Complete Mitochondrial Genomes of Nedyopus patrioticus: New Insights into the Color Polymorphism of Millipedes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Sequence Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
3.1. Genome Organization and Composition
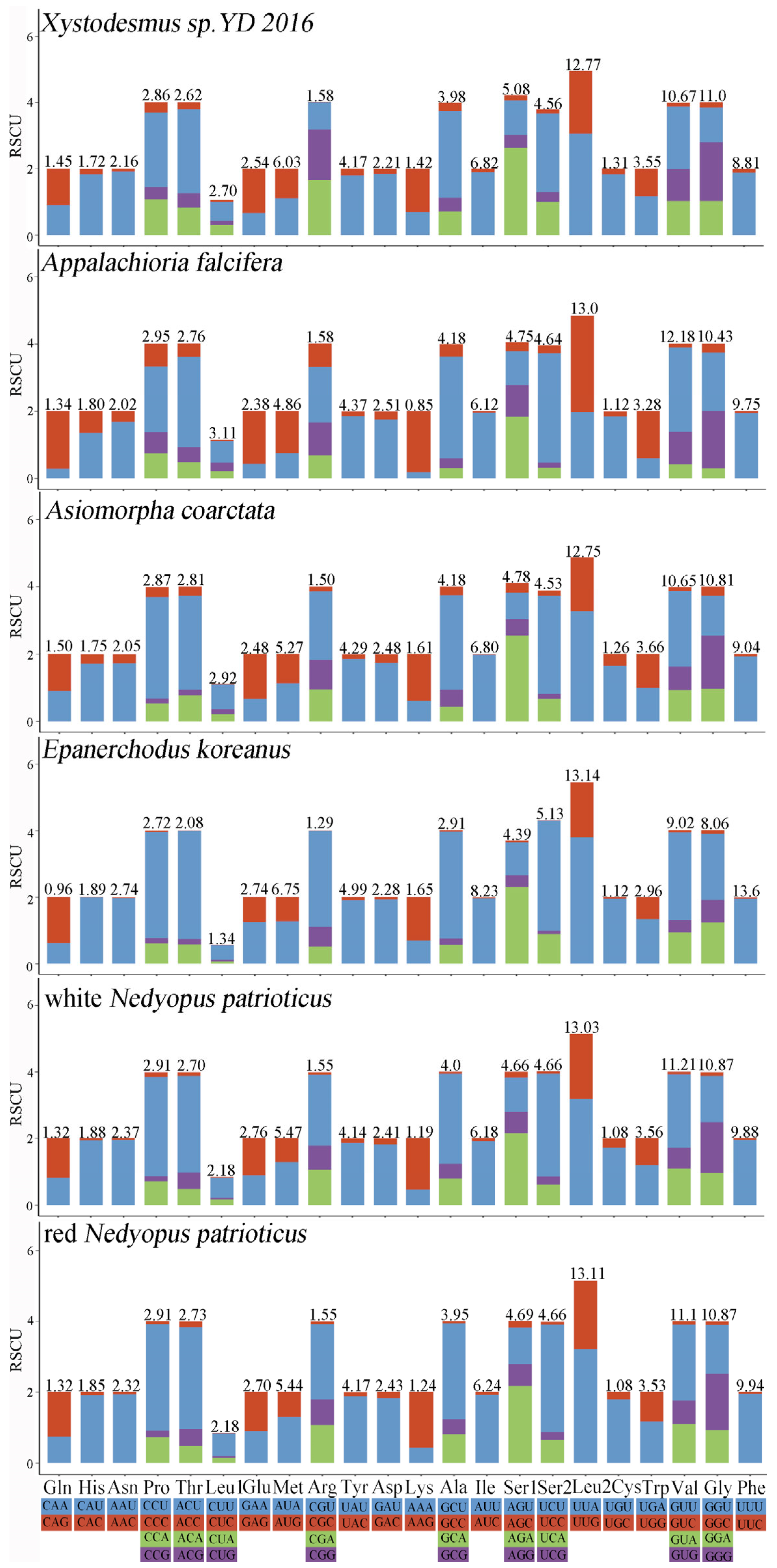
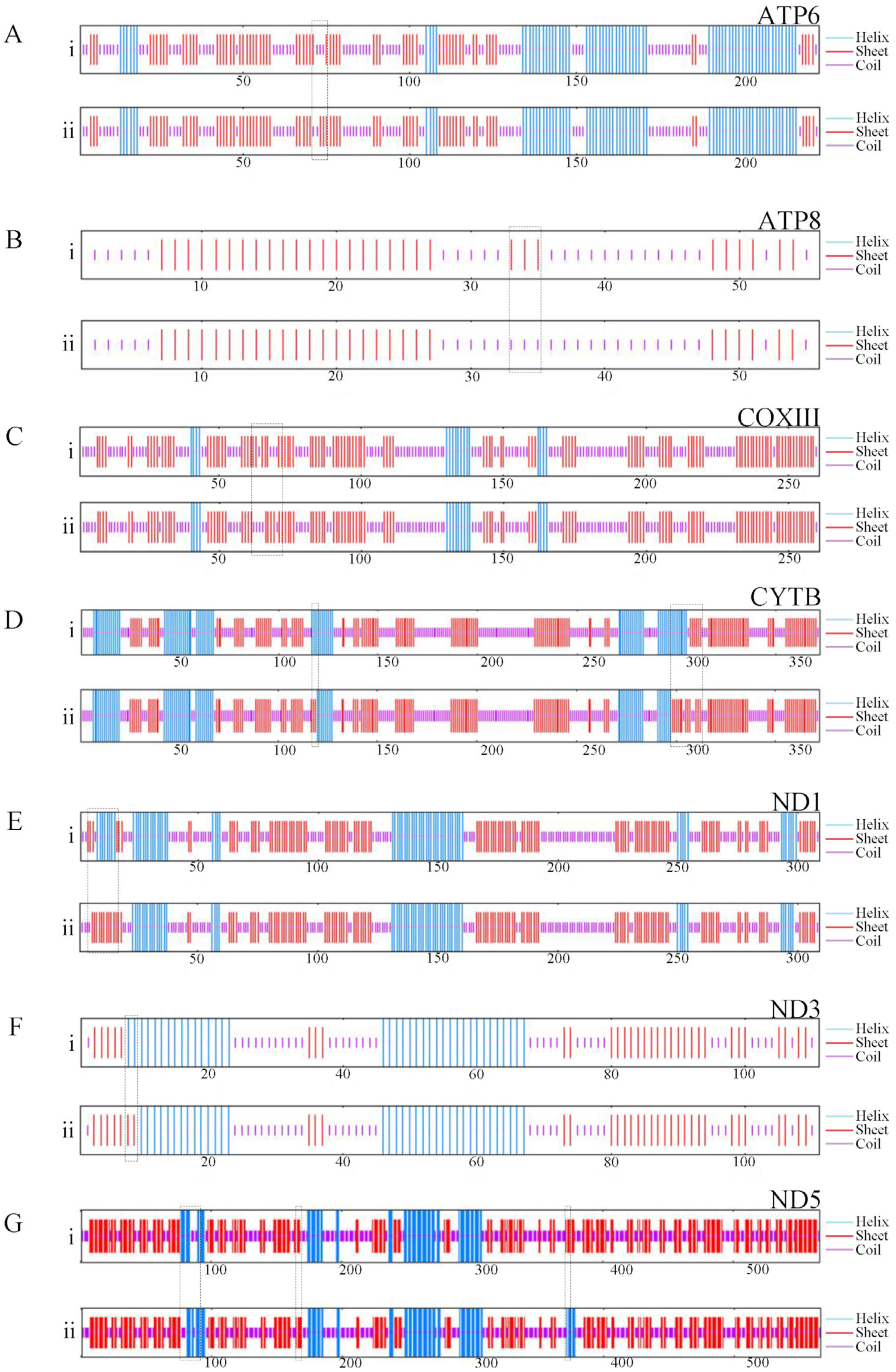
3.2. Protein-Coding Genes
3.3. rRNAs, tRNAs, and CR
3.4. Gene Order
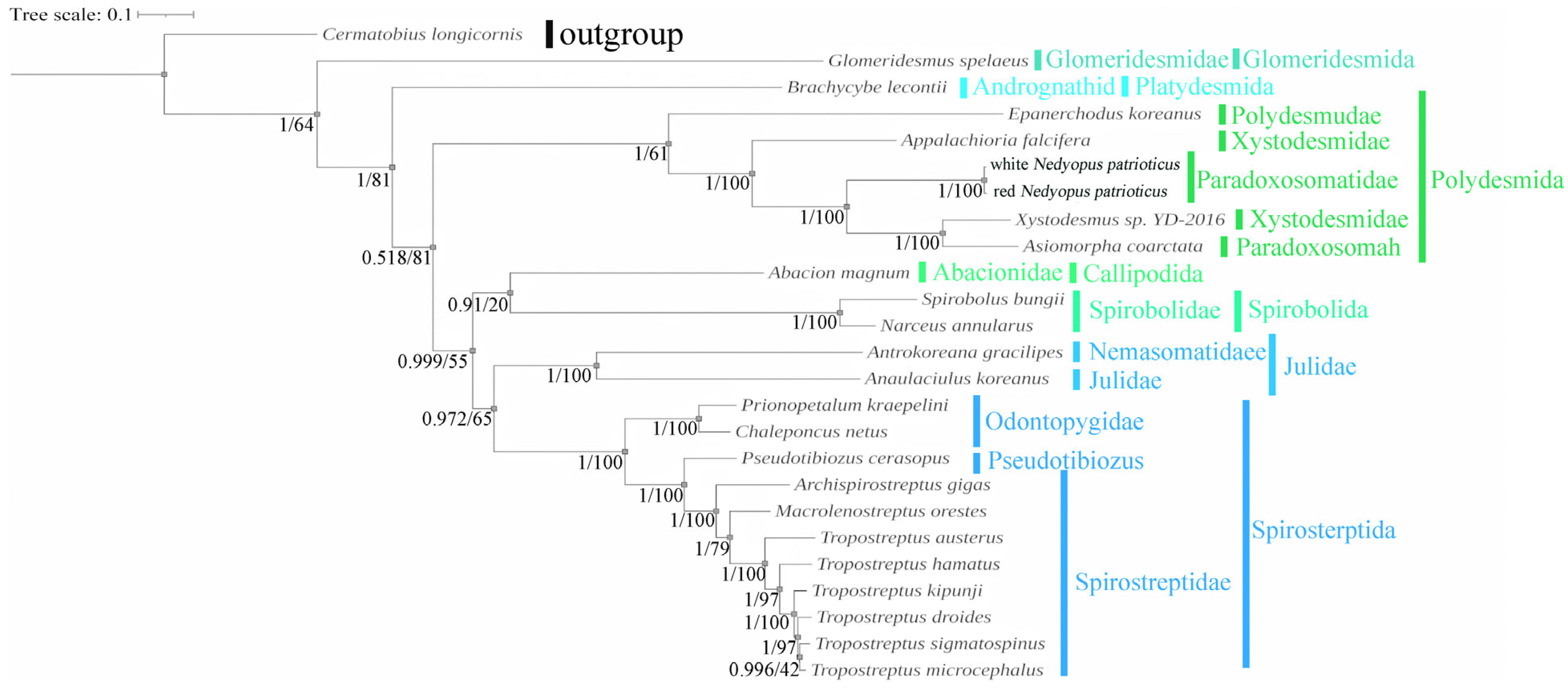
3.5. Phylogenetic Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beukema, W.; Nicieza, A.G.; Lourenço, A.; Velo-Antón, G. Colour polymorphism in Salamandra salamandra (Amphibia: Urodela), revealed by a lack of genetic and environmental differentiation between distinct phenotypes. J. Zool. Syst. Evol. Res. 2016, 54, 127–136. [Google Scholar] [CrossRef]
- Galeotti, P.; Rubolini, D.; Dunn, P.O.; Fasola, M. Colour polymorphism in birds: Causes and functions. J. Evol. Biol. 2003, 16, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.F. Revision of the genus Arthrotus Motschulsky, 1858 (Coleoptera, Chrysomelidae, Galerucinae) of Taiwan, with notes on color polymorphism. Zookeys 2022, 1091, 161–208. [Google Scholar] [CrossRef]
- Mulder, K.P.; Alarcón-Ríos, L.; Nicieza, A.G.; Fleischer, R.C.; Bell, R.C.; Velo-Antón, G. Independent evolutionary transitions to pueriparity across multiple timescales in the viviparous genus Salamandra. Mol. Phylogenet. Evol. 2022, 167, 107347. [Google Scholar] [CrossRef] [PubMed]
- Wellenreuther, M.; Svensson, E.I.; Hansson, B. Sexual selection and genetic colour polymorphisms in animals. Mol. Ecol. 2014, 23, 5398–5414. [Google Scholar] [CrossRef]
- Sun, Z.; Zhang, J.; Wang, R.; Xu, Y.; Zhang, D. Progress of insect mitochondrial genome. J. Insp. Quar. 2010, 3, 69–73. (In Chinese) [Google Scholar]
- Wei, M.; Liu, Y.; Guo, H.; Zhao, F.; Chen, S. Characterization of the complete mitochondrial genome of Cynoglossus gracilis and a comparative analysis with other Cynoglossinae fishes. Gene 2016, 591, 369–375. [Google Scholar] [CrossRef]
- Xu, W.; Lin, S.P.; Liu, H.Y. Mitochondrial genomes of five Hyphessobrycon tetras and their phylogenetic implications. Ecol. Evol. 2021, 11, 12754–12764. [Google Scholar] [CrossRef]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M. Mitofish and Mitoannotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Beckenbach, A.T.; Joy, J.B. Evolution of the mitochondrial genomes of Gall Midges (Diptera: Cecidomyiidae): Rearrangement and severe truncation of tRNA genes. Genome Biol. Evol. 2009, 1, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.F.; Kirkness, E.F.; Barker, S.C. The single mitochondrial chromosome typical of animals has evolved into 18 minichromsomes in the human body louse, Pediculus humanus. Genome Res. 2009, 19, 904–912. [Google Scholar] [CrossRef]
- Franck, P.; Garnery, L.; Solignac, M.; Cornuet, J.M. Molecular confirmation of a fourth lineage in honeybees from the near east. Apidologie 2000, 31, 167–180. [Google Scholar] [CrossRef]
- Yu, Y.; Zhou, S.; Zhu, X.; Xu, X.; Wang, W.; Zha, L.; Wang, P.; Wang, J.; Lai, K.; Wang, S.; et al. Genetic differentiation of eastern honey bee (Apis cerana) populations across Qinghai-Tibet plateau-valley landforms. Front. Genet. 2019, 10, 483. [Google Scholar] [CrossRef]
- Chen, C.C.; Golovatch, S.I.; Chang, H.W. The millipede tribe Nedyopodini, with special reference to the fauna of Taiwan (Diplopoda: Polydesmida: Paradoxosomatidae). J. Nat. Hist. 2005, 39, 3997–4030. [Google Scholar] [CrossRef]
- Wang, Y.M. Serica la: Records of Myriapods on Formosa with descriptions of new species. Q. J. Taiwan Mus. 1955, 8, 13–16. [Google Scholar]
- Ambriz-Morales, P.; Rosa-Reyna, X.F.D.L.; Sifuentes-Rincon, A.M.; Parra-Bracamonte, G.M.; Villa-Melchor, A.; Chassin-Noria, O.; Arellano-Vera, W. The complete mitochondrial genomes of nine white-tailed deer subspecies and their genomic differences. J. Mammal. 2016, 97, 234–245. [Google Scholar] [CrossRef]
- Noguchi, S.; Mori, N.; Higa, Y.; Kuwahara, Y. Identification of Nedyopus patrioticus (ATTEMS, 1898) (Polydesmida: Paradoxosomatidae) Secretions as Possible Defense Substances. Appl. Entomol. Zool. 1997, 32, 447–452. [Google Scholar] [CrossRef]
- García-Roselló, E.; Guisande, C.; Manjarrés-Hernández, A.; González-Dacosta, J.; Heine, J.; Pelayo-Villamil, P.; González-Vilas, L.; Vari, R.P.; Vaamonde, A.; Granado-Lorencio, C.; et al. Can we derive macroecological patterns from primary Global Biodiversity Information Facility data? Glob. Ecol. Biogeogr. 2015, 24, 335–347. [Google Scholar] [CrossRef]
- Dong, Y.; Zhu, L.X.; Bai, Y.; Ou, Y.Y.; Wang, C.B. Complete mitochondrial genomes of two flat-backed millipedes by next-generation sequencing (Diplopoda, Polydesmida). Zookeys 2016, 637, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Clewley, J.P. Macintosh sequence analysis software. Mol. Biotechnol. 1995, 3, 221–224. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.H.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Murphy, D. Application of ggplot2 to Pharmacometric Graphics. CPT-Pharmacomet. Syst. Pharmacol. 2013, 2, 1–16. [Google Scholar] [CrossRef]
- Suresh, V.; Parthasarathy, S. SVM-PB-Pred: SVM Based Protein Block Prediction Method Using Sequence Profiles and Secondary Structures. Protein Pept. Lett. 2014, 21, 736–742. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.; Lee, J.; Lee, D.Y.; Xi, H.; Park, J. The complete mitochondrial genome of the millipede Epanerchodus koreanus Verhoeff, 1937 collected in limestone cave of Korea (Polydesmidae: Polydesmida). Mitochondrial DNA Part B-Resour. 2020, 5, 3845–3847. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Fang, Y.; Cao, G.; Shen, C.; Liu, H.; Ruan, H. The Complete Mitochondrial Genome of Spirobolus bungii (Diplopoda, Spirobolidae): The First Sequence for the Genus Spirobolus. Genes 2022, 13, 1587. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, X.J.; Yuan, H.; Shen, J.; Yang, M.X. Complete mitochondrial genome of Sellanucheza jaegeri Golovatch, 2013 by next generation sequencing (Polydesmida: Paradoxosomatidae) and phylogenetic analysis in Diplopoda. Mitochondrial DNA Part B-Resour. 2018, 3, 603. [Google Scholar] [CrossRef]
- Dai, X.; Deng, L.F.; Xiao, X.; Yu, X.J.; Lv, J.X.; Xiong, H.Y.; Ma, L.; Chen, Q.; Yang, L.Y.; Wang, X. First record of the complete mitochondrial genome of Botyodes diniasalis (Walker, 1859) (Lepidoptera: Crambidae). Mitochondrial DNA Part B-Resour. 2023, 8, 1401–1405. [Google Scholar] [CrossRef]
- Woo, H.J.; Lee, Y.S.; Park, S.J.; Lim, J.T.; Jang, K.H.; Choi, E.H.; Choi, Y.G.; Hwang, U.W. Complete mitochondrial genome of a troglobite millipede Antrokoreana gracilipes (Diplopoda, Juliformia, Julida), and juliformian phylogeny. Mol. Cells 2007, 23, 182–191. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Wu, X.; Wang, L.; Chen, S.; Zan, R.; Xiao, H.; Zhang, Y.P. The complete mitochondrial genomes of two species from Sinocyclocheilus (Cypriniformes: Cyprinidae) and a phylogenetic analysis within Cyprininae. Mol. Biol. Rep. 2010, 37, 2163–2171. [Google Scholar] [CrossRef]
- Sun, Y.B.; Shen, Y.Y.; Irwin, D.M.; Zhang, Y.P. Evaluating the roles of energetic functional constraints on teleost mitochondrial-encoded protein evolution. Mol. Biol. Evol. 2011, 28, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Castro, L.R.; Dowton, M. Mitochondrial genomes in the Hymenoptera and their utility as phylogenetic markers. Syst. Ento. 2007, 32, 60–69. [Google Scholar] [CrossRef]
- Dowton, M.; Castro, R.L.; Austin, D.A. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: The examination of genome ‘morphology’. Invertebr. Syst. 2002, 16, 345–356. [Google Scholar] [CrossRef]
- Blanke, A.; Wesener, T. Revival of forgotten characters and modern imaging techniques help to produce a robust phylogeny of the Diplopoda (Arthropoda, Myriapoda). Arthropod Struct. Dev. 2014, 43, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Zhang, Z.S.; Shen, Y.J. Novel mitochondrial gene rearrangements pattern in the millipede Polydesmus sp. GZCS-2019 and phylogenetic analysis of the Myriapoda. Ecol. Evol. 2022, 12, e8764. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, G.; Tan, S.; Gong, S.; Yang, M.; Peng, Q.; Peng, R.; Zou, F. The complete mitochondrial genome sequence analysis of Tibetan argali (Ovis ammon hodgsoni): Implications of Tibetan argali and Gansu argali as the same subspecies. Gene 2013, 521, 24–31. [Google Scholar] [CrossRef]



| Class | Order | Family | Species | Length (bp) | Accession No. |
|---|---|---|---|---|---|
| Dilopoda | Callipodida | Callipodidae | Abacion magnum | 15,160 | JX437062.1 |
| Glomeridesmida | Glomeridesmidae | Glomeridesmus spelaeus | 14,863 | MH590615.1 | |
| Julida | Nemasomatidae | Antrokoreana gracilipes | 14,747 | DQ344025.1 | |
| Julidae | Anaulaciulus koreanus | 14,916 | KX096886.1 | ||
| Playtdesmida | Andrognathidae | Brachycybe lecontii | 15,115 | JX437064.1 | |
| Polydesmida | Paradoxosomatidae | Asiomorpha coarctata | 15,644 | KU721885.1 | |
| red Nedyopus patrioticus | 15,814 | OR755973.1 | |||
| white Nedyopus patrioticus | 15,798 | OR777861.1 | |||
| Polydesmidae | Epanerchodus koreanus | 15,581 | MT898420.1 | ||
| Xystodesmidae | Appalachioria falcifera | 15,282 | JX437063.1 | ||
| Xystodesmus sp. YD-2016 | 15,791 | KU721886.1 | |||
| Spirobolida | Spirobolidae | Narceus annularus | 14,868 | AY055727.1 | |
| Spirobolus bungii | 14,879 | MT767838.1 | |||
| Spirostreptida | Odontopygidae | Chaleponcus netus | 15,093 | MT394513.1 | |
| Prionopetalum kraepelini | 15,114 | MT394524.1 | |||
| Spirostreptidae | Archispirostreptus gigas | 15,177 | MT394525.1 | ||
| Macrolenostreptus orestes | 15,367 | MT394512.1 | |||
| Pseudotibiozus cerasopus | 15,121 | MT394506.1 | |||
| Tropostreptus austerus | 15,261 | MT394523.1 | |||
| Tropostreptus droides | 15,172 | MT394522.1 | |||
| Tropostreptus hamatus | 15,156 | MT394508.1 | |||
| Tropostreptus kipunji | 15,170 | MT394503.1 | |||
| Tropostreptus microcephalus | 15,169 | MT394516.1 | |||
| Tropostreptus sigmatospinus | 15,176 | MT394504.1 | |||
| Chilooda | Lithobiomorpha | Henicopidae | Cermatobius longicornis | 16,833 | KC155628.1 |
| Gene | Location | Length (bp) | Intergenic Region | Codon | Stand | ||
|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||
| COXI | 1/1 | 1533/1533 | 1533/1533 | ATG/ATG | TAG/TAG | N/N | |
| COXII | 1541/1541 | 2218/2218 | 678/678 | 7/7 | ATG/ATG | TAA/TAA | N/N |
| trnK | 2219/2219 | 2281/2281 | 63/63 | N/N | |||
| trnD | 2282/2282 | 2351/2351 | 70/70 | N/N | |||
| ATP8 | 2352/2352 | 2522/2522 | 171/171 | ATG/ATG | TAG/TAG | N/N | |
| ATP6 | 2519/2519 | 3190/3190 | 672/672 | −5/−5 | ATA/ATA | TAA/TAA | N/N |
| COXIII | 3192/3183 | 3968/3968 | 786/786 | 1/−9 | ATG/ATG | TAA/TAA | N/N |
| trnG | 3969/3969 | 4032/4032 | 64/64 | N/N | |||
| ND3 | 4048/4048 | 4381/4381 | 334/334 | 15/15 | ATT/ATT | T/T | N/N |
| trnA | 4382/4382 | 4444/4444 | 63/63 | N/N | |||
| trnR | 4445/4445 | 4507/4507 | 63/63 | N/N | |||
| trnN | 4508/4508 | 4575/4575 | 68/68 | N/N | |||
| trnS1 | 4576/4576 | 4634/4634 | 59/59 | N/N | |||
| trnE | 4637/4637 | 4697/4697 | 61/61 | 2/2 | N/N | ||
| ND6 | 4728/4728 | 5195/5195 | 468/468 | 30/30 | ATA/ATA | TAA/TAA | N/N |
| CYTB | 5170/5170 | 6288/6288 | 1119/1119 | −26/−26 | ATA/ATA | TAG/TAG | N/N |
| trnS2 | 6291/6291 | 6350/6351 | 60/61 | 2/2 | N/N | ||
| CR | 6351/6352 | 7518/7505 | 1168/1154 | / | |||
| trnH | 7519/7506 | 7581/7568 | 63/63 | N/N | |||
| rrnS | 7703/7690 | 8439/8426 | 737/737 | 121/121 | N/N | ||
| rrnL | 8450/8437 | 9809/9795 | 1360/1359 | 10/10 | N/N | ||
| trnL1 | 9799/9786 | 9858/9844 | 60/59 | −11/−11 | N/N | ||
| ND1 | 9859/9845 | 10,786/10,772 | 928/928 | ATA/GTA | T/T | N/N | |
| trnP | 10,787/10,773 | 10,849/10,835 | 63/63 | N/N | |||
| ND4L | 10,851/10,837 | 11,132/11,118 | 282/282 | 1/1 | ATG/ATG | TAG/TAG | N/N |
| ND4 | 11,222/11,208 | 12,457/12,443 | 1236/1236 | 89/89 | ATG/ATG | TAG/TAG | N/N |
| trnT | 12,458/12,444 | 12,523/12,509 | 66/66 | N/N | |||
| ND5 | 12,665/12,650 | 14,354/14,339 | 1690/1690 | 141/141 | ATG/ATA | T/T | N/N |
| trnF | 14,364/14,349 | 14,425/14,410 | 62/62 | 9/9 | N/N | ||
| trnY | 14,426/14,411 | 14,488/14,473 | 63/63 | N/N | |||
| trnQ | 14,500/14,485 | 14,562/14,547 | 63/63 | 11/11 | N/N | ||
| trnC | 14,563/14,548 | 14,624/14,609 | 62/62 | N/N | |||
| trnM | 14,679/14,664 | 14,742/14,727 | 64/64 | 54/54 | N/N | ||
| ND2 | 14,752/14,737 | 15,747/15,732 | 996/996 | 9/9 | ATT/ATA | TAG/TAG | N/N |
| trnW | 15,748/15,733 | 15,813/15,798 | 66/66 | N/N | |||
| Region | Species | Length (bp) | A + T% | AT-Skew | GC-Skew |
|---|---|---|---|---|---|
| Whole mitogenome | red Nedyopus patrioticus | 15,834 | 68.23 | −0.264 | 0.462 |
| white Nedyopus patrioticus | 15,798 | 68.28 | −0.262 | 0.463 | |
| Xystodesmus sp. YD-2016 | 15,791 | 67.01 | −0.217 | 0.471 | |
| Asiomorpha coarctata | 15,644 | 67.45 | −0.235 | 0.429 | |
| Epanerchodus koreanus | 15,581 | 75.11 | −0.260 | 0.446 | |
| Appalachioria falcifera | 15,282 | 64.04 | −0.368 | 0.441 | |
| PCGs | red Nedyopus patrioticus | 10,767 | 67.93 | −0.346 | 0.464 |
| white Nedyopus patrioticus | 10,767 | 67.00 | −0.344 | 0.464 | |
| Xystodesmus sp. YD-2016 | 10,995 | 65.57 | −0.301 | 0.460 | |
| Asiomorpha coarctata | 11,019 | 66.11 | −0.331 | 0.428 | |
| Epanerchodus koreanus | 10,959 | 73.87 | −0.357 | 0.435 | |
| Appalachioria falcifera | 10,998 | 63.08 | −0.447 | 0.451 | |
| rRNAs | red Nedyopus patrioticus | 2097 | 72.53 | −0.073 | 0.434 |
| white Nedyopus patrioticus | 2096 | 72.62 | −0.074 | 0.446 | |
| Xystodesmus sp. YD-2016 | 2007 | 69.20 | −0.042 | 0.511 | |
| Asiomorpha coarctata | 2016 | 69.34 | −0.044 | 0.421 | |
| Epanerchodus koreanus | 2082 | 79.06 | −0.066 | 0.450 | |
| Appalachioria falcifera | 2025 | 68.75 | −0.221 | 0.472 | |
| tRNAs | red Nedyopus patrioticus | 1203 | 67.67 | −0.091 | 0.414 |
| white Nedyopus patrioticus | 1208 | 67.31 | −0.082 | 0.428 | |
| Xystodesmus sp. YD-2016 | 1430 | 71.05 | −0.069 | 0.338 | |
| Asiomorpha coarctata | 1437 | 69.52 | −0.083 | 0.361 | |
| Epanerchodus koreanus | 1378 | 77.14 | −0.091 | 0.435 | |
| Appalachioria falcifera | 1363 | 66.47 | −0.157 | 0.343 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Xu, T.; Chen, Y.; Xu, W.; Wang, Y.; Li, Y.; Zhu, F.; Liu, H.; Ruan, H. Complete Mitochondrial Genomes of Nedyopus patrioticus: New Insights into the Color Polymorphism of Millipedes. Curr. Issues Mol. Biol. 2024, 46, 2514-2527. https://doi.org/10.3390/cimb46030159
Zhang G, Xu T, Chen Y, Xu W, Wang Y, Li Y, Zhu F, Liu H, Ruan H. Complete Mitochondrial Genomes of Nedyopus patrioticus: New Insights into the Color Polymorphism of Millipedes. Current Issues in Molecular Biology. 2024; 46(3):2514-2527. https://doi.org/10.3390/cimb46030159
Chicago/Turabian StyleZhang, Gaoji, Tangjun Xu, Yukun Chen, Wei Xu, Yinuo Wang, Yuanyuan Li, Fuyuan Zhu, Hongyi Liu, and Honghua Ruan. 2024. "Complete Mitochondrial Genomes of Nedyopus patrioticus: New Insights into the Color Polymorphism of Millipedes" Current Issues in Molecular Biology 46, no. 3: 2514-2527. https://doi.org/10.3390/cimb46030159
APA StyleZhang, G., Xu, T., Chen, Y., Xu, W., Wang, Y., Li, Y., Zhu, F., Liu, H., & Ruan, H. (2024). Complete Mitochondrial Genomes of Nedyopus patrioticus: New Insights into the Color Polymorphism of Millipedes. Current Issues in Molecular Biology, 46(3), 2514-2527. https://doi.org/10.3390/cimb46030159





