MicroRNAs Associated with IgLON Cell Adhesion Molecule Expression
Abstract
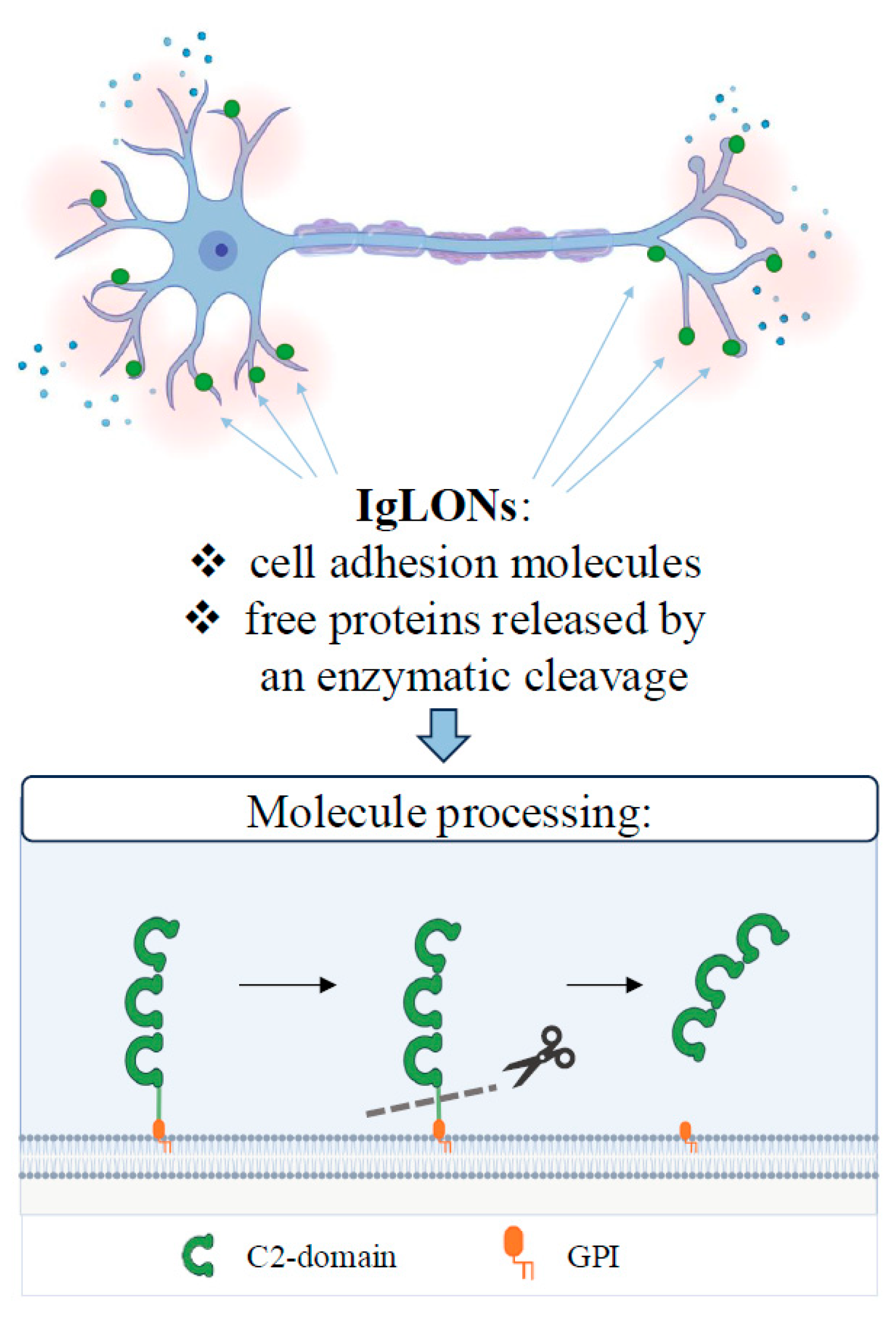
:1. Introduction
2. MicroRNA Overview: Biogenesis, Regulatory Properties, and Nomenclature Tips
3. LSAMP
4. OPCML
5. NTM
6. NEGR1
7. IgLON5
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McNamee, C.J.; Youssef, S.; Moss, D. IgLONs form heterodimeric complexes on forebrain neurons. Cell Biochem. Funct. 2011, 29, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Levitt, P. A monoclonal antibody to limbic system neurons. Science 1984, 223, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Zacco, A.; Cooper, V.; Chantler, P.; Fisher-Hyland, S.; Horton, H.; Levitt, P. Isolation, biochemical characterization and ultrastructural analysis of the limbic system-associated membrane protein (LAMP), a protein expressed by neurons comprising functional neural circuits. J. Neurosci. 1990, 10, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, A.F.; Zhukareva, V.; Barbe, M.F.; Reinoso, B.S.; Grimley, C.; Henzel, W.; Fischer, I.; Levitt, P. The limbic system-associated membrane protein is an Ig superfamily member that mediates selective neuronal growth and axon targeting. Neuron 1995, 15, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Schofield, P.R.; McFarland, K.C.; Hayflick, J.S.; Wilcox, J.N.; Cho, T.M.; Roy, S.; Lee, N.M.; Loh, H.H.; Seeburg, P.H. Molecular characterization of a new immunoglobulin superfamily protein with potential roles in opioid binding and cell contact. EMBO J. 1989, 8, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Lippman, D.A.; Lee, N.M.; Loh, H.H. Opioid-binding cell adhesion molecule (OBCAM)-related clones from a rat brain cDNA library. Gene 1992, 117, 249–254. [Google Scholar] [CrossRef]
- Struyk, A.F.; Canoll, P.D.; Wolfgang, M.J.; Rosen, C.L.; D’Eustachio, P.; Salzer, J.L. Cloning of neurotrimin defines a new subfamily of differentially expressed neural cell adhesion molecules. J. Neurosci. 1995, 15, 2141–2156. [Google Scholar] [CrossRef] [PubMed]
- Sanz, R.; Ferraro, G.B.; Fournier, A.E. IgLON Cell Adhesion Molecules Are Shed from the Cell Surface of Cortical Neurons to Promote Neuronal Growth. J. Biol. Chem. 2015, 290, 4330–4342. [Google Scholar] [CrossRef] [PubMed]
- Funatsu, N.; Miyata, S.; Kumanogoh, H.; Shigeta, M.; Hamada, K.; Endo, Y.; Sokawa, Y.; Maekawa, S. Characterization of a novel rat brain glycosylphosphatidylinositol- anchored protein (Kilon), a member of the IgLON cell adhesion molecule family. J. Biol. Chem. 1999, 274, 8224–8230. [Google Scholar] [CrossRef]
- Marg, A.; Sirim, P.; Spaltmann, F.; Plagge, A.; Kauselmann, G.; Buck, F.; Rathjen, F.G.; Brümmendorf, T. Neurotractin, a novel neurite outgrowth-promoting Ig-like protein that interacts with CEPU-1 and LAMP. J. Cell Biol. 1999, 145, 865–876. [Google Scholar] [CrossRef]
- Pischedda, F.; Piccoli, G. The IgLON Family Member Negr1 Promotes Neuronal Arborization Acting as Soluble Factor via FGFR2. Front. Mol. Neurosci. 2016, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Sabater, L.; Planagumà, J.; Dalmau, J.; Graus, F. Cellular investigations with human antibodies associated with the anti-IgLON5 syndrome. J. Neuroinflamm. 2016, 13, 226. [Google Scholar] [CrossRef] [PubMed]
- Noh, K.; Park, J.C.; Han, J.S.; Lee, S.J. From bound cells comes a sound mind: The role of neuronal growth regulator 1 in psychiatric disorders. Exp. Neurobiol. 2020, 29, 1–10. [Google Scholar] [CrossRef]
- Salluzzo, M.; Vianello, C.; Abdullatef, S.; Rimondini, R.; Piccoli, G.; Carboni, L. The Role of IgLON Cell Adhesion Molecules in Neurodegenerative Diseases. Genes 2023, 14, 1886. [Google Scholar] [CrossRef] [PubMed]
- Antony, J.; Zanini, E.; Birtley, J.R.; Gabra, H.; Recchi, C. Emerging roles for the GPI-anchored tumor suppressor OPCML in cancers. Cancer Gene Ther. 2021, 28, 18–26. [Google Scholar] [CrossRef]
- Martinez-Monleon, A.; Gaarder, J.; Djos, A.; Kogner, P.; Fransson, S. Identification of recurrent 3q13.31 chromosomal rearrangement indicates LSAMP as a tumor suppressor gene in neuroblastoma. Int. J. Oncol. 2023, 62, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Fareh, M.; Yeom, K.-H.; Haagsma, A.C.; Chauhan, S.; Heo, I.; Joo, C. TRBP ensures efficient Dicer processing of precursor microRNA in RNA-crowded environments. Nat. Commun. 2016, 7, 13694. [Google Scholar] [CrossRef] [PubMed]
- Letelier, P.; Riquelme, I.; Hernández, A.H.; Guzmán, N.; Farías, J.G.; Roa, J.C. Circulating MicroRNAs as Biomarkers in Biliary Tract Cancers. Int. J. Mol. Sci. 2016, 17, 791. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Bulut, R.; Kantar, M.; Alptekin, B. MicroRNA nomenclature and the need for a revised naming prescription. Brief. Funct. Genom. 2016, 15, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, T.; Batzel, P.; Berezikov, E.; Eilbeck, K.; Eppig, J.T.; McAndrews, M.S.; Singer, A.; Postlethwait, J.H. miRNA Nomenclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends Genet. 2015, 31, 613–626. [Google Scholar] [CrossRef]
- Meyers, B.C.; Axtell, M.J.; Bartel, B.; Bartel, D.P.; Baulcombe, D.; Bowman, J.L.; Cao, X.; Carrington, J.C.; Chen, X.; Green, P.J.; et al. Criteria for annotation of plant MicroRNAs. Plant Cell 2008, 20, 3186–3190. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Jeng, S.F.; Rau, C.S.; Liliang, P.C.; Wu, C.J.; Lu, T.H.; Chen, Y.C.; Lin, C.J.; Hsieh, C.H. Profiling muscle-specific microrna expression after peripheral denervation and reinnervation in a rat model. J. Neurotrauma 2009, 26, 2345–2353. [Google Scholar] [CrossRef] [PubMed]
- Latgé, G.; Poulet, C.; Bours, V.; Josse, C.; Jerusalem, G. Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers. Int. J. Mol. Sci. 2018, 19, 123. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Panda, A.; Kang, M.J.; Xu, J.; Selimyan, R.; Yoon, J.H.; Martindale, J.L.; De, S.; Wood, W.H.; Becker, K.G.; et al. Senescence-associated lncRNAs: Senescence-associated long noncoding RNAs. Aging Cell 2013, 12, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Gao, W.; Zhang, Z.; Xue, X.; Mijiti, M.; Guo, Q.; Wusiman, D.; Wang, K.; Zeng, X.; Xue, L.; et al. Identification of a seven-lncRNAs panel that serves as a prognosis predictor and contributes to the malignant progression of laryngeal squamous cell carcinoma. Front. Oncol. 2023, 13, 1106249. [Google Scholar] [CrossRef]
- Nie, Q.; Cao, H.; Yang, J.W.; Liu, T.; Wang, B. PI3K/Akt signalling pathway-associated long noncoding RNA signature predicts the prognosis of laryngeal cancer patients. Sci. Rep. 2023, 13, 14764. [Google Scholar] [CrossRef]
- Li, N.; Shi, K.; Li, W. TUSC7: A novel tumor suppressor long non-coding RNA in human cancers. J. Cell. Physiol. 2018, 233, 6401–6407. [Google Scholar] [CrossRef]
- Luo, L.; Wang, M.; Li, X.; Tian, J.; Zhang, K.; Tan, S.; Luo, C. Long non-coding RNA LOC285194 in cancer. Clin. Chim. Acta 2020, 502, 1–8. [Google Scholar] [CrossRef]
- Chen, J.; Lui, W.O.; Vos, M.D.; Clark, G.J.; Takahashi, M.; Schoumans, J.; Khoo, S.K.; Petillo, D.; Lavery, T.; Sugimura, J.; et al. The t(1;3) breakpoint-spanning genes LSAMP and NORE1 are involved in clear cell renal cell carcinomas. Cancer Cell 2003, 4, 405–413. [Google Scholar] [CrossRef]
- Petrovics, G.; Li, H.; Stümpel, T.; Tan, S.H.; Young, D.; Katta, S.; Li, Q.; Ying, K.; Klocke, B.; Ravindranath, L.; et al. A novel genomic alteration of LSAMP associates with aggressive prostate cancer in African American men. eBioMedicine 2015, 2, 1957–1964. [Google Scholar] [CrossRef]
- Nobusawa, S.; Hirato, J.; Kurihara, H.; Ogawa, A.; Okura, N.; Nagaishi, M.; Ikota, H.; Yokoo, H.; Nakazato, Y. Intratumoral heterogeneity of genomic imbalance in a case of epithelioid glioblastoma with BRAF V600E mutation. Brain Pathol. 2014, 24, 239–246. [Google Scholar] [CrossRef]
- Chang, C.Y.; Wu, K.L.; Chang, Y.Y.; Liu, Y.W.; Huang, Y.C.; Jian, S.F.; Lin, Y.S.; Tsai, P.H.; Hung, J.Y.; Tsai, Y.M.; et al. The downregulation of lsamp expression promotes lung cancer progression and is associated with poor survival prognosis. J. Pers. Med. 2021, 11, 578. [Google Scholar] [CrossRef]
- Yen, C.-C.; Chen, W.-M.; Chen, T.-H.; Chen, W.Y.-K.; Chen, P.C.-H.; Chiou, H.-J.; Hung, G.-Y.; Wu, H.-T.H.; Wei, C.-J.; Shiau, C.-Y.; et al. Identification of chromosomal aberrations associated with disease progression and a novel 3q13.31 deletion involving LSAMP gene in osteosarcoma. Int. J. Oncol. 2009, 35, 775–788. [Google Scholar] [CrossRef]
- Barøy, T.; Kresse, S.H.; Skårn, M.; Stabell, M.; Castro, R.; Lauvrak, S.; Llombart-Bosch, A.; Myklebost, O.; Meza-Zepeda, L.A. Reexpression of LSAMP inhibits tumor growth in a preclinical osteosarcoma model. Mol. Cancer 2014, 13, 93. [Google Scholar] [CrossRef]
- Wang, J.; Zuo, Z.; Yu, Z.; Chen, Z.; Tran, L.J.; Zhang, J.; Ao, J.; Ye, F.; Sun, Z. Collaborating single-cell and bulk RNA sequencing for comprehensive characterization of the intratumor heterogeneity and prognostic model development for bladder cancer. Aging 2023, 15, 12104–12119. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; Jaratlerdsiri, W.; Jiang, J.; Willet, C.; Chew, T.; Patrick, S.M.; Lyons, R.J.; Haynes, A.M.; Pasqualim, G.; Brum, I.S.; et al. Genome-wide interrogation of structural variation reveals novel African-specific prostate cancer oncogenic drivers. Genome Med. 2022, 14, 100. [Google Scholar] [CrossRef]
- Huang, S.P.; Lin, V.C.; Lee, Y.C.; Yu, C.C.; Huang, C.Y.; Chang, T.Y.; Lee, H.Z.; Juang, S.H.; Lu, T.L.; Bao, B.Y. Genetic variants in nuclear factor-kappa B binding sites are associated with clinical outcomes in prostate cancer patients. Eur. J. Cancer 2013, 49, 3729–3737. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Liu, Y.; Ye, F. Construction and integrated analysis of crosstalking ceRNAs networks in laryngeal squamous cell carcinoma. PeerJ 2019, 2019, e7380. [Google Scholar] [CrossRef] [PubMed]
- Dell’orco, M.; Elyaderani, A.; Vannan, A.; Sekar, S.; Powell, G.; Liang, W.S.; Neisewander, J.L.; Perrone-Bizzozero, N.I. HuD regulates mRNA-circRNA-miRNA networks in the mouse striatum linked to neuronal development and drug addiction. Biology 2021, 10, 939. [Google Scholar] [CrossRef] [PubMed]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of circ RNA s. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Niu, Y.; Luo, L.; Lu, Z.; Chen, Q.; Zhang, S.; Guo, Q.; Li, L.; Gou, D. Decoding ceRNA regulatory network in the pulmonary artery of hypoxia-induced pulmonary hypertension (HPH) rat model. Cell Biosci. 2022, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zou, W.; Xiao, Z.; Deng, Z.; Liu, R. Hypoxia-induced Long Non-coding RNA LSAMP-AS1 Regulates ceRNA Network to Predict Prognosis for Pancreatic Cancer. Comb. Chem. High Throughput Screen. 2023, 26, 2358–2371. [Google Scholar] [CrossRef]
- Berillo, O.; Régnier, M.; Ivashchenko, A. Binding of intronic miRNAs to the mRNAs of host genes encoding intronic miRNAs and proteins that participate in tumourigenesis. Comput. Biol. Med. 2013, 43, 1374–1381. [Google Scholar] [CrossRef]
- Zhou, S.L.; Yin, D.; Hu, Z.Q.; Luo, C.B.; Zhou, Z.J.; Xin, H.Y.; Yang, X.R.; Shi, Y.H.; Wang, Z.; Huang, X.W.; et al. A Positive Feedback Loop Between Cancer Stem-Like Cells and Tumor-Associated Neutrophils Controls Hepatocellular Carcinoma Progression. Hepatology 2019, 70, 1214–1230. [Google Scholar] [CrossRef] [PubMed]
- Ghobadi, M.Z.; Afsaneh, E.; Emamzadeh, R.; Soroush, M. Potential miRNA-gene interactions determining progression of various ATLL cancer subtypes after infection by HTLV-1 oncovirus. BMC Med. Genom. 2023, 16, 62. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Gong, C.; Wang, Y.; Yin, Z.; Wang, X.; Hu, Y.; Fang, Z. Bioinformatics analysis and experimental validation identified HMGA2/microRNA-200c-3p/LSAMP/Wnt axis as an immunological factor of patients with colorectal cancer. Am. J. Cancer Res. 2023, 13, 3898–3920. [Google Scholar] [PubMed]
- Fang, X.; Dong, Y.; Yang, R.; Wei, L. LINC00619 restricts gastric cancer progression by preventing microRNA-224-5p-mediated inhibition of OPCML. Arch. Biochem. Biophys. 2020, 689, 108390. [Google Scholar] [CrossRef]
- Tang, S.; Liao, K.; Shi, Y.; Tang, T.; Cui, B.; Huang, Z. Bioinformatics analysis of potential Key lncRNA-miRNA-mRNA molecules as prognostic markers and important ceRNA axes in gastric cancer. Am. J. Cancer Res. 2022, 12, 2397–2418. [Google Scholar]
- He, Y.; Liu, C.; Song, P.; Pang, Z.; Mo, Z.; Huang, C.; Yan, T.; Sun, M.; Fa, X. Investigation of miRNA- And lncRNA-mediated competing endogenous RNA network in cholangiocarcinoma. Oncol. Lett. 2019, 18, 5283–5293. [Google Scholar] [CrossRef]
- Niskakoski, A.; Kaur, S.; Staff, S.; Renkonen-Sinisalo, L.; Lassus, H.; Järvinen, H.J.; Mecklin, J.-P.; Bützow, R.; Peltomäki, P. Epigenetic analysis of sporadic and Lynch-associated ovarian cancers reveals histology-specific patterns of DNA methylation. Epigenetics 2014, 9, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Houshdaran, S.; Hawley, S.; Palmer, C.; Campan, M.; Olsen, M.N.; Ventura, A.P.; Knudsen, B.S.; Drescher, C.W.; Urban, N.D.; Brown, P.O.; et al. DNA Methylation Profiles of Ovarian Epithelial Carcinoma Tumors and Cell Lines. PLoS ONE 2010, 5, e9359. [Google Scholar] [CrossRef]
- Kolbe, D.L.; DeLoia, J.A.; Porter-Gill, P.; Strange, M.; Petrykowska, H.M.; Guirguis, A.; Krivak, T.C.; Brody, L.C.; Elnitski, L. Differential Analysis of Ovarian and Endometrial Cancers Identifies a Methylator Phenotype. PLoS ONE 2012, 7, e32941. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, J.; Yang, Z.; Xu, L. Identification of RNA expression profiles in thyroid cancer to construct a competing endogenous RNA (ceRNA) network of mRNAs, long noncoding RNAs (lncRNAs), and microRNAs (miRNAs). Med. Sci. Monit. 2019, 25, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Davison, J.; Qu, X.; Morrissey, C.; Storer, B.; Brown, L.; Vessella, R.; Nelson, P.; Fang, M. Methylation profiling identified novel differentially methylated markers including OPCML and FLRT2 in prostate cancer. Epigenetics 2016, 11, 247–258. [Google Scholar] [CrossRef]
- Duarte-Pereira, S.; Paiva, F.; Costa, V.L.; Ramalho-Carvalho, J.; Savva-Bordalo, J.; Rodrigues, Â.; Ribeiro, F.R.; Silva, V.M.; Oliveira, J.; Henrique, R.; et al. Prognostic value of opioid binding protein/cell adhesion molecule-like promoter methylation in bladder carcinoma. Eur. J. Cancer 2011, 47, 1106–1114. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Ying, Y.; van Hasselt, A.; Ng, K.M.; Yu, J.; Zhang, Q.; Jin, J.; Liu, D.; Rhim, J.S.; Rha, S.Y.; et al. OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation. PLoS ONE 2008, 3, e2990. [Google Scholar] [CrossRef]
- Ramirez, C.M.; Dávalos, A.; Goedeke, L.; Salerno, A.G.; Warrier, N.; Cirera-Salinas, D.; Suárez, Y.; Fernández-Hernando, C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2707–2714. [Google Scholar] [CrossRef]
- Janowski, B.A.; Willy, P.J.; Devi, T.R.; Falck, J.R.; Mangelsdorf, D.J. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 1996, 383, 728–731. [Google Scholar] [CrossRef]
- Wang, X.; Collins, H.L.; Ranalletta, M.; Fuki, I.V.; Billheimer, J.T.; Rothblat, G.H.; Tall, A.R.; Rader, D.J. Macrophage ABCA1 and ABCG1, but not SR-BI, promote macrophage reverse cholesterol transport in vivo. J. Clin. Investig. 2007, 117, 2216–2224. [Google Scholar] [CrossRef]
- Yu, B.; Qian, T.; Wang, Y.; Zhou, S.; Ding, G.; Ding, F.; Gu, X. MiR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res. 2012, 40, 10356–10365. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, G.; Liu, J.; Zhang, Y. MicroRNA-182 promotes proliferation and metastasis by targeting FOXF2 in triple-negative breast cancer. Oncol. Lett. 2017, 14, 4805–4811. [Google Scholar] [CrossRef]
- Kimura, Y.; Katoh, A.; Kaneko, T.; Takahama, K.; Tanaka, H. Two members of the IgLON family are expressed in a restricted region of the developing chick brain and neural crest. Dev. Growth Differ. 2001, 43, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Karolina, D.S.; Sepramaniam, S.; Armugam, A.; Jeyaseelan, K. Expression profiling of RNA transcripts during neuronal maturation and ischemic injury. PLoS ONE 2014, 9, e103525. [Google Scholar] [CrossRef]
- Kaur, P.; Tan, J.R.; Karolina, D.S.; Sepramaniam, S.; Armugam, A.; Wong, P.T.-H.; Jeyaseelan, K. A long non-coding RNA, BC048612 and a microRNA, miR-203 coordinate the gene expression of neuronal growth regulator 1 (NEGR1) adhesion protein. Biochim. Biophys. Acta 2016, 1863, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Costain, W.J.; Haqqani, A.S.; Rasquinha, I.; Giguere, M.; Slinn, J.; Zurakowski, B.; Stanimirovic, D.B. Proteomic analysis of synaptosomal protein expression reveals that cerebral ischemia alters lysosomal Psap processing. Proteomics 2010, 10, 3272–3291. [Google Scholar] [CrossRef]
- Yan, Y.; Chen, L.; Zhou, J.; Xie, L. SNHG12 inhibits oxygen-glucose deprivation-induced neuronal apoptosis via the miR-181a-5p/NEGR1 axis. Mol. Med. Rep. 2020, 22, 3886–3894. [Google Scholar] [CrossRef]
- Qiu, L.; He, J.; Chen, H.; Xu, X.; Tao, Y. CircDLGAP4 overexpression relieves oxygen-glucose deprivation—Induced neuronal injury by elevating NEGR1 through sponging miR-503-3p. J. Mol. Histol. 2022, 53, 321–332. [Google Scholar] [CrossRef]
- Jin, L.; Li, X.; Zhao, Y.; Zhu, G.; Shen, W. miR-576-5p Facilitates Aggressive Cell Behaviors in Colon Adenocarcinoma via Targeting NEGR1. Crit. Rev. Eukaryot. Gene Expr. 2022, 32, 25–33. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, C.; Li, Q.; Li, J.; Wu, Y.; Liu, J. Biochimie MicroRNA-25 e 5p counteracts oxidized LDL-induced pathological changes by targeting neuronal growth regulator 1 (NEGR1) in human brain micro-vessel endothelial cells. Biochimie 2019, 165, 141–149. [Google Scholar] [CrossRef]
- Dong, Y.; Fan, X.; Wang, Z.; Zhang, L.; Guo, S. Circ_HECW2 functions as a miR-30e-5p sponge to regulate LPS-induced endothelial-mesenchymal transition by mediating NEGR1 expression. Brain Res. 2020, 1748, 147114. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, J.; Yang, Q.; Ma, M. LncRNA MIAT Upregulates NEGR1 by Competing for miR-150-5p as a Competitive Endogenous RNA in SCIRI Rats. Genet. Res. 2022, 2022, 2942633. [Google Scholar] [CrossRef]
- Gulluoglu, S.; Tuysuz, E.C.; Sahin, M.; Kuskucu, A.; Kaan Yaltirik, C.; Ture, U.; Kucukkaraduman, B.; Akbar, M.W.; Gure, A.O.; Bayrak, O.F.; et al. Simultaneous miRNA and mRNA transcriptome profiling of glioblastoma samples reveals a novel set of OncomiR candidates and their target genes. Brain Res. 2018, 1700, 199–210. [Google Scholar] [CrossRef]
- Zeng, Y.; Liu, J.-X.; Yan, Z.-P.; Yao, X.-H.; Liu, X.-H. Potential microRNA biomarkers for acute ischemic stroke. Int. J. Mol. Med. 2015, 36, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Jiang, H.; Li, X.; Wu, P.; Liu, J.; Wang, T.; Zhou, X.; Xiong, J.; Li, W. Bioinformatics analysis on the differentiation of bone mesenchymal stem cells into osteoblasts and adipocytes. Mol. Med. Rep. 2017, 15, 1571–1576. [Google Scholar] [CrossRef]
- Ruan, L.; Xie, Y.; Liu, F.; Chen, X. Serum miR-1181 and miR-4314 associated with ovarian cancer: MiRNA microarray data analysis for a pilot study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2018, 222, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, X.; Zhou, L.; Luo, Y.; Zhan, N.; Ye, Y.; Liu, Z.; Zhang, X.; Qiu, T.; Lin, L.; et al. Ferroptosis-related lncRNAs: Distinguishing heterogeneity of the tumour microenvironment and predicting immunotherapy response in bladder cancer. Heliyon 2024, 10, e32018. [Google Scholar] [CrossRef]
- Xiong, W.; Feng, S.; Wang, H.; Qing, S.; Yang, Y.; Zhao, Y.; Zeng, Z.; Gong, J. Identification of candidate genes and pathways in limonin-mediated cardiac repair after myocardial infarction. Biomed. Pharmacother. 2021, 142, 112088. [Google Scholar] [CrossRef]
- Velle-Forbord, T.; Eidlaug, M.; Debik, J.; Sæther, J.C.; Follestad, T.; Nauman, J.; Gigante, B.; Røsjø, H.; Omland, T.; Langaas, M.; et al. Circulating microRNAs as predictive biomarkers of myocardial infarction: Evidence from the HUNT study. Atherosclerosis 2019, 289, 1–7. [Google Scholar] [CrossRef]

| IgLON Name | Alias | Expression | Role |
|---|---|---|---|
| LSAMP | IgLON3, LAMP | Brain, retina-enriched, cancer-enriched | Cell adhesion molecule |
| OPCML | IgLON1, OBCAM | Brain, parathyroid gland, retina-enriched, cancer-enriched | Cell adhesion molecule, neuronal signalling |
| NTM | IgLON2, neurotrimin, CEPU-1 | Brain, retina-enriched, cancer-enhanced | Cell adhesion molecule |
| NEGR1 | IgLON4, kilon, neurotractin | Brain-enriched | Cell adhesion molecule, synaptic function |
| IgLON5 | Brain, testis-enriched, cancer-enriched | Cell adhesion molecule, brain development |
| miRNA | Target | Model | Target Variation |
|---|---|---|---|
| miR-206 | LSAMP | Nerve lesion | Potential regulatory role |
| miR-375 | LSAMP-AS1 | Tumour samples | Inhibition of miR-375 by upregulated LSAMP-AS1 |
| miR-1193-3p; miR-129-1-3p; miR-129-2-3p; miR-132-3p; miR-1930-5p; miR-212-3p; miR-297a-3p; miR-297b-3p; miR-297c-3p; miR-3068-3p; miR-3069-3p; miR-3072-3p; miR-3102-3p; miR-326-3p; miR-329-3p; miR-350-3p; miR-369-3p; miR-434-3p; miR-450b-3p; miR-466a-3p; miR-466b-3p; miR-466c-3p; miR-466e-3p; miR-466f-3p; miR-466h-5p; miR-466p-3p; miR-467d-3p; miR-488-3p; miR-700-5p; miR-874-5p | LSAMP | HuD KO mice | Predicted LSAMP downregulation |
| miR-541-5p | LSAMP | Hypoxia-induced pulmonary hypertension | Predicted LSAMP downregulation |
| miR-129-5p | LSAMP-AS1 | Pancreatic cancer | Regulating hypoxic tumoural environment |
| miR-4447 | LSAMP | Tumorigenesis | miRNA encoded within LSAMP |
| miR-301b-3p | LSAMP | Hepatocellular carcinoma | Suppression of LSAMP gene expression |
| miR-29b-2-5p and miR-342-3p | LSAMP | Leukaemia/lymphoma | Interaction in the pathogenetic mechanism of the disease |
| miRNA-200c-3p | LSAMP | Colorectal cancer | Inhibition of LSAMP expression |
| miR-224-5p | OPCML | Gastric cancer | Downregulates OPCML |
| hsa-mir-137 | OPCML | Gastric cancer | Downregulates OPCML via POU6F2-AS2 |
| hsa-mir-372, hsa-mir-373, hsa-mir-519d, hsa-mir-184, hsa-mir-205, hsa-mir-506, hsa-mir-375 | OPCML-iT1 | Cholangiocarcinoma (CCA) | Predicted by miRcode database |
| MIR34B, Let-7a-3 | OPCML | Epithelial ovarian cancer | Potentially influencing OPCML expression levels as methylation-sensitive |
| miR-184, miR-372, miR-373, miR-519d, miR-205, miR-375 | OPCML-iT1 | Thyroid cancer | Predicted by competing endogenous RNA (ceRNA) network |
| miR-758 | NTM | Glioblastoma | Downregulation of NTM mRNA expression |
| miR-182 | NTM | Peripheral nerve injury | Downregulates mRNA and NTM protein levels |
| miR-377 | NEGR1 | Neurodevelopment of primary cells/OGD | Upregulation of NEGR1 in neurodevelopment primary cells. Predicted interaction in OGD |
| miR-203 | NEGR1 | Neurodevelopment of primary cells/OGD | Downregulation of NEGR1 in OGD. Predicted interaction in neurodevelopment of primary cells |
| miR-181a-5p | NEGR1 | OGD | Downregulation of NEGR1 |
| miR-181a-5p | NEGR1 | Bladder cancer | NEGR1 regulation |
| miR-503-3p | NEGR1 | OGD | Downregulation of NEGR1 |
| miR-576-5p | NEGR1 | Colon adenocarcinoma | Downregulation of NEGR1 |
| miR-25-5p | NEGR1 | HBMEC treated with OX-LDL | Downregulation of NEGR1 |
| miR-30e-5p | NEGR1 | HBMEC treated with LPS | Downregulation of NEGR1 |
| miR-150-5p | NEGR1 | Spinal cord ischaemia–reperfusion injury | Downregulation of NEGR1 |
| miR-21-5p | NEGR1 | Glioblastoma | Predicted negative correlation with NEGR1 |
| miR-9, miR-181, miR-124 | NEGR1 | Acute ischaemic stroke | Predicted interaction with NEGR1 |
| miR-203, miR-382 | NEGR1 | Mesenchymal stem cells | Predicted interaction with NEGR1 |
| miR-4314 | NEGR1 | Ovarian cancer | Predicted downregulation of NEGR1 |
| rno-miR-10a-5p | IGLON5 | Myocardial infarction (MI) | MI caused decreased miR-10a-5p expression and increased IGLON5 expression |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salluzzo, M.; Vianello, C.; Flotta, F.; Rimondini, R.; Carboni, L. MicroRNAs Associated with IgLON Cell Adhesion Molecule Expression. Curr. Issues Mol. Biol. 2024, 46, 7702-7718. https://doi.org/10.3390/cimb46070456
Salluzzo M, Vianello C, Flotta F, Rimondini R, Carboni L. MicroRNAs Associated with IgLON Cell Adhesion Molecule Expression. Current Issues in Molecular Biology. 2024; 46(7):7702-7718. https://doi.org/10.3390/cimb46070456
Chicago/Turabian StyleSalluzzo, Marco, Clara Vianello, Francesca Flotta, Roberto Rimondini, and Lucia Carboni. 2024. "MicroRNAs Associated with IgLON Cell Adhesion Molecule Expression" Current Issues in Molecular Biology 46, no. 7: 7702-7718. https://doi.org/10.3390/cimb46070456





