The Potential MicroRNA Diagnostic Biomarkers in Oral Squamous Cell Carcinoma of the Tongue
Abstract
:1. Introduction
2. Materials and Methods
2.1. Acquisition of TCGA Data
2.2. Selection of Candidate Diagnostic miRNAs
2.3. Prediction of microRNA Targets
2.4. Functional Enrichment Analysis
2.5. Protein–Protein Interaction Network
2.6. Statistical Analyses
3. Results
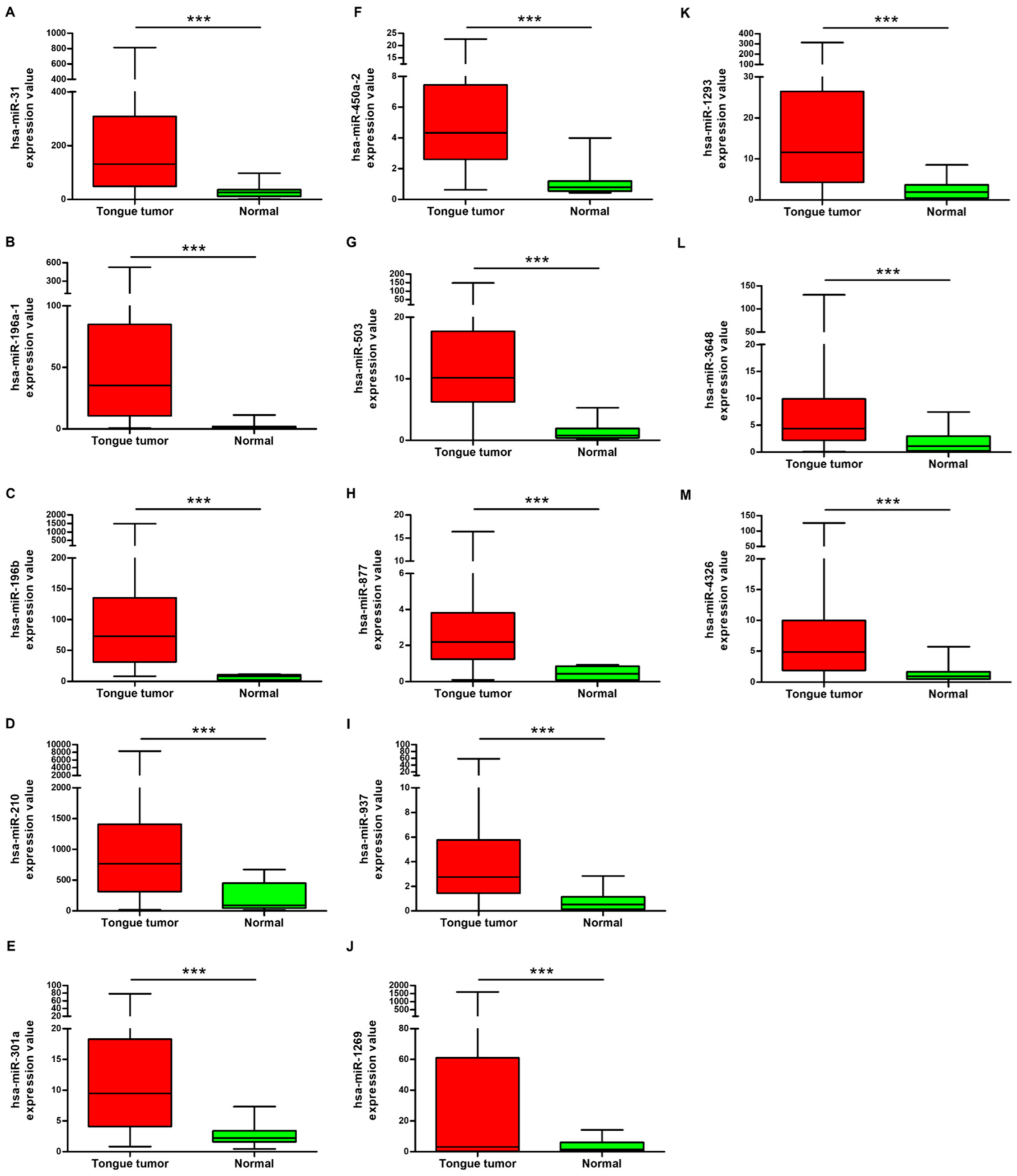
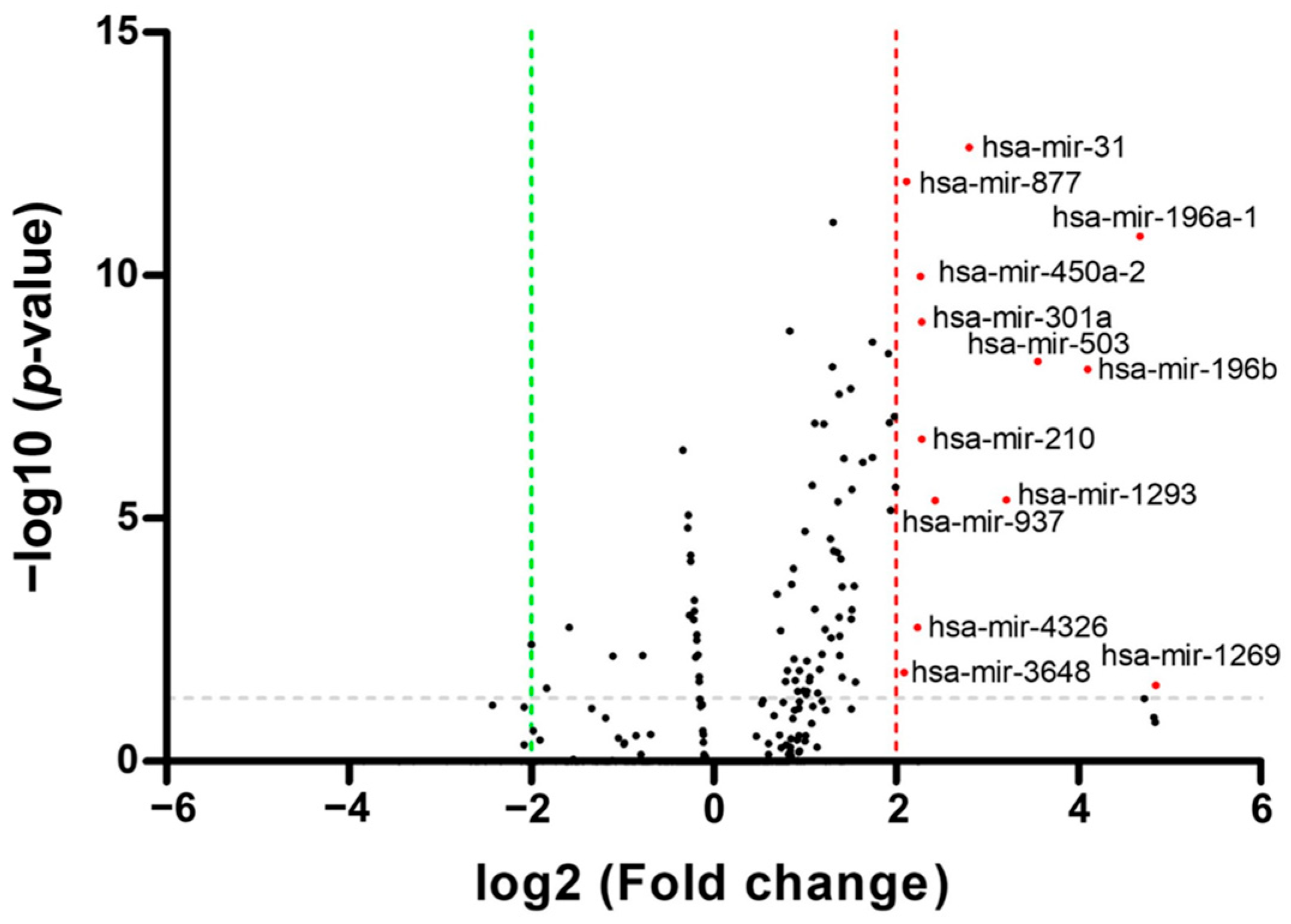
3.1. Identification of Differentially Expressed miRNAs in OSCC Patients from TCGA Database
3.2. Evaluation of the Diagnostic Values of the Five Potential miRNAs
3.3. Gene Ontology and KEGG Pathway Enrichment Analysis
3.4. Identification of the Hub Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liang, L.; Li, Y.; Ying, B.; Huang, X.; Liao, S.; Yang, J.; Liao, G. Mutation-associated transcripts reconstruct the prognostic features of oral tongue squamous cell carcinoma. Int. J. Oral Sci. 2023, 15, 1. [Google Scholar] [CrossRef]
- Wang, S.; Li, K.; Zhao, T.; Sun, Y.; Zeng, T.; Wu, Y.; Ding, L.; Huang, X.; Celentano, A.; Yang, X.; et al. Oral tongue squamous cell carcinoma diagnosis from tissue metabolic profiling. Oral Dis. 2023, 30, 2158–2165. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Xiao, Z.; Ren, J.; Xia, R.H.; Wu, Y.; Yuan, Y.; Tao, X. Machine learning based on magnetic resonance imaging and clinical parameters helps predict mesenchymal-epithelial transition factor expression in oral tongue squamous cell carcinoma: A pilot study. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2024, 137, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhang, L.; Guo, L.; Yang, S. PKM2 regulates proliferation and apoptosis through the Hippo pathway in oral tongue squamous cell carcinoma. Oncol Lett. 2021, 21, 461. [Google Scholar] [CrossRef]
- Lin, Y.W.; Kang, W.P.; Hong, C.Q.; Huang, B.L.; Qiu, Z.H.; Liu, C.T.; Chu, L.Y.; Xu, Y.W.; Guo, F.C. Nutritional and immune-related indicators-based Nomogram for predicting overall survival of surgical oral tongue squamous cell carcinoma. Sci. Rep. 2023, 13, 8525. [Google Scholar] [CrossRef]
- Tang, W.; Wang, Y.; Yuan, Y.; Tao, X. Assessment of tumor depth in oral tongue squamous cell carcinoma with multiparametric MRI: Correlation with pathology. Eur. Radiol. 2022, 32, 254–261. [Google Scholar] [CrossRef]
- Almangush, A.; Heikkinen, I.A.; Mäkitie, A.; Coletta, R.D.; Läärä, E.; Leivo, I.; Salo, T. Prognostic biomarkers for oral tongue squamous cell carcinoma: A systematic review and meta-analysis. Br. J. Cancer 2017, 117, 856–866. [Google Scholar] [CrossRef]
- Crosby, D.; Bhatia, S.; Brindle, K.M.; Coussens, L.M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald, R.C.; Gambhir, S.S.; Kuhn, P.; et al. Early Detection of Cancer. Science 2022, 375, eaay9040. [Google Scholar] [CrossRef] [PubMed]
- Myers, D.; Allen, E.; Essa, A.; Gbadamosi-Akindele, M. Rapidly Growing Squamous Cell Carcinoma of the Tongue. Cureus 2020, 12, e7164. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Naeli, P.; Winter, T.; Hackett, A.P.; Alboushi, L.; Jafarnejad, S.M. The intricate balance between microRNA-induced mRNA decay and translational repression. FEBS J. 2023, 290, 2508–2524. [Google Scholar] [CrossRef] [PubMed]
- Dillhoff, M.; Wojcik, S.E.; Bloomston, M. MicroRNAs in Solid Tumors. J. Surg. Res. 2009, 154, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Croce, C.M.; Bloomston, M. MicroRNA: Trends in clinical trials of cancer diagnosis and therapy strategies. Exp. Mol. Med. 2023, 55, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Boichard, A.; Wagner, M.J.; Kurzrock, R. Angiosarcoma heterogeneity and potential therapeutic vulnerability to immune checkpoint blockade: Insights from genomic sequencing. Genome Med. 2020, 12, 61. [Google Scholar] [CrossRef]
- Chen, G.; Luo, D.; Zhong, N.; Li, D.; Zheng, J.; Liao, H.; Li, Z.; Lin, X.; Chen, Q.; Zhang, C.; et al. GPC2 Is a Potential Diagnostic, Immunological, and Prognostic Biomarker in Pan-Cancer. Front. Immunol. 2022, 13, 857308. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Azmi, A.S.; Moore, A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Genome Med. 2021, 12, e1662. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.X. An Integrative Analysis of microRNA and mRNA Expression—A Case Study. Cancer Inform. 2008, 6, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zheng, C.; Shen, H.; Zhou, Z.; Lei, Y. MicroRNAs-mRNAs Expression Profile and Their Potential Role in Malignant Transformation of Human Bronchial Epithelial Cells Induced by Cadmium. Biomed. Res. Int. 2015, 2015, 902025. [Google Scholar] [CrossRef]
- Zou, K.H.; O’Malley, A.J.; Mauri, L. Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation 2007, 115, 654–657. [Google Scholar] [CrossRef]
- Walter, S. The partial area under the summary ROC curve. Stat. Med. 2005, 24, 2025–2040. [Google Scholar] [CrossRef] [PubMed]
- Mandrekar, J.N. Receiver Operating Characteristic Curve in Diagnostic Test Assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed]
- Çorbacıoğlu, S.K.; Aksel, G. Receiver operating characteristic curve analysis in diagnostic accuracy studies: A guide to interpreting the area under the curve value. Turk. J. Emerg. Med. 2023, 23, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Unal, I. Defining an Optimal Cut-Point Value in ROC Analysis: An Alternative Approach. Comput. Math Methods Med. 2017, 2017, 3762651. [Google Scholar] [CrossRef]
- Jorge, A.L.; Pereira, E.R.; Oliveira, C.S.; Ferreira, E.D.S.; Menon, E.T.N.; Diniz, S.N.; Pezuk, J.A. MicroRNAs: Understanding their role in gene expression and cancer. Einstein 2021, 19, eRB5996. [Google Scholar] [CrossRef]
- Liu, X.J.; Yin, H.L.; Li, Y.; Hao, H.; Liu, Y.; Zhao, Q.L. The Construction and Analysis of a ceRNA Network Related to Salt-Sensitivity Hypertensives. Biomed. Res. Int. 2022, 2022, 8258351. [Google Scholar] [CrossRef] [PubMed]
- Althubaiti, S.; Karwath, A.; Dallol, A.; Noor, A.; Alkhayyat, S.S.; Alwassia, R.; Mineta, K.; Gojobori, T.; Beggs, A.D.; Schofield, P.N.; et al. Ontology-based prediction of cancer driver genes. Sci. Rep. 2019, 9, 17405. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acid Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ruffalo, M.; Bar-Joseph, Z. Protein interaction disruption in cancer. BMC Cancer 2019, 19, 370. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Kalia, M. Feature selection with the Fisher score followed by the Maximal Clique Centrality algorithm can accurately identify the hub genes of hepatocellular carcinoma. Sci. Rep. 2019, 9, 17283. [Google Scholar]
- Ding, W.; Xin, J.; Jiang, L.; Zhou, Q.; Wu, T.; Shi, D.; Lin, B.; Li, L.; Li, J. Characterisation of peripheral blood mononuclear cell microRNA in hepatitis B-related acute-on-chronic liver failure. Sci. Rep. 2015, 5, 13098. [Google Scholar] [CrossRef] [PubMed]
- Kalia, M. Biomarkers for personalized oncology: Recent advances and future challenges. Metabolism 2015, 64, S16–S21. [Google Scholar] [CrossRef] [PubMed]
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Hartl, D.; de Luca, V.D.; Kostikova, A.; Laramin, J.; Kennedy, S.; Ferrero, E.; Siegel, R.; Fink, M.; Ahmed, S.; Millholland, J.; et al. Translational precision medicine: An industry perspective. J. Transl. Med. 2021, 19, 245. [Google Scholar] [CrossRef] [PubMed]
- Lussier, Y.A.; Stadler, W.M.; Chen, J.L. Advantages of genomic complexity: Bioinformatics opportunities in microrna cancer signatures. J. Am. Med. Inform. Assoc. 2012, 19, 156–160. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Soga, D.; Yoshiba, S.; Shiogama, S.; Miyazaki, H.; Kondo, S.; Shintani, S. microRNA expression profiles in oral squamous cell carcinoma. Oncol. Rep. 2013, 30, 579–583. [Google Scholar] [CrossRef]
- Gombos, K.; Horváth, R.; Szele, E.; Juhász, K.; Gocze, K.; Somlai, K.; Pajkos, G.; Ember, I.; Olasz, L. miRNA expression profiles of oral squamous cell carcinomas. Anticancer Res. 2013, 33, 1511–1517. [Google Scholar] [PubMed]
- Tarrad, N.A.F.; Hassan, S.; Shaker, O.G.; AbdelKawy, M. “Salivary LINC00657 and miRNA-106a as diagnostic biomarkers for oral squamous cell carcinoma, an observational diagnostic study”. BMC Oral Health 2023, 23, 994. [Google Scholar] [CrossRef] [PubMed]
- Vimal, J.; George, N.A.; Kumar, R.R.; Kattoor, J.; Kannan, S. Identification of salivary metabolic biomarker signatures for oral tongue squamous cell carcinoma. Arch. Oral Biol. 2023, 155, 105780. [Google Scholar] [CrossRef]
- Mehrjoei, B.; Haghnazari, L.; Bashiri, H.; Rezvani, N. The diagnostic potential of miR-196a-1 in colorectal cancer. BMC Cancer 2024, 24, 162. [Google Scholar] [CrossRef] [PubMed]
- Raei, N.; Safaralizadeh, R.; Hesseinpourfeizi, M.; Yazdanbod, A.; Pourfarzi, F.; Latifi-Navid, S. Crosstalk between lncRNAs and miRNAs in gastrointestinal cancer drug resistance. Life Sci. 2021, 284, 119933. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Cheng, J.; Li, A. hsa_circ_0072387 Suppresses Proliferation, Metastasis, and Glycolysis of Oral Squamous Cell Carcinoma Cells by Downregulating miR-503-5p. Cancer Biother. Radiopharm. 2021, 36, 84–94. [Google Scholar] [CrossRef]
- Shao, Y.; Song, Y.; Xu, S.; Li, S.; Zhou, H. Expression Profile of Circular RNAs in Oral Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 533616. [Google Scholar] [CrossRef] [PubMed]
- Rajan, C.; Roshan, V.G.D.; Khan, I.; Manasa, V.G.; Himal, I.; Kattoor, J.; Thomas, S.; Kondaiah, P.; Kannan, S. MiRNA expression profiling and emergence of new prognostic signature for oral squamous cell carcinoma. Sci. Rep. 2021, 11, 7298. [Google Scholar] [CrossRef]
- Álvarez-Teijeiro, S.; Menéndez, S.T.; Villaronga, M.; Rodrigo, J.P.; Manterola, L.; de Villalaín, L.; de Vicente, J.C.; Alonso-Durán, L.; Fernández, M.P.; Lawrie, C.H.; et al. Dysregulation of Mir-196b in Head and Neck Cancers Leads to Pleiotropic Effects in the Tumor Cells and Surrounding Stromal Fibroblasts. Sci. Rep. 2017, 7, 17785. [Google Scholar] [CrossRef]
- Lu, Y.C.; Chang, J.T.C.; Huang, Y.C.; Huang, C.C.; Chen, W.H.; Lee, L.Y.; Huang, B.S.; Chen, Y.J.; Li, H.F.; Cheng, A.J. Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin. Biochem. 2015, 48, 115–121. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Janal, M.N.; Veeramachaneni, R.; Dolgalev, I.; Dubeykovskaya, Z.; Tu, N.H.; Kim, H.; Zhang, S.; Wu, A.K.; Hagiwara, M.; et al. Oncogenes overexpressed in metastatic oral cancers from patients with pain: Potential pain mediators released in exosomes. Sci. Rep. 2020, 10, 14724. [Google Scholar] [CrossRef] [PubMed]
- Rub, U.; Brunt, E.R.; Petrasch-Parwez, E.; Schols, L.; Theegarten, D.; Auburger, G.; Seidel, K.; Schultz, C.; Gierga, K.; Paulson, H.; et al. Degeneration of ingestion-related brainstem nuclei in spinocerebellar ataxia type 2, 3, 6 and 7. Neuropathol. Appl. Neurobiol. 2006, 32, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.R.; Choi, K.H.; Kim, T.G. Dysphagia in Tongue Cancer Patients. Ann. Rehabil. Med. 2015, 39, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Chen, J.; Shi, Y.; Fang, X.; Tang, Z. MAPK Signaling Pathway in Oral Squamous Cell Carcinoma: Biological Function and Targeted Therapy. Cancers 2022, 14, 4625. [Google Scholar] [CrossRef] [PubMed]
- Bongiorno, R.; Colombo, M.P.; Lecis, D. Deciphering the nonsense-mediated mRNA decay pathway to identify cancer cell vulnerabilities for effective cancer therapy. J. Exp. Clin. Cancer Res. 2021, 40, 376. [Google Scholar] [CrossRef] [PubMed]
- Youle, R.J.; Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011, 12, 9–14. [Google Scholar] [CrossRef]
- Bai, J.; Wu, L.; Wang, X.; Wang, Y.; Shang, Z.; Jiang, E.; Shao, Z. Roles of mitochondria in oral squamous cell carcinoma therapy: Friend or foe? Cancers 2022, 14, 5723. [Google Scholar] [CrossRef]
- Rodríguez-Molinero, J.; Migueláñez-Medrán, B.D.C.; Puente-Gutiérrez, C.; Delgado-Somolinos, E.; Martín Carreras-Presas, C.; Fernández-Farhall, J.; López-Sánchez, A.F. Association between Oral Cancer and Diet: An Update. Nutrients 2021, 13, 1299. [Google Scholar] [CrossRef]
- Padam, K.S.R.; Morgan, R.; Hunter, K.; Chakrabarty, S.; Kumar, N.A.N.; Radhakrishnan, R. Identification of HOX signatures contributing to oral cancer phenotype. Sci. Rep. 2022, 12, 10123. [Google Scholar] [CrossRef]
- Rodini, C.O.; Xavier, F.C.; Paiva, K.B.; De Souza Setúbal Destro, M.F.; Moyses, R.A.; Michaluarte, P.; Carvalho, M.B.; Fukuyama, E.E.; Head and Neck Genome Project Gencapo; Tajara, E.H.; et al. Head and Neck Genome Project Gencapo. Int. J. Oncol. 2012, 40, 1180–1188. [Google Scholar] [CrossRef]
- Duan, X.; Chen, H.; Ma, H.; Song, Y. The expression and significance of the HOXA7 gene in oral squamous cell carcinoma. J. Oral Sci. 2017, 59, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Xavier, F.C.; Destro, M.F.; Duarte, C.M.; Nunes, F.D. Epigenetic repression of HOXB cluster in oral cancer cell lines. Arch. Oral Biol. 2014, 59, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Marcinkiewicz, K.M.; Gudas, L.J. Altered epigenetic regulation of homeobox genes in human oral squamous cell carcinoma cells. Exp. Cell Res. 2014, 320, 128–143. [Google Scholar] [CrossRef] [PubMed]
- Platais, C.; Radhakrishnan, R.; Ebensberger, S.N.; Morgan, R.; Lambert, D.W.; Hunter, K.D. Targeting HOX-PBX interactions causes death in oral potentially malignant and squamous carcinoma cells but not normal oral keratinocytes. BMC Cancer 2018, 18, 723. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, P.; Wang, X.; Duan, Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci. Rep. 2014, 4, 6802. [Google Scholar] [CrossRef]
- Hu, Y.; Dingerdissen, H.; Gupta, S.; Kahsay, R.; Shanker, V.; Wan, Q.; Yan, C.; Mazumder, R. Identification of key differentially expressed MicroRNAs in cancer patients through pan-cancer analysis. Comput. Biol. Med. 2018, 103, 183–197. [Google Scholar] [CrossRef]

| Characteristic | Overall |
|---|---|
| Gender, n (%) | |
| Male | 82 (64.1) |
| Female | 46 (35.9) |
| Age (years) | |
| Mean ± SD | 58.17 ± 13.27 |
| NA | 1 |
| Pathologic stage, n (%) | |
| Stage I | 15 (11.72) |
| Stage II | 22 (17.19) |
| Stage III | 30 (23.44) |
| Stage IV | 51 (39.84) |
| Not Available | 10 (7.81) |
| Pathological, T, n (%) | |
| T1 | 22 (17.19) |
| T2 | 45 (35.16) |
| T3 | 34 (26.56) |
| T4 | 19 (14.84) |
| TX | 6 (4.69) |
| Not Available | 2 (1.56) |
| Pathological, N, n (%) | |
| N0 | 53 (41.41) |
| N1 | 18 (14.06) |
| N2 | 44 (34.38) |
| NX | 11 (8.59) |
| Not Available | 2 (1.56) |
| Pathological, M, n (%) | |
| M0 | 43 (33.59) |
| MX | 13 (10.16) |
| Not Available | 72 (56.25) |
| miRNAs | AUC | 95% CI | Cutoff | Sensitivity (%) | Specificity (%) | p-Value |
|---|---|---|---|---|---|---|
| miR-196a-1 | 0.9447 | 0.8991–0.9903 | <2.319 | 84.62 | 92.97 | <0.0001 |
| miR-196b | 0.9838 | 0.9661–1.001 | <11.47 | 92.31 | 95.31 | <0.0001 |
| miR-450a-2 | 0.9303 | 0.8588–1.002 | <1.284 | 84.62 | 93.75 | <0.0001 |
| miR-503 | 0.9549 | 0.9153–0.9946 | <2.26 | 84.62 | 93.75 | <0.0001 |
| miR-877 | 0.9285 | 0.8791–0.9779 | <0.8552 | 84.62 | 83.59 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, Y.-N.; Ryu, J.-K.; Ju, Y. The Potential MicroRNA Diagnostic Biomarkers in Oral Squamous Cell Carcinoma of the Tongue. Curr. Issues Mol. Biol. 2024, 46, 6746-6756. https://doi.org/10.3390/cimb46070402
Park Y-N, Ryu J-K, Ju Y. The Potential MicroRNA Diagnostic Biomarkers in Oral Squamous Cell Carcinoma of the Tongue. Current Issues in Molecular Biology. 2024; 46(7):6746-6756. https://doi.org/10.3390/cimb46070402
Chicago/Turabian StylePark, Young-Nam, Jae-Ki Ryu, and Yeongdon Ju. 2024. "The Potential MicroRNA Diagnostic Biomarkers in Oral Squamous Cell Carcinoma of the Tongue" Current Issues in Molecular Biology 46, no. 7: 6746-6756. https://doi.org/10.3390/cimb46070402
APA StylePark, Y.-N., Ryu, J.-K., & Ju, Y. (2024). The Potential MicroRNA Diagnostic Biomarkers in Oral Squamous Cell Carcinoma of the Tongue. Current Issues in Molecular Biology, 46(7), 6746-6756. https://doi.org/10.3390/cimb46070402







