Effects of rpl1001 Gene Deletion on Cell Division of Fission Yeast and Its Molecular Mechanism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Strains
2.2. Cell Growth Analysis
2.3. Spore Production Analysis
2.4. Construction of Fluorescent Protein Tags
2.5. Microscopy and Data Analysis
2.6. Bioinformatics Analysis
2.7. Quantitative Real-Time PCR
3. Results
3.1. Changes of Cell Growth and Spore Production in the rpl1001Δ Strain
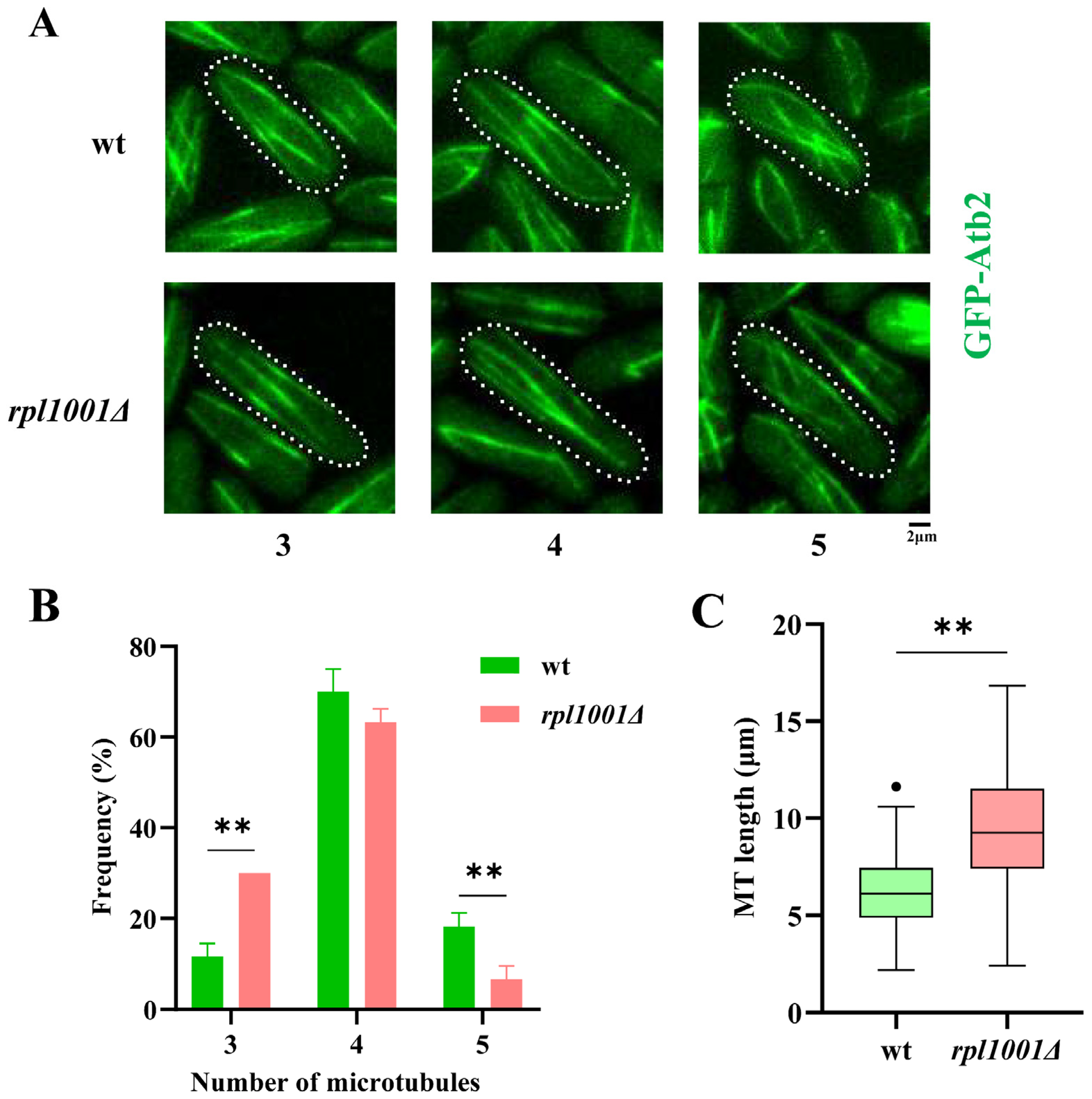
3.2. Microtubule Alterations in the rpl1001Δ Strain during Interphase
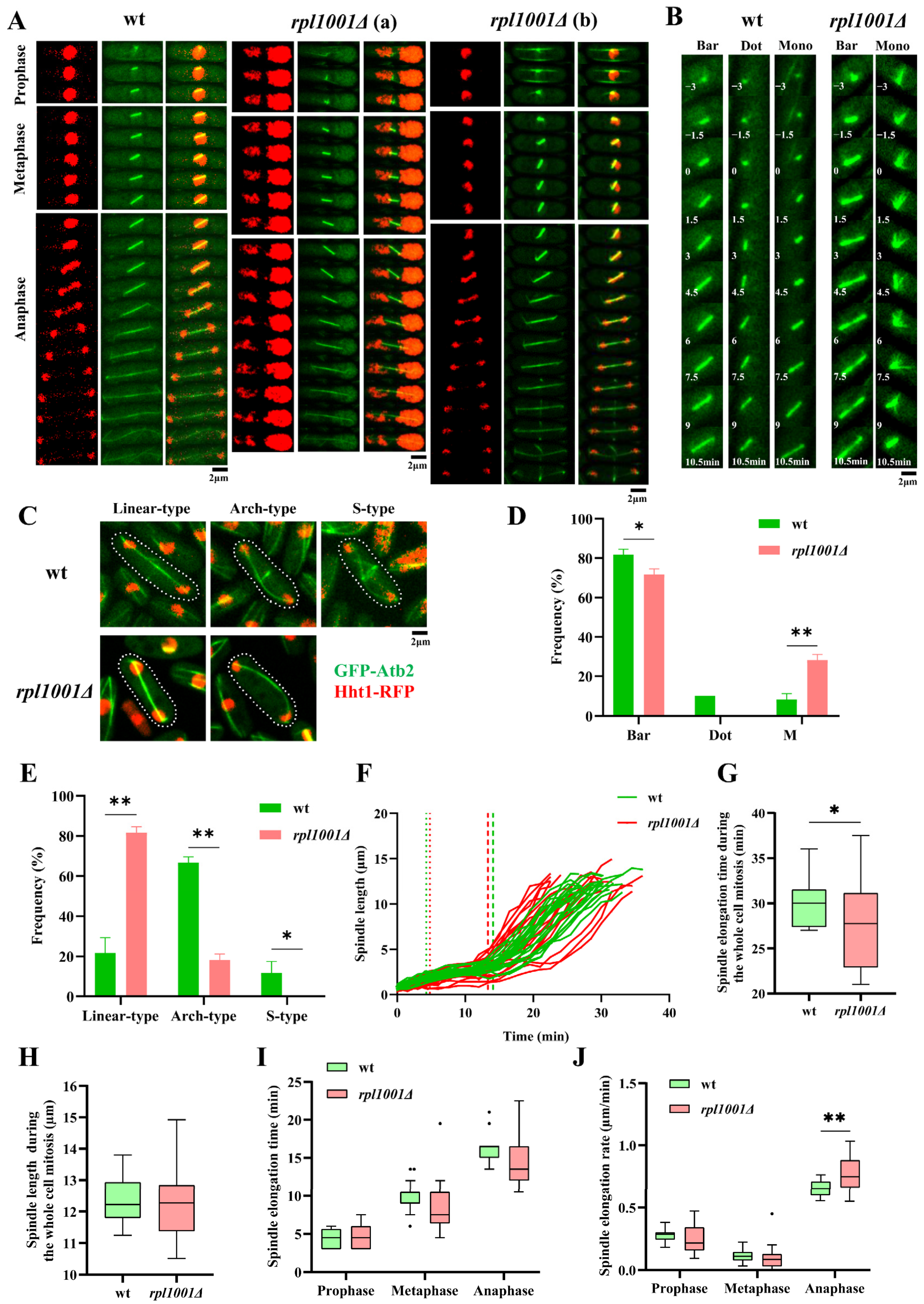
3.3. Changes in Chromosome and Spindle in the rpl1001Δ Strain during Mitosis
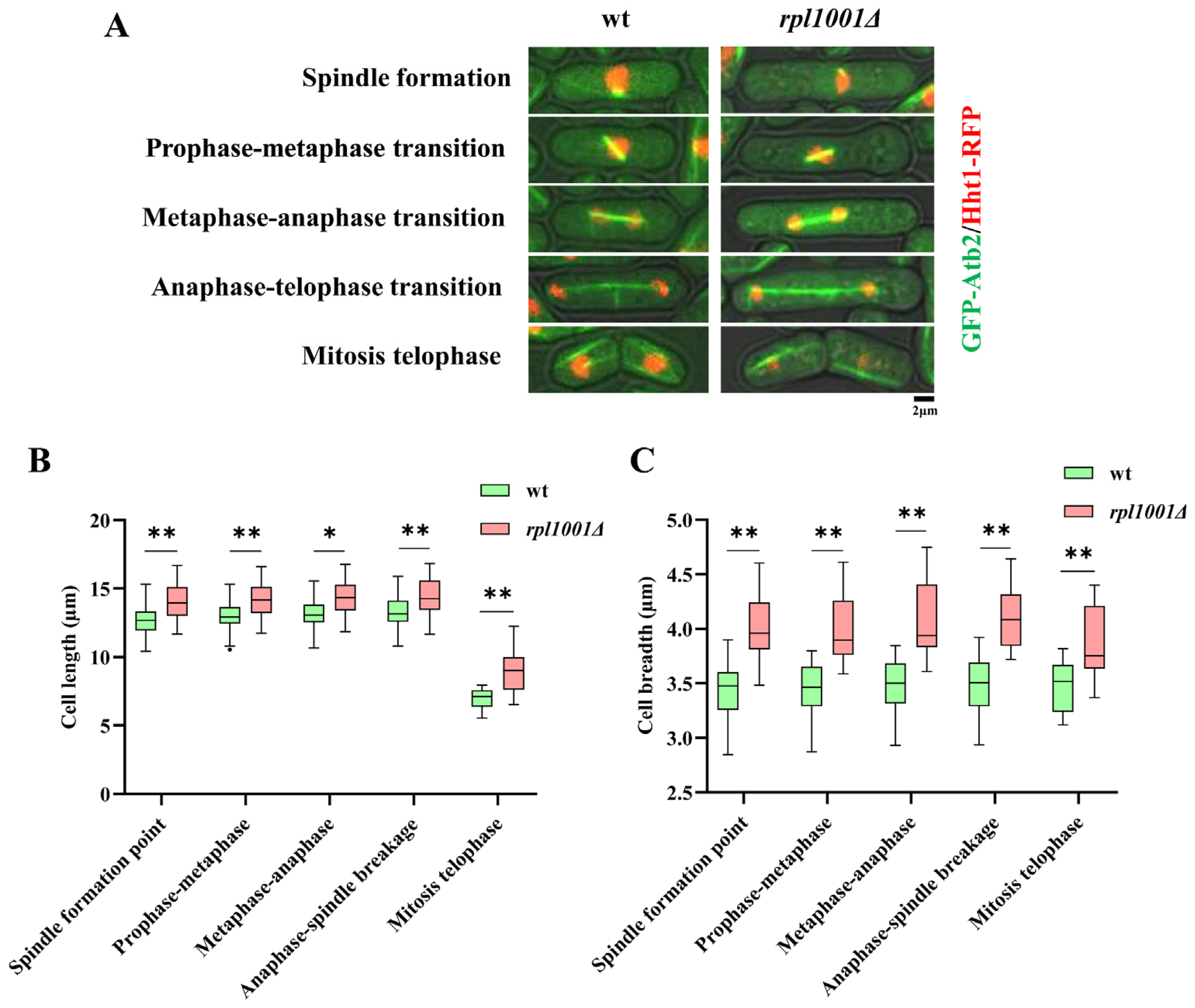
3.4. Changes of Cell Morphology in the rpl1001Δ Strain
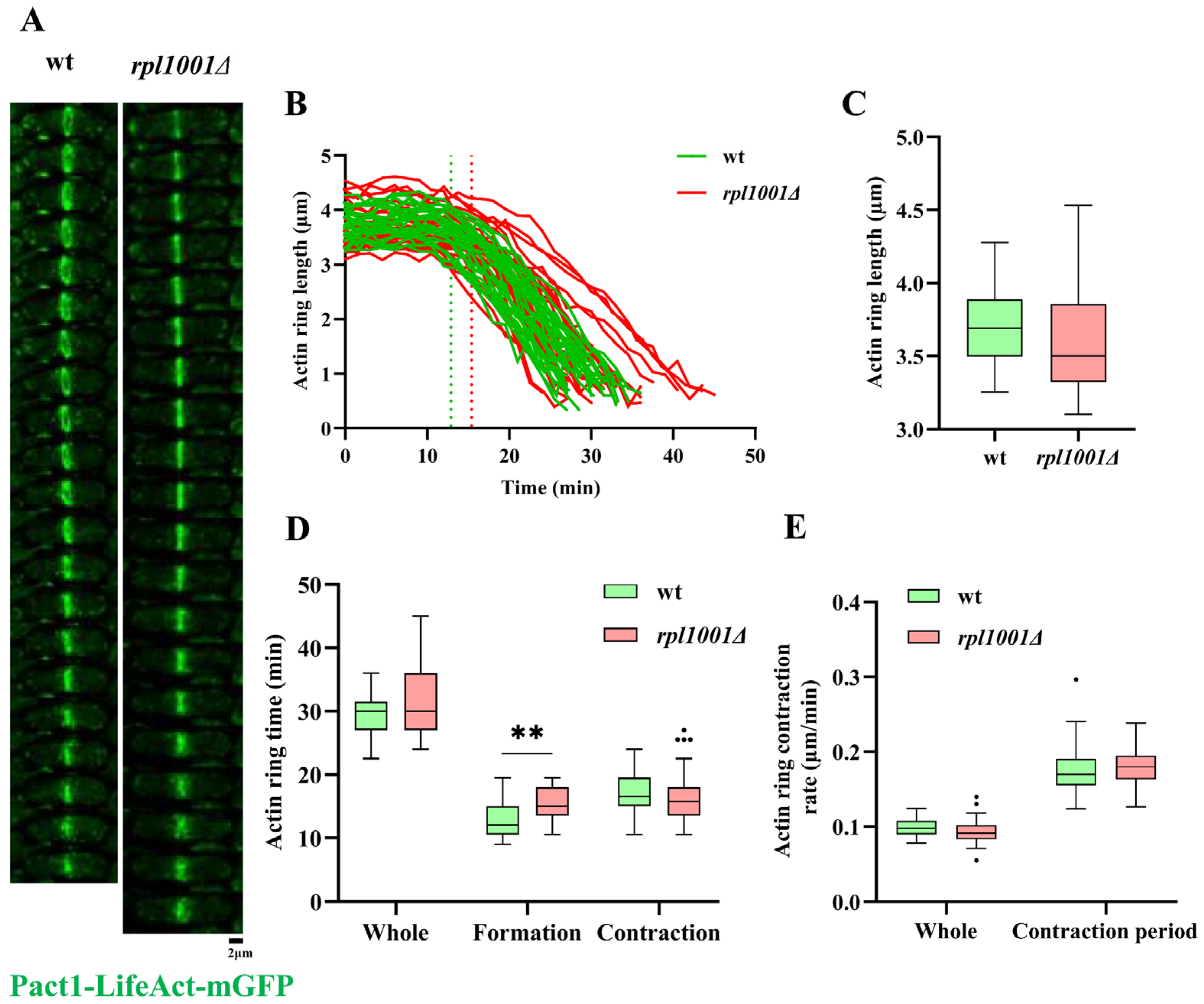
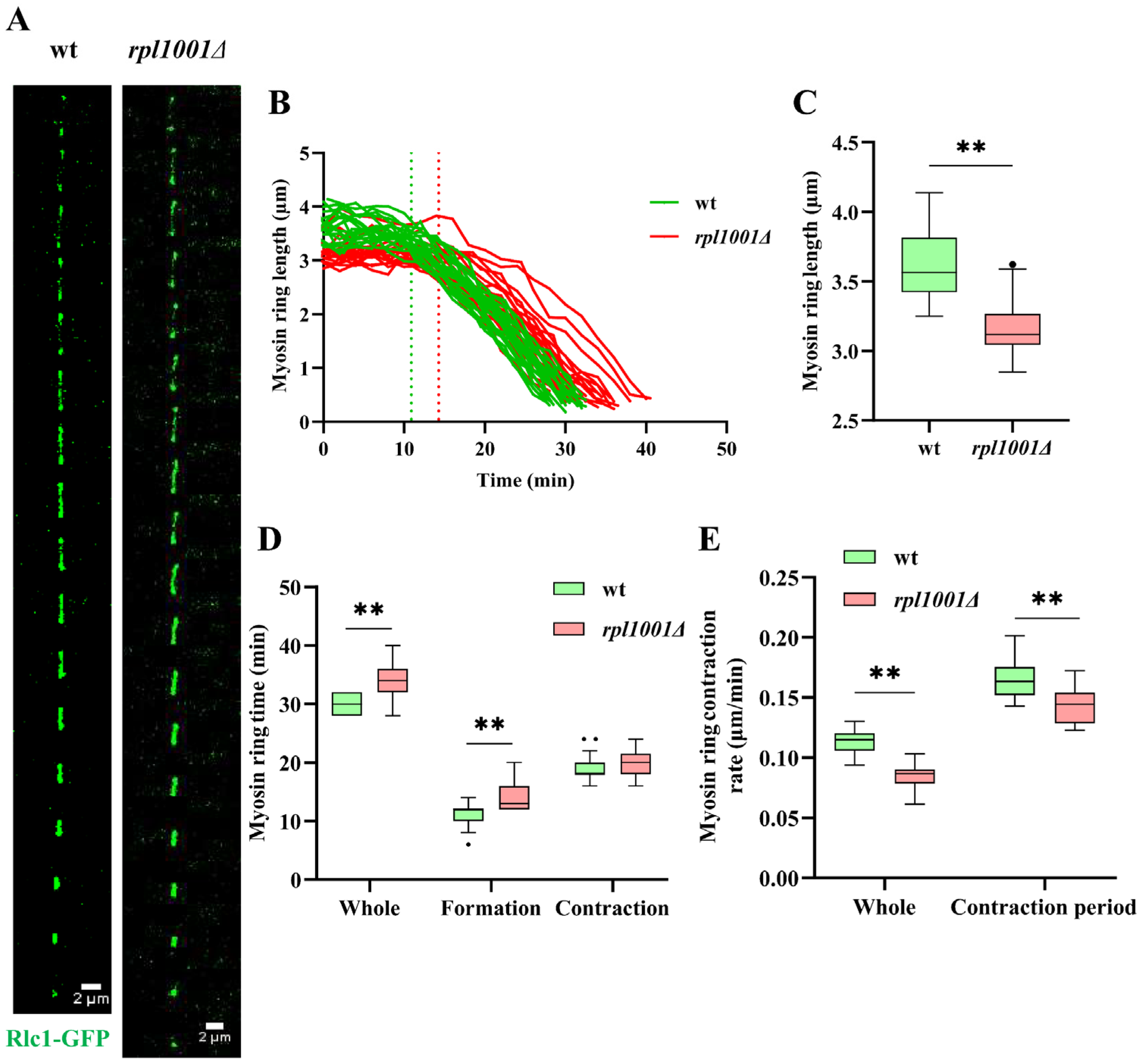
3.5. Changes of the Actin and Myosin Ring in the rpl1001Δ Strain during Mitosis
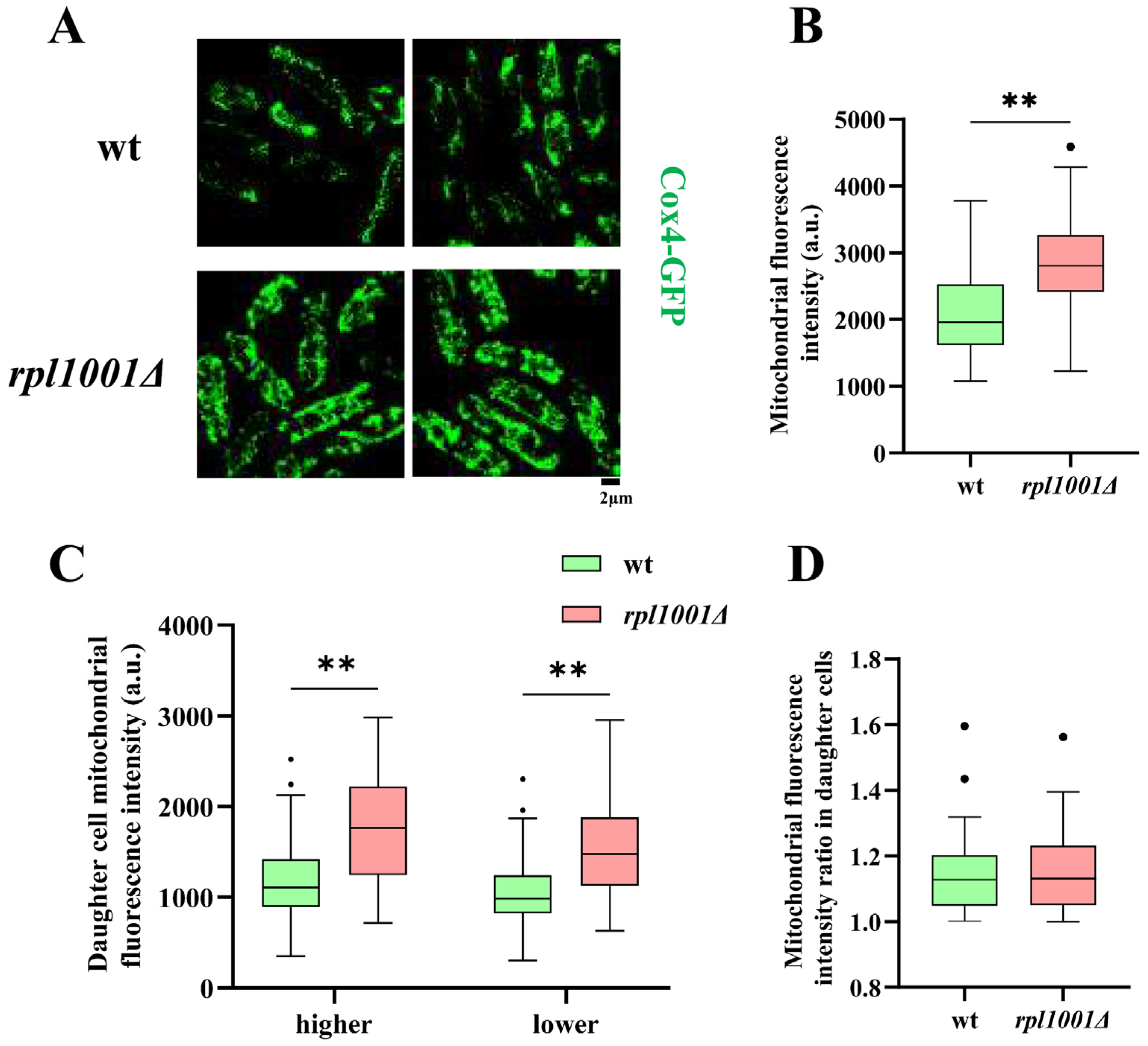
3.6. Mitochondrial Changes in the rpl1001Δ Strain
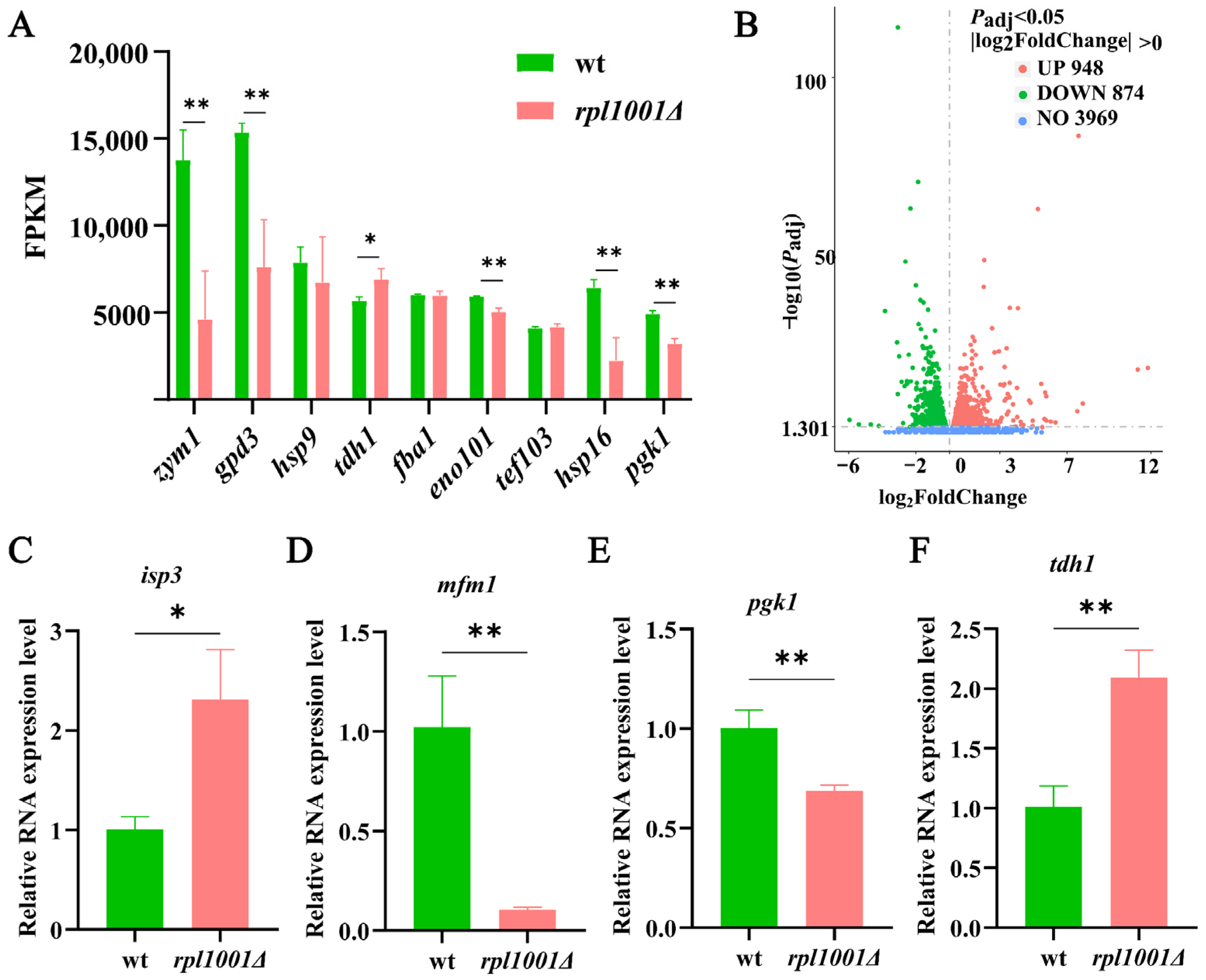
3.7. Analysis of Highly Expressed Genes and Differentially Expressed Genes
3.8. Differentially Expressed Gene Validation by qRT-PCR
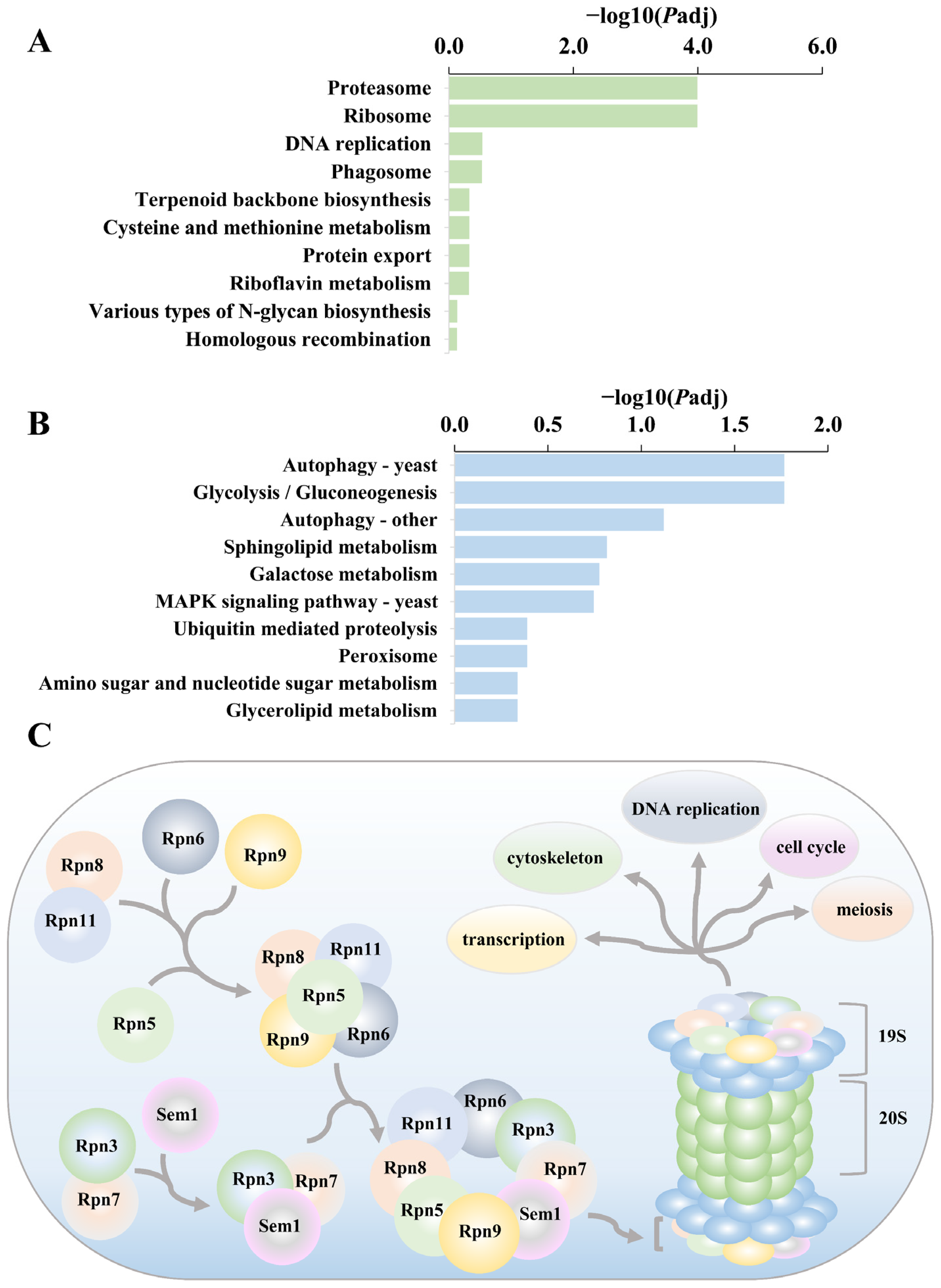
3.9. Analysis of Differential Genes for GO and KEGG Enrichment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, R.; Li, X.; Sun, L.; Chen, F.; Liu, Z.; Gu, Y.; Gong, X.; Liu, Z.; Wei, H.; Huang, Y.; et al. Reduction of ribosome level triggers flocculation of fission yeast cells. Eukaryot. Cell 2013, 12, 450–459. [Google Scholar] [CrossRef]
- Kim, S.J.; Strich, R. Rpl22 is required for IME1 mRNA translation and meiotic induction in S. cerevisiae. Cell Div. 2016, 11, 10. [Google Scholar] [CrossRef]
- Jang, C.Y.; Kim, H.D.; Zhang, X.; Chang, J.S.; Kim, J. Ribosomal protein S3 localizes on the mitotic spindle and functions as a microtubule associated protein in mitosis. Biochem. Biophys. Res. Commun. 2012, 429, 57–62. [Google Scholar] [CrossRef]
- Aronova, S.; Wedaman, K.; Anderson, S.; Yates, J., 3rd; Powers, T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 2779–2794. [Google Scholar] [CrossRef]
- Mok, K.W.; Mruk, D.D.; Silvestrini, B.; Cheng, C.Y. rpS6 Regulates blood-testis barrier dynamics by affecting F-actin organization and protein recruitment. Endocrinology 2012, 153, 5036–5048. [Google Scholar] [CrossRef]
- Spahn, C.M.; Beckmann, R.; Eswar, N.; Penczek, P.A.; Sali, A.; Blobel, G.; Frank, J. Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell 2001, 107, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Pollutri, D.; Penzo, M. Ribosomal Protein L10: From Function to Dysfunction. Cells 2020, 9, 2503. [Google Scholar] [CrossRef] [PubMed]
- Zuk, D.; Belk, J.P.; Jacobson, A. Temperature-sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover. Genetics 1999, 153, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Eisinger, D.P.; Dick, F.A.; Trumpower, B.L. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol. Cell. Biol. 1997, 17, 5136–5145. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-López, M.; Bordin, N.; Lees, J.; Scholes, H.; Hassan, S.; Saintain, Q.; Kamrad, S.; Orengo, C.; Bähler, J. Broad functional profiling of fission yeast proteins using phenomics and machine learning. eLife 2023, 12, RP88229. [Google Scholar] [CrossRef]
- Sajiki, K.; Tahara, Y.; Uehara, L.; Sasaki, T.; Pluskal, T.; Yanagida, M. Genetic regulation of mitotic competence in G(0) quiescent cells. Sci. Adv. 2018, 4, eaat5685. [Google Scholar] [CrossRef]
- Chan, Y.L.; Diaz, J.J.; Denoroy, L.; Madjar, J.J.; Wool, I.G. The primary structure of rat ribosomal protein L10: Relationship to a Jun-binding protein and to a putative Wilms’ tumor suppressor. Biochem. Biophys. Res. Commun. 1996, 225, 952–956. [Google Scholar] [CrossRef]
- Brooks, S.S.; Wall, A.L.; Golzio, C.; Reid, D.W.; Kondyles, A.; Willer, J.R.; Botti, C.; Nicchitta, C.V.; Katsanis, N.; Davis, E.E. A novel ribosomopathy caused by dysfunction of RPL10 disrupts neurodevelopment and causes X-linked microcephaly in humans. Genetics 2014, 198, 723–733. [Google Scholar] [CrossRef]
- Chiocchetti, A.; Pakalapati, G.; Duketis, E.; Wiemann, S.; Poustka, A.; Poustka, F.; Klauck, S.M. Mutation and expression analyses of the ribosomal protein gene RPL10 in an extended German sample of patients with autism spectrum disorder. Am. J. Med. Genet. Part A 2011, 155a, 1472–1475. [Google Scholar] [CrossRef]
- Rincon, S.A.; Paoletti, A. Molecular control of fission yeast cytokinesis. Semin. Cell Dev. Biol. 2016, 53, 28–38. [Google Scholar] [CrossRef]
- Hayles, J.; Nurse, P. Introduction to Fission Yeast as a Model System. Cold Spring Harb. Protoc. 2018, 2018, pdb-top079749. [Google Scholar] [CrossRef]
- Hales, K.G.; Fuller, M.T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 1997, 90, 121–129. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Z.; Zhu, R.; Wang, F.; Cheng, Y.; Liu, Y. Three Differential Expression Analysis Methods for RNA Sequencing: Limma, EdgeR, DESeq2. J. Vis. Exp. JoVE 2021, 18, e62528. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Morigasaki, S.; Shimada, K.; Ikner, A.; Yanagida, M.; Shiozaki, K. Glycolytic enzyme GAPDH promotes peroxide stress signaling through multistep phosphorelay to a MAPK cascade. Mol. Cell 2008, 30, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Blyth, J.; Makrantoni, V.; Barton, R.E.; Spanos, C.; Rappsilber, J.; Marston, A.L. Genes Important for Schizosaccharomyces pombe Meiosis Identified Through a Functional Genomics Screen. Genetics 2018, 208, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Dudin, O.; Merlini, L.; Bendezú, F.O.; Groux, R.; Vincenzetti, V.; Martin, S.G. A systematic screen for morphological abnormalities during fission yeast sexual reproduction identifies a mechanism of actin aster formation for cell fusion. PLoS Genet. 2017, 13, e1006721. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Won, M.; Choi, J.H.; Kyun, M.L.; Cho, H.S.; Park, H.M.; Kang, C.M.; Chung, K.S. Small heat-shock protein Hsp9 has dual functions in stress adaptation and stress-induced G2-M checkpoint regulation via Cdc25 inactivation in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 2012, 417, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Hayles, J.; Wood, V.; Jeffery, L.; Hoe, K.L.; Kim, D.U.; Park, H.O.; Salas-Pino, S.; Heichinger, C.; Nurse, P. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol. 2013, 3, 130053. [Google Scholar] [CrossRef] [PubMed]
- Vještica, A.; Merlini, L.; Nkosi, P.J.; Martin, S.G. Gamete fusion triggers bipartite transcription factor assembly to block re-fertilization. Nature 2018, 560, 397–400. [Google Scholar] [CrossRef]
- Duncan, C.D.S.; Rodríguez-López, M.; Ruis, P.; Bähler, J.; Mata, J. General amino acid control in fission yeast is regulated by a nonconserved transcription factor, with functions analogous to Gcn4/Atf4. Proc. Natl. Acad. Sci. USA 2018, 115, E1829–E1838. [Google Scholar] [CrossRef]
- Fukunishi, K.; Miyakubi, K.; Hatanaka, M.; Otsuru, N.; Hirata, A.; Shimoda, C.; Nakamura, T. The fission yeast spore is coated by a proteinaceous surface layer comprising mainly Isp3. Mol. Biol. Cell 2014, 25, 1549–1559. [Google Scholar] [CrossRef]
- Kjaerulff, S.; Davey, J.; Nielsen, O. Analysis of the structural genes encoding M-factor in the fission yeast Schizosaccharomyces pombe: Identification of a third gene, mfm3. Mol. Cell. Biol. 1994, 14, 3895–3905. [Google Scholar] [CrossRef]
- Murase, Y.; Yamagishi, M.; Okada, N.; Toya, M.; Yajima, J.; Hamada, T.; Sato, M. Fission yeast Dis1 is an unconventional TOG/XMAP215 that induces microtubule catastrophe to drive chromosome pulling. Commun. Biol. 2022, 5, 1298. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Dynamic Regulation of the 26S Proteasome: From Synthesis to Degradation. Front. Mol. Biosci. 2019, 6, 40. [Google Scholar] [CrossRef]
- Yen, H.C.; Espiritu, C.; Chang, E.C. Rpn5 is a conserved proteasome subunit and required for proper proteasome localization and assembly. J. Biol. Chem. 2003, 278, 30669–30676. [Google Scholar] [CrossRef]
- Lucena, R.; Dephoure, N.; Gygi, S.P.; Kellogg, D.R.; Tallada, V.A.; Daga, R.R.; Jimenez, J. Nucleocytoplasmic transport in the midzone membrane domain controls yeast mitotic spindle disassembly. J. Cell Biol. 2015, 209, 387–402. [Google Scholar] [CrossRef]
- Yang, H.J.; Asakawa, H.; Ohtsuki, C.; Haraguchi, T.; Hiraoka, Y. Transient Breakage of the Nucleocytoplasmic Barrier Controls Spore Maturation via Mobilizing the Proteasome Subunit Rpn11 in the Fission Yeast Schizosaccharomyces pombe. J. Fungi 2020, 6, 242. [Google Scholar] [CrossRef]
- Seeger, M.; Gordon, C.; Ferrell, K.; Dubiel, W. Characteristics of 26 S proteases from fission yeast mutants, which arrest in mitosis. J. Mol. Biol. 1996, 263, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Itadani, A.; Nakamura, T.; Hirata, A.; Shimoda, C. Schizosaccharomyces pombe calmodulin, Cam1, plays a crucial role in sporulation by recruiting and stabilizing the spindle pole body components responsible for assembly of the forespore membrane. Eukaryot. Cell 2010, 9, 1925–1935. [Google Scholar] [CrossRef] [PubMed]
- Aoi, Y.; Arai, K.; Miyamoto, M.; Katsuta, Y.; Yamashita, A.; Sato, M.; Yamamoto, M. Cuf2 boosts the transcription of APC/C activator Fzr1 to terminate the meiotic division cycle. EMBO Rep. 2013, 14, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Fujita, Y.; Yamamoto, M. Proper microtubule structure is vital for timely progression through meiosis in fission yeast. PLoS ONE 2013, 8, e65082. [Google Scholar] [CrossRef] [PubMed]
- Merlini, L.; Dudin, O.; Martin, S.G. Mate and fuse: How yeast cells do it. Open Biol. 2013, 3, 130008. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.P.; Guo, Y.S.; Hu, Y.; Liu, W.X.; Wang, Q.; Wang, Y.T.; Yu, H.Y.; Tang, C.M.; Yang, J.; Zhou, T.; et al. Distinct temporal requirements for autophagy and the proteasome in yeast meiosis. Autophagy 2016, 12, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Song, M.; Wang, Y.; Tan, T.; Tian, Z.; Zhai, B.; Yang, X.; Tan, Y.; Cao, Y.; Dai, S.; et al. The ubiquitin-proteasome system regulates meiotic chromosome organization. Proc. Natl. Acad. Sci. USA 2022, 119, e2106902119. [Google Scholar] [CrossRef] [PubMed]
- Tanenbaum, M.E.; Medema, R.H. Mechanisms of centrosome separation and bipolar spindle assembly. Dev. Cell 2010, 19, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Walczak, C.E.; Heald, R. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 2008, 265, 111–158. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Li, T.; Jin, D.Y.; Syrovatkina, V.; Scheffler, K.; Tran, P.T.; Fu, C. Csi1p recruits alp7p/TACC to the spindle pole bodies for bipolar spindle formation. Mol. Biol. Cell 2014, 25, 2750–2760. [Google Scholar] [CrossRef] [PubMed]
- Bridge, A.J.; Morphew, M.; Bartlett, R.; Hagan, I.M. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 1998, 12, 927–942. [Google Scholar] [CrossRef]
- Aoki, K.; Nakaseko, Y.; Kinoshita, K.; Goshima, G.; Yanagida, M. CDC2 phosphorylation of the fission yeast dis1 ensures accurate chromosome segregation. Curr. Biol. 2006, 16, 1627–1635. [Google Scholar] [CrossRef]
- Garcia, M.A.; Vardy, L.; Koonrugsa, N.; Toda, T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001, 20, 3389–3401. [Google Scholar] [CrossRef]
- Garcia, M.A.; Koonrugsa, N.; Toda, T. Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 2002, 21, 6015–6024. [Google Scholar] [CrossRef]
- East, D.A.; Mulvihill, D.P. Regulation and function of the fission yeast myosins. J. Cell Sci. 2011, 124, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Walther, A. ATP-Responsive and ATP-Fueled Self-Assembling Systems and Materials. Adv. Mater. 2020, 32, e2002629. [Google Scholar] [CrossRef] [PubMed]
- Sladewski, T.E.; Previs, M.J.; Lord, M. Regulation of fission yeast myosin-II function and contractile ring dynamics by regulatory light-chain and heavy-chain phosphorylation. Mol. Biol. Cell 2009, 20, 3941–3952. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Balasubramanian, M.K. Pxl1p, a paxillin-related protein, stabilizes the actomyosin ring during cytokinesis in fission yeast. Mol. Biol. Cell 2008, 19, 1680–1692. [Google Scholar] [CrossRef]
- Laporte, D.; Ojkic, N.; Vavylonis, D.; Wu, J.Q. α-Actinin and fimbrin cooperate with myosin II to organize actomyosin bundles during contractile-ring assembly. Mol. Biol. Cell 2012, 23, 3094–3110. [Google Scholar] [CrossRef]
- Pivovarova, A.V.; Mikhailova, V.V.; Chernik, I.S.; Chebotareva, N.A.; Levitsky, D.I.; Gusev, N.B. Effects of small heat shock proteins on the thermal denaturation and aggregation of F-actin. Biochem. Biophys. Res. Commun. 2005, 331, 1548–1553. [Google Scholar] [CrossRef]
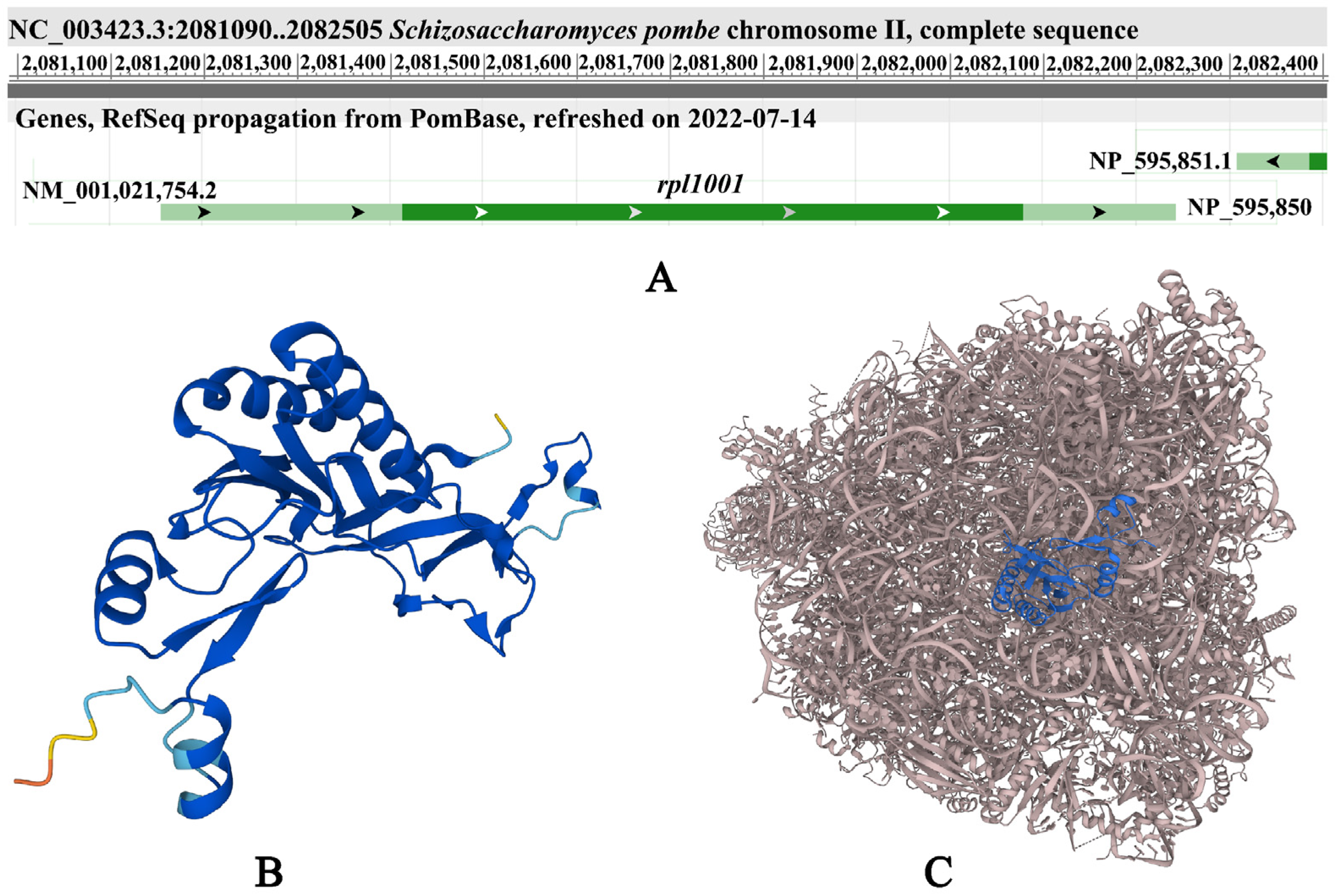


| Strains | Genotypes | Source |
|---|---|---|
| PT286 | h- wt ade6-210 leu1-32 ura4-D18 | Lab. Trans 1 |
| PT287 | h+ wt ade6-210 leu1-32 ura4-D18 | Lab. Trans |
| PT4436 | h- Hht1-RFP-KanR GFP-Atb2-KanR ade6-210 leu1-32 ura4-D18 | Lab. Trans |
| PT3850 | h+ Pact1-LifeAct-mGFP::leu1 | Lab. Trans |
| PT2006 | h- Rlc1-GFP::ura4 leu1-32 | Lab. Trans |
| YL18 | h- mCherry-Atb2-HygR Cox4-GFP::leu2 ade6-210 leu1-32 ura4-D18 | This study |
| 2599-1 | h+ rpl1001Δ::kanR | This study |
| 2599-2 | h- rpl1001Δ::kanR | This study |
| 2599-3 | h? rpl1001Δ::kanR Hht1-RFP-KanR GFP-Atb2-KanR ade6-210 leu1-32 ura4-D18 | This study |
| 2599-4 | h? rpl1001Δ:: kanR Pact1-LifeAct-mGFP::leu1 | This study |
| 2599-5 | h? rpl1001Δ:: kanR Rlc1-GFP::ura4 | This study |
| 2599-6 | h? rpl1001Δ:: kanR mCherry-Atb2-HygR Cox4-GFP::leu2 ade6-210 leu1-32 ura4-D18 | This study |
| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| isp3 | TTT GGT GAT GGT GAC TGC GA | GAG TGG ATC AGG GCA GGT TC |
| mfm1 | TG GAC TCA ATG GCT AAC TCC G | A ACG TAG GCA AGA AAA GTG GC |
| pgk1 | C TTG AAG CCT GTT GCT GCT G | TC CTC GAT GTG GAA ACG CAA |
| tdh1 | TC GTC AAG CTC GTC TCT TGG | G CAT TGC CTT TAA GCA CCC A |
| act1 | CCC AAA TCC AAC CGT GAG AAG | CC AGA GTC CAA GAC GAT ACC AGT G |
| Gene | FPKM (wt) | FPKM (rpl1001Δ) | Description |
|---|---|---|---|
| zym1 | 13,760.9 | 4601.6 | Metallothionein, Zym1 |
| gpd3 | 15,353.5 | 6102.4 | Glyceraldehyde 3-phosphate dehydrogenase, Gpd3 |
| hsp9 | 7879.5 | 7169.7 | Heat shock protein, Hsp9 |
| fba1 | 6012.5 | 6445.7 | Fructose-bisphosphate aldolase, Fba1 |
| tdh1 | 5679.2 | 6817.2 | Glyceraldehyde-3-phosphate dehydrogenase, Tdh1 |
| eno101 | 5927.2 | 5517.0 | Enolase |
| tef103 | 4107.5 | 4608.1 | Translation elongation factor EF-1 alpha Ef1a-c |
| hsp16 | 6429.3 | 3209.0 | Heat shock protein, Hsp16 |
| pgk1 | 4919.3 | 2722.2 | Phosphoglycerate kinase, Pgk1 |
| Gene | log2 Fold Change | Padj | Description |
|---|---|---|---|
| mat2-Pc | 11.8 | 1.3 × 10−18 | Silenced P-specific polypeptide Pc |
| Tf2-13 | 7.9 | 1.4 × 10−8 | Retrotransposable element/transposon Tf2-type |
| mat2-Pi | 7.6 | 2.2 × 10−6 | Silenced P-specific polypeptide Pi |
| aim27 | 5.8 | 7.4 × 10−4 | ER membrane protein complex subunit Aim27 |
| pdc102 | 5.8 | 1.4 × 10−10 | Pyruvate decarboxylase |
| str3 | 5.7 | 1.2 × 10−11 | Siderophore-iron transporter Str3 |
| ftm4 | 5.3 | 1.6 × 10−63 | S. pombe specific 5Tm protein family |
| shu1 | 4.7 | 2.0 × 10−9 | Heme import protein Shu1 |
| isp3 | 4.0 | 5.4 × 10−13 | Schizosaccharomyces pombe specific protein Isp3 |
| fmp40 | 3.8 | 4.3 × 10−2 | UPF0061 family mitochondrial protein |
| Gene | log2 Fold Change | Padj | Description |
|---|---|---|---|
| mat3-Mc | −6.0 | 6.8 × 10−5 | Mating-type m-specific HMG-box transcription factor Mc at silenced MAT3 locus |
| mfm1 | −3.1 | 3.3 × 10−28 | M-factor precursor Mfm1 |
| mfm2 | −2.5 | 9.2 × 10−17 | M-factor precursor Mfm2 |
| mam1 | −2.4 | 1.5 × 10−24 | M-factor transmembrane transporter Mam1 |
| mel1 | −2.4 | 4.7 × 10−9 | Alpha-galactosidase, melibiase |
| pcm2 | −2.3 | 7.8 × 10−67 | Protein-L-isoaspartate O-methyltransferase Pcm2 |
| ftm7 | −2.2 | 2.7 × 10−3 | S. pombe specific 5Tm protein family |
| hry1 | −2.1 | 4.6 × 10−6 | HHE domain cation binding protein |
| arg4 | −2.1 | 1.4 × 10−3 | Arginine specific carbamoyl-phosphate synthase Arg4 |
| dis1 | −2.0 | 8.2 × 10−45 | TOG/XMAP14 microtubule-associated protein Dis1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, W.; Yuan, R.; Liu, M.; Liu, K.; Ding, X.; Hou, Y. Effects of rpl1001 Gene Deletion on Cell Division of Fission Yeast and Its Molecular Mechanism. Curr. Issues Mol. Biol. 2024, 46, 2576-2597. https://doi.org/10.3390/cimb46030164
Yu W, Yuan R, Liu M, Liu K, Ding X, Hou Y. Effects of rpl1001 Gene Deletion on Cell Division of Fission Yeast and Its Molecular Mechanism. Current Issues in Molecular Biology. 2024; 46(3):2576-2597. https://doi.org/10.3390/cimb46030164
Chicago/Turabian StyleYu, Wen, Rongmei Yuan, Mengnan Liu, Ke Liu, Xiang Ding, and Yiling Hou. 2024. "Effects of rpl1001 Gene Deletion on Cell Division of Fission Yeast and Its Molecular Mechanism" Current Issues in Molecular Biology 46, no. 3: 2576-2597. https://doi.org/10.3390/cimb46030164
APA StyleYu, W., Yuan, R., Liu, M., Liu, K., Ding, X., & Hou, Y. (2024). Effects of rpl1001 Gene Deletion on Cell Division of Fission Yeast and Its Molecular Mechanism. Current Issues in Molecular Biology, 46(3), 2576-2597. https://doi.org/10.3390/cimb46030164




