Resolution. Science 2001, 292, 1876–1882. [Google Scholar] [CrossRef]
- Korzheva, N.; Mustaev, A.; Kozlov, M.; Malhotra, A.; Nikiforov, V.; Goldfarb, A.; Darst, S.A. A Structural Model of Transcription Elongation. Science 2000, 289, 619–625. [Google Scholar] [CrossRef]
- Xu, J.; Chong, J.; Wang, D. Opposite roles of transcription elongation factors Spt4/5 and Elf1 in RNA polymerase II transcription through B-form versus non-B DNA structures. Nucleic Acids Res. 2021, 49, 4944–4953. [Google Scholar] [CrossRef]
- Yu, J.; Retamal, J.C.; Sanz, M.; Solano, E.; Albarrán-Arriagada, F. Superconducting circuit architecture for digital-analog quantum computing. EPJ Quantum Technol. 2022, 9, 9. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, X.; Li, W.; Fang, X.; Li, T. Phase-Slip Based SQUID Used as a Photon Switch in Superconducting Quantum Computation Architectures. Electronics 2024, 13, 2380. [Google Scholar] [CrossRef]
- Wu, K.; Bozzi, M.; Fonseca, N.J.G. Substrate Integrated Transmission Lines: Review and Applications. IEEE J. Microw. 2021, 1, 345–363. [Google Scholar] [CrossRef]
- Prostejovsky, A.M.; Gehrke, O.; Kosek, A.M.; Strasser, T.; Bindner, H.W. Distribution Line Parameter Estimation Under Consideration of Measurement Tolerances. IEEE Trans. Ind. Inf. 2016, 12, 726–735. [Google Scholar] [CrossRef]
- Yun, K. Two types of electric field enhancements by infinitely many circular conductors arranged closely in two parallel lines. Q. Appl. Math. 2017, 75, 649–676. [Google Scholar] [CrossRef]
- Coufal, O.; Radil, L.; Toman, P. Magnetic field and forces in a pair of parallel conductors. Int. J. Appl. Electromagn. Mech. 2018, 56, 243–261. [Google Scholar] [CrossRef]
- Fedoseyev, V.G. Conservation laws and transverse motion of energy on reflection and transmission of electromagnetic waves. J. Phys. A Math. Gen. 1988, 21, 2045–2059. [Google Scholar] [CrossRef]
- Kundu, S.; Simserides, C. Charge transport in a double-stranded DNA: Effects of helical symmetry and long-range hopping. Phys. Rev. E 2024, 109, 014401. [Google Scholar] [CrossRef]
- Triberis, G.P.; Simserides, C.; Karavolas, V.C. Small polaron hopping transport along DNA molecules. J. Phys. Condens. Matter 2005, 17, 2681–2690. [Google Scholar] [CrossRef]
- Zhuravel, R.; Huang, H.; Polycarpou, G.; Polydorides, S.; Motamarri, P.; Katrivas, L.; Rotem, D.; Sperling, J.; Zotti, L.A.; Kotlyar, A.B.; et al. Backbone charge transport in double-stranded DNA. Nat. Nanotechnol. 2020, 15, 836–840. [Google Scholar] [CrossRef]
- Schilling, O.F.; Sugui, S.S. Faraday’s law and perfect (super) conductivity. Eur. J. Phys. 2004, 25, 337–341. [Google Scholar] [CrossRef]
- Kühn, S. General Analytic Solution of the Telegrapher’s Equations and the Resulting Consequences for Electrically Short Transmission Lines. J. Electromagn. Anal. Appl. 2020, 12, 71–87. [Google Scholar] [CrossRef]
- Reagor, M.J. Superconducting Cavities for Circuit Quantum Electrodynamics; Yale University: Connecticut, CT, USA, 2015. [Google Scholar]
- Frimmer, M.; Novotny, L. The classical Bloch equations. Am. J. Phys. 2014, 82, 947–954. [Google Scholar] [CrossRef]
- Stassi, R.; Cirio, M.; Nori, F. Scalable quantum computer with superconducting circuits in the ultrastrong coupling regime. NPJ Quantum Inf. 2020, 6, 67. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Young, R.A. Biomolecular Condensates in the Nucleus. Trends Biochem. Sci. 2020, 45, 961–977. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.A.; Chakraborty, A.K.; Henninger, J.E.; Young, R.A. RNA in formation and regulation of transcriptional condensates. RNA 2022, 28, 52–57. [Google Scholar] [CrossRef]
- Du, M.; Stitzinger, S.H.; Spille, J.H.; Cho, W.K.; Lee, C.; Hijaz, M.; Quintana, A.; Cissé, I.I. Direct observation of a condensate effect on super-enhancer controlled gene bursting. Cell 2024, 187, 331–344.e17. [Google Scholar] [CrossRef]
- Richter, W.F.; Nayak, S.; Iwasa, J.; Taatjes, D.J. The Mediator complex as a master regulator of transcription by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2022, 23, 732–749. [Google Scholar] [CrossRef]
- Mann, R.; Notani, D. Transcription factor condensates and signaling driven transcription. Nucleus 2023, 14, 2205758. [Google Scholar] [CrossRef]
- Salari, H.; Fourel, G.; Jost, D. Transcription regulates the spatio-temporal dynamics of genes through micro-compartmentalization. Nat. Commun. 2024, 15, 5393. [Google Scholar] [CrossRef]
- Li, C.; Li, Z.; Wu, Z.; Lu, H. Phase separation in gene transcription control. Acta Biochim. Biophys. Sin. 2023, 55, 1052–1063. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, Y.; McCammon, J.A. How Does the cAMP-Dependent Protein Kinase Catalyze the Phosphorylation Reaction: An ab Initio QM/MM Study. J. Am. Chem. Soc. 2005, 127, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Greiwe, J.F.; Miller, T.C.R.; Locke, J.; Martino, F.; Howell, S.; Schreiber, A.; Nans, A.; Diffley, J.F.X.; Costa, A. Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase. Nat. Struct. Mol. Biol. 2022, 29, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.; Senior, A.E. ATP synthase: What we know about ATP hydrolysis and what we do not know about ATP synthesis. Biochim. Et. Biophys. Acta (BBA) Bioenerg. 2000, 1458, 300–309. [Google Scholar] [CrossRef]
- Kamerlin, S.C.L.; Warshel, A. On the Energetics of ATP Hydrolysis in Solution. J. Phys. Chem. B 2009, 113, 15692–15698. [Google Scholar] [CrossRef]
- Meurer, F.; Do, H.T.; Sadowski, G.; Held, C. Standard Gibbs energy of metabolic reactions: II. Glucose-6-phosphatase reaction and ATP hydrolysis. Biophys. Chem. 2017, 223, 30–38. [Google Scholar] [CrossRef]
- Fagan, S.P.; Mukherjee, P.; Jaremko, W.J.; Nelson-Rigg, R.; Wilson, R.C.; Dangerfield, T.L.; Johnson, A.K.; Lahiri, I.; Pata, J.D. Pyrophosphate release acts as a kinetic checkpoint during high-fidelity DNA replication by the Staphylococcus aureus replicative polymerase PolC. Nucleic Acids Res. 2021, 49, 8324–8338. [Google Scholar] [CrossRef]
- Da, L.T.; Chao, E.; Duan, B.; Zhang, C.; Zhou, X.; Yu, J. A Jump-from-Cavity Pyrophosphate Ion Release Assisted by a Key Lysine Residue in T7 RNA Polymerase Transcription Elongation. PLoS Comput. Biol. 2015, 11, e1004624. [Google Scholar] [CrossRef]
- Niepce, D.; Burnett, J.J.; Kudra, M.; Cole, J.H.; Bylander, J. Stability of superconducting resonators: Motional narrowing and the role of Landau-Zener driving of two-level defects. Sci. Adv. 2021, 7, eabh0462. [Google Scholar] [CrossRef]
- Boguslavski, K.; Lappi, T.; Peuron, J.; Singh, P. Conserved energy–momentum tensor for real-time lattice simulations. Eur. Phys. J. C 2024, 84, 368. [Google Scholar] [CrossRef]
- Haroche, S. Cavity quantum electrodynamics: The strange properties of photons and atoms confined in a box. In Proceedings of the Quantum Electronics and Laser Science Conference 1992, Anaheim, CA, USA, 10–15 May 1992; Volume 13. [Google Scholar]
- Boyer, M.; Liss, R.; Mor, T. Geometry of entanglement in the Bloch sphere. Phys. Rev. A 2017, 95, 032308. [Google Scholar] [CrossRef]
- McKay, D.C.; Wood, C.J.; Sheldon, S.; Chow, J.M.; Gambetta, J.M. Efficient Z-Gates for Quantum Computing. Phys. Rev. A 2017, 96, 022330. [Google Scholar] [CrossRef]
- Bluvstein, D.; Levine, H.; Semeghini, G.; Wang, T.T.; Ebadi, S.; Kalinowski, M.; Keesling, A.; Maskara, N.; Pichler, H.; Greiner, M.; et al. A quantum processor based on coherent transport of entangled atom arrays. Nature 2022, 604, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Li, Y.; Mong, R.S.K.; Singh, C. Student Understanding of the Bloch Sphere. Eur. J. Phys. 2024, 45, 025705. [Google Scholar] [CrossRef]
- Carbonell-Ballestero, M.; Garcia-Ramallo, E.; Montañez, R.; Rodriguez-Caso, C.; Macía, J. Dealing with the genetic load in bacterial synthetic biology circuits: Convergences with the Ohm’s law. Nucleic Acids Res. 2016, 44, 496–507. [Google Scholar] [CrossRef]
- Petersson, K.D.; McFaul, L.W.; Schroer, M.D.; Jung, M.; Taylor, J.M.; Houck, A.A.; Petta, J.R. Circuit quantum electrodynamics with a spin qubit. Nature 2012, 490, 380–383. [Google Scholar] [CrossRef]
- Hubač, I.; Švec, M.; Wilson, S. Quantum entanglement and quantum information in biological systems (DNA). In Proceedings of the Proceedings, Workshop on Black Holes and Neutron Stars (RAGtime 17–19), Opava, Czech Republic, 23–26 October 2017; Volume 1, pp. 61–84. [Google Scholar]
- Venta, R.; Valk, E.; Örd, M.; Košik, O.; Pääbo, K.; Maljavin, A.; Kivi, R.; Faustova, I.; Shtaida, N.; Lepiku, M.; et al. A processive phosphorylation circuit with multiple kinase inputs and mutually diversional routes controls G1/S decision. Nat. Commun. 2020, 11, 1836. [Google Scholar] [CrossRef]
- Ye, B.; Zheng, Z.-F.; Yang, C.-P. Multiplex-controlled phase gate with qubits distributed in a multicavity system. Phys. Rev. A 2018, 97, 062336. [Google Scholar] [CrossRef]
- Liu, T.; Cao, X.-Z.; Su, Q.-P.; Xiong, S.-J.; Yang, C.-P. Multi-target-qubit unconventional geometric phase gate in a multi-cavity system. Sci. Rep. 2016, 6, 21562. [Google Scholar] [CrossRef]
- Yang, C.-P.; Chu, S.-I.; Han, S. Quantum Information Transfer and Entanglement with SQUID Qubits in Cavity QED: A Dark-State Scheme with Tolerance for Nonuniform Device Parameter. Phys. Rev. Lett. 2004, 92, 117902. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riera Aroche, R.; Ortiz García, Y.M.; Sánchez Moreno, E.C.; Enriquez Cervantes, J.S.; Machado Sulbaran, A.C.; Riera Leal, A. DNA Gene’s Basic Structure as a Nonperturbative Circuit Quantum Electrodynamics: Is RNA Polymerase II the Quantum Bus of Transcription? Curr. Issues Mol. Biol. 2024, 46, 12152-12173. https://doi.org/10.3390/cimb46110721
Riera Aroche R, Ortiz García YM, Sánchez Moreno EC, Enriquez Cervantes JS, Machado Sulbaran AC, Riera Leal A. DNA Gene’s Basic Structure as a Nonperturbative Circuit Quantum Electrodynamics: Is RNA Polymerase II the Quantum Bus of Transcription? Current Issues in Molecular Biology. 2024; 46(11):12152-12173. https://doi.org/10.3390/cimb46110721
Chicago/Turabian StyleRiera Aroche, Raul, Yveth M. Ortiz García, Esli C. Sánchez Moreno, José S. Enriquez Cervantes, Andrea C. Machado Sulbaran, and Annie Riera Leal. 2024. "DNA Gene’s Basic Structure as a Nonperturbative Circuit Quantum Electrodynamics: Is RNA Polymerase II the Quantum Bus of Transcription?" Current Issues in Molecular Biology 46, no. 11: 12152-12173. https://doi.org/10.3390/cimb46110721
APA StyleRiera Aroche, R., Ortiz García, Y. M., Sánchez Moreno, E. C., Enriquez Cervantes, J. S., Machado Sulbaran, A. C., & Riera Leal, A. (2024). DNA Gene’s Basic Structure as a Nonperturbative Circuit Quantum Electrodynamics: Is RNA Polymerase II the Quantum Bus of Transcription? Current Issues in Molecular Biology, 46(11), 12152-12173. https://doi.org/10.3390/cimb46110721







