ALA Promotes Sucrose Accumulation in Early Peach Fruit by Regulating SPS Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Chemical Treatments
2.2. The Determination of Physiological and Biochemical Indicators of Peach Fruits
2.3. Sucrose Determination
2.4. Determination of SPS Activity
2.5. Identification of SPS Genes in Peaches and Other Plants
2.6. Bioinformatics Analysis
2.7. RNA Isolation and Quantitative Real-Time Fluorescence PCR (qRT-PCR)
2.8. Subcellular Localization of PpSPS2 and PpSPS3 in N. benthamiana
2.9. Statistics and Reproducibility
3. Results
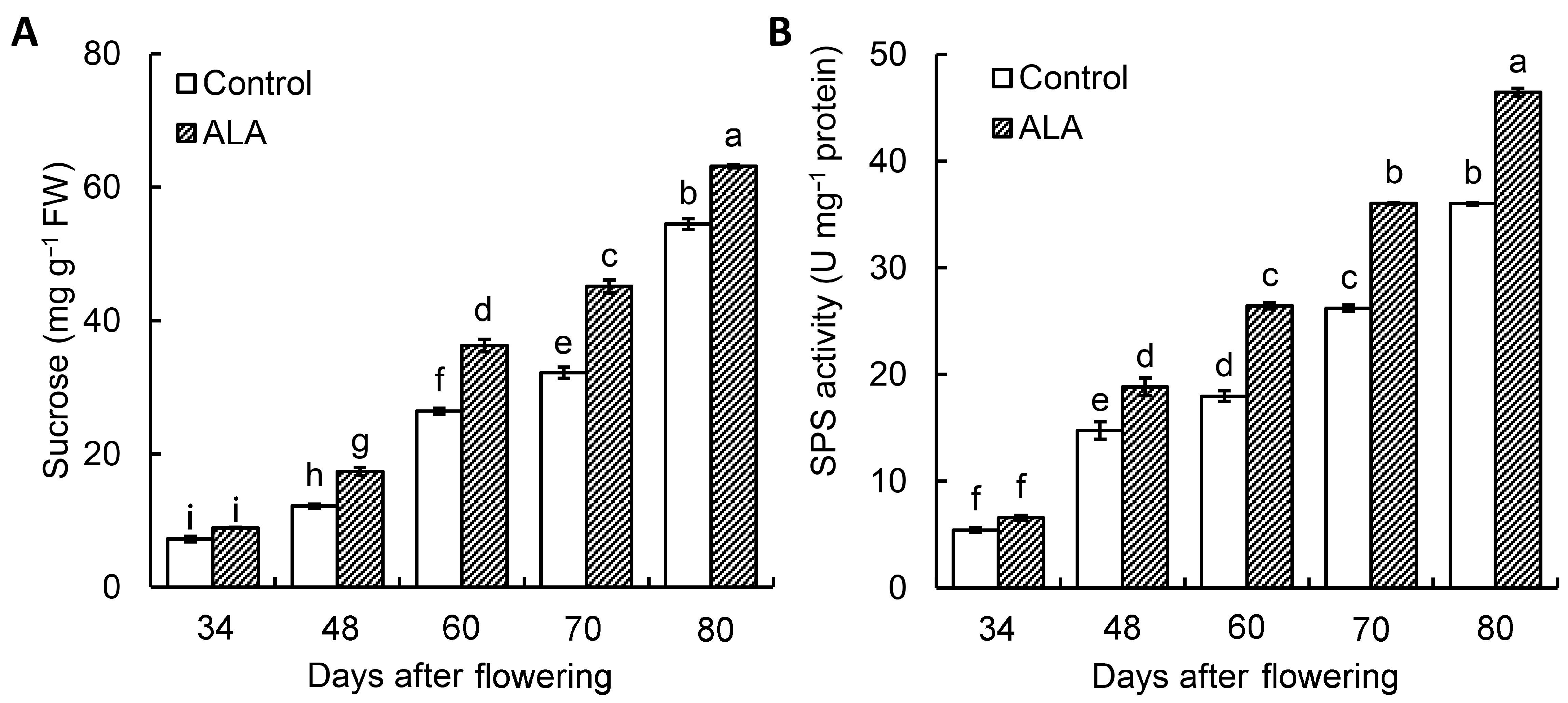
3.1. ALA Promoted the Sucrose Content and SPS Activity of Peach Fruit during Fruit Development
3.2. ALA Promoted the Sucrose Content and SPS Activity of Peach Fruit during Fruit Development
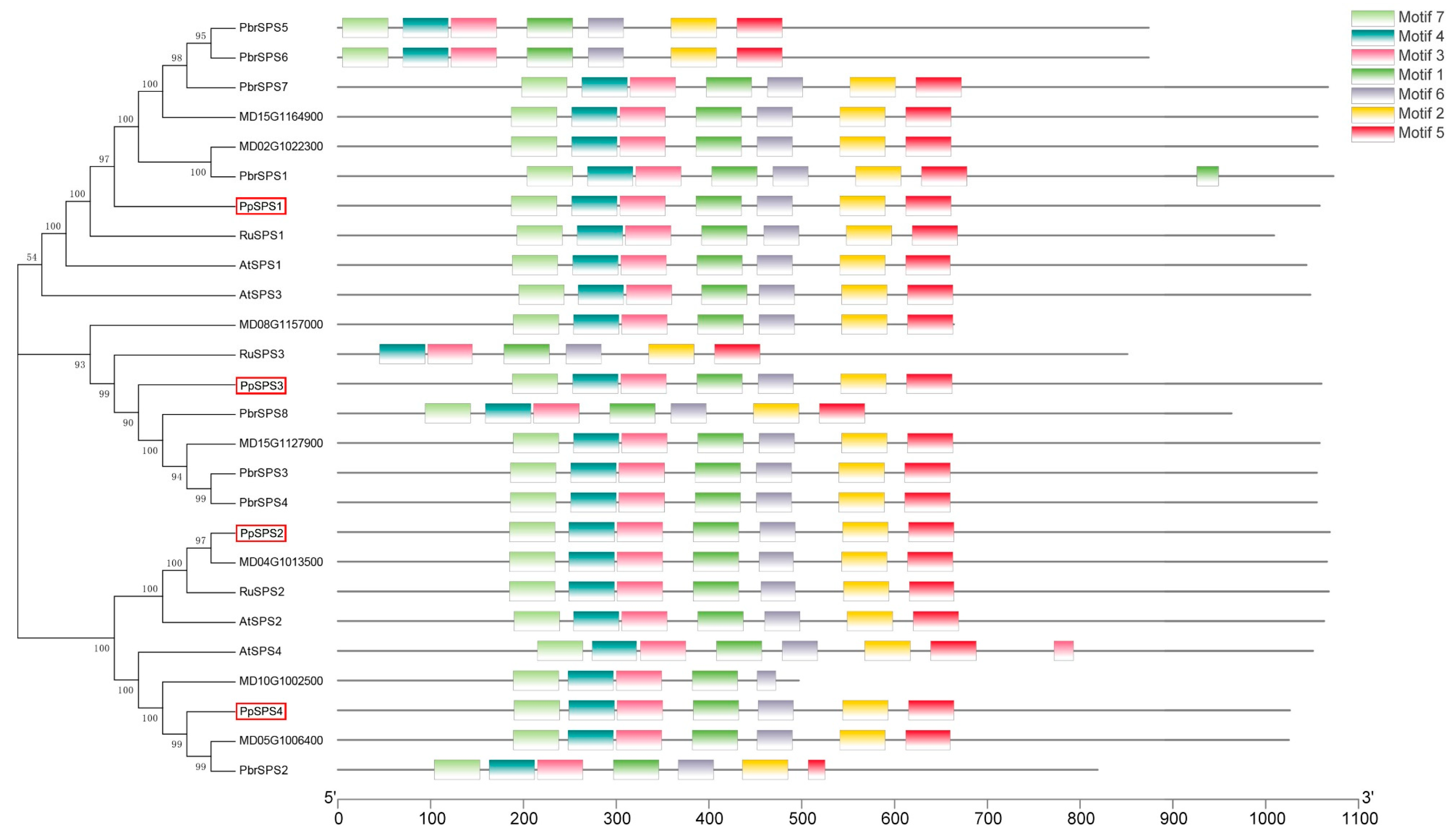
3.3. Identification and Characterization of the PpSPS Gene Family
3.4. Chromosomal Location, Gene Structure and Promoter Analysis of PpSPS Family Genes
3.5. ALA-Regulated PpSPS-Related Genes
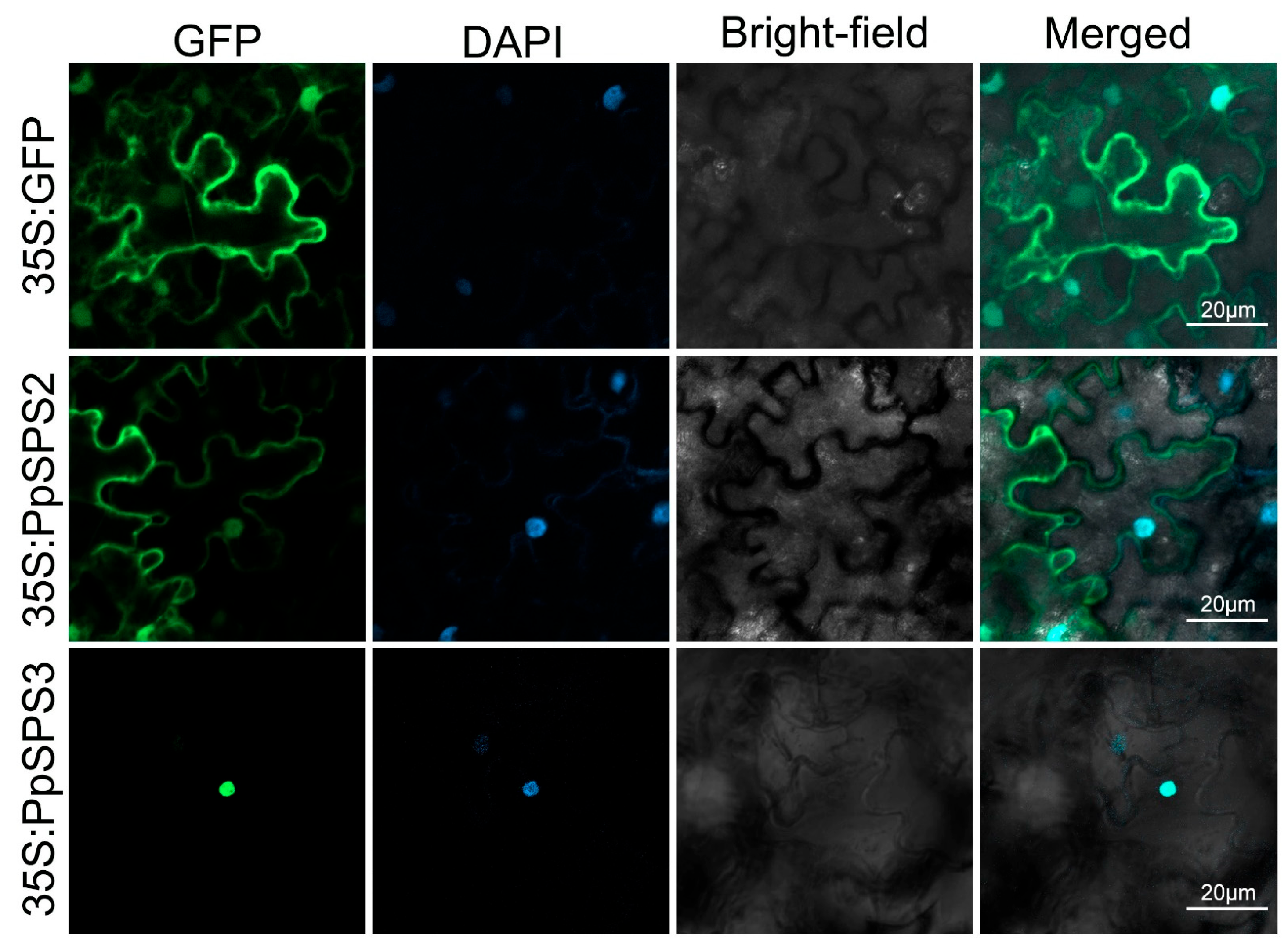
3.6. Subcellular Localization of PpSPS2 and PpSPS3
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liang, R.L.; Wang, L.J.; Wang, X.Q.; Zhang, J.T.; Gan, X. Effects of Exogenous ALA on Leaf Photosynthesis, Photosynthate Transport, and Sugar Accumulation in Prunus persica L. Forests 2023, 14, 723. [Google Scholar] [CrossRef]
- Lei, R.L.; Qu, W.G.; Fan, Y.J. Problems and countermeasures of nectarine industry development. Fruit Grow. Friend 2016, 1, 38–39. [Google Scholar]
- Wang, C.; Fang, J.G.; Wang, T.; Tan, H.H. The sugar metabolism in fruits. Acta Agric. Zhejiangensis 2009, 21, 529–534. [Google Scholar]
- Vimolmangkang, S.; Zheng, H.; Peng, Q.; Jiang, Q.; Wang, H.; Fang, T.; Liao, L.; Wang, L.; He, H.; Han, Y. Assessment of Sugar Components and Genes Involved in the Regulation of Sucrose Accumulation in Peach Fruit. J. Agric. Food Chem. 2016, 64, 6723–6729. [Google Scholar] [CrossRef] [PubMed]
- Cirilli, M.; Bassi, D.; Ciacciulli, A. Sugars in peach fruit: A breeding perspective. Hortic. Res. 2016, 3, 15067. [Google Scholar] [CrossRef] [PubMed]
- Stein, O.; Granot, D. An overview of sucrose synthases in plants. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.C.; Huber, J.L. Role and regulation of sucrose-phosphate synthase in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Shen, Z.J.; Zhang, Y.P.; Han, J.; Ma, R.J.; Korir, N.K.; Yu, M.L. Cloning and expression of genes related to the sucrose-metabolizing enzymes and carbohydrate changes in peach. Acta Physiol. Plant. 2013, 35, 589–602. [Google Scholar] [CrossRef]
- Zhang, C.H.; Yu, M.L.; Ma, R.J.; Shen, Z.J.; Zhang, B.B.; Korir, N.K. Structure, expression profile, and evolution of the sucrose synthase gene family in peach (Prunus persica). Acta Physiol. Plant. 2015, 37, 1–15. [Google Scholar] [CrossRef]
- Kobashi, K.; Sugaya, S.; Gemma, H.; Iwahori, S. Effect of abscisic acid (ABA) on sugar accumulation in the flesh tissue of peach fruit at the start of the maturation stage. Plant Growth Regul. 2001, 35, 215–223. [Google Scholar] [CrossRef]
- Li, X.; Liu, P.; Zhou, J.; Su, M.; Ma, Y.; Jia, H.; Du, J.; Gao, Z.; Ye, Z. Effects of exogenous application of GA4+7 and NAA on sugar accumulation and related gene expression in peach fruits during developing and ripening stages. J. Plant Growth Regul. 2021, 40, 962–973. [Google Scholar] [CrossRef]
- Kumar, M.; Rawat, V.; Rawat, J.M.S.; Tomar, Y.K. Effect of pruning intensity on peach yield and fruit quality. Sci. Hortic. 2010, 125, 218–221. [Google Scholar] [CrossRef]
- Akram, N.A.; Ashraf, M. Regulation in Plant Stress Tolerance by a Potential Plant Growth Regulator, 5-Aminolevulinic Acid. J. Plant Growth Regul. 2013, 32, 663–679. [Google Scholar] [CrossRef]
- Wu, W.; He, S.; An, Y.; Cao, R.; Sun, Y.; Tang, Q.; Wang, L. Hydrogen peroxide as a mediator of 5-aminolevulinic acid-induced Na+ retention in roots for improving salt tolerance of strawberries. Physiol. Plant. 2019, 167, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.Y.; He, S.S.; An, Y.Y.; Wang, L.J. Exogenous 5-aminolevulinic acid improves strawberry tolerance to osmotic stress and its possible mechanisms. Physiol. Plant. 2020, 168, 948–962. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lou, Y.R.; Wang, L.J. MdDGK3-like as a Negative Regulator Participates in ALA-Induced PP2AC to Promote Stomatal Opening in Apple leaves. Hortic. Plant J. 2023, 9, 898–908. [Google Scholar] [CrossRef]
- Fang, X.; Zhang, L.Z.; Wang, L.J. The Transcription Factor MdERF78 Is Involved in ALA-Induced Anthocyanin Accumulation in Apples. Front. Plant Sci. 2022, 13, 915197. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Cai, Z.X.; Zhang, B.B.; Xu, J.L.; Song, H.F.; Ma, R.J. The Mechanism Analysis of Anthocyanin Accumulation in Peach Accelerated by ALA. Acta Hortic. Sin. 2013, 40, 1043–1050. [Google Scholar]
- Li, H.S. Principles and Techniques of Plant Physiological and Biochemical Experiments; Higher Education Press: Beijing, China, 2000; pp. 190–202. [Google Scholar]
- Zhang, Z.L.; Zhai, W.J.; Li, X.F. Experimental guidance of Plant Physiology; Higher Education Press: Beijing, China, 2009; pp. 244–246. [Google Scholar]
- Luo, Z.H.; Chen, L.N.; Chen, G.D.; Wei, Q.; Li, N. A novel UV-visible method for the detection of vitamin C in Shatian pumelo. J. Food Saf. Qual. 2018, 9, 2321–2325. [Google Scholar]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2010, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, Z.; An, Y.Y.; Wang, L.J. ALA reverses ABA-induced stomatal closure by modulating PP2AC and SnRK2.6 activity in apple leaves. Hortic. Res. 2023, 10, uhad067. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Foyer, C.H.; Huber, J.; Voelker, T.A.; Huber, S.C. Effects of Elevated Sucrose-Phosphate Synthase Activity on Photosynthesis, Assimilate Partitioning, and Growth in Tomato (Lycopersicon esculentum var UC82B). Plant Physiol. 1993, 101, 535–543. [Google Scholar] [CrossRef]
- Ma, M.Y.; Zhu, T.; Cheng, X.Y.; Li, M.Y.; Yuan, G.L.; Li, C.B.; Zhang, A.H.; Lu, C.M.; Fang, Y.; Zhang, Y. Sucrose phosphate synthase 8 is required for the remobilization of carbon reserves in rice stems during grain filling. J. Exp. Bot. 2023, 75, erad375. [Google Scholar] [CrossRef]
- Hirose, T.; Hashida, Y.; Aoki, N.; Okamura, M.; Yonekura, M.; Ohto, C.; Terao, T.; Ohsugi, R. Analysis of gene-disruption mutants of a sucrose phosphate synthase gene in rice, OsSPS1, shows the importance of sucrose synthesis in pollen germination. Plant Sci. 2014, 225, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Bahaji, A.; Baroja-Fernández, E.; Ricarte-Bermejo, A.; Sánchez-López, Á.M.; Muñoz, F.J.; Romero, J.M.; Ruiz, M.T.; Baslam, M.; Almagro, G.; Sesma, M.T.; et al. Characterization of multiple SPS knockout mutants reveals redundant functions of the four Arabidopsis sucrose phosphate synthase isoforms in plant viability, and strongly indicates that enhanced respiration and accelerated starch turnover can alleviate the blockage of sucrose biosynthesis. Plant Sci. 2015, 238, 135–147. [Google Scholar]
- Haigler, C.H.; Singh, B.; Zhang, D.; Hwang, S.; Wu, C.; Cai, W.X.; Hozain, M.; Kang, W.; Kiedaisch, B.; Strauss, R.E.; et al. Transgenic cotton over-producing spinach sucrose phosphate synthase showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant Mol. Biol. 2007, 63, 815–832. [Google Scholar] [CrossRef]
- Wen, X.X.; Zhang, W.Q.; Feng, Y.Q.; Yu, X.Y. Cloning and characterization of a sucrose synthase-encoding gene from muskmelon. Mol. Biol. Rep. 2010, 37, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Solís-Guzmán, M.G.; Argüello-Astorga, G.; López-Bucio, J.; Ruiz-Herrera, L.F.; López-Meza, J.E.; Sánchez-Calderón, L.; Carreón-Abud, Y.; Martínez-Trujillo, M. Arabidopsis thaliana sucrose phosphate synthase (sps) genes are expressed differentially in organs and tissues, and their transcription is regulated by osmotic stress. Gene Expr. Patterns 2017, 25–26, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Wang, J.Q.; Huang, Z.L.; Mi, L.; Xu, K.F.; Wu, J.J.; Fan, Y.H.; Ma, S.Y.; Jiang, D.G. Effects of Low Temperature at Booting Stage on Sucrose Metabolism and Endogenous Hormone Contents in Winter Wheat Spikelet. Front. Plant Sci. 2019, 10, 498. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P.; Reimholz, R.; Deiting, U.; Sonnewald, U.; Stitt, M. Decreased expression of sucrose phosphate synthase strongly inhibits the water stress-induced synthesis of sucrose in growing potato tubers. Plant J. 1999, 19, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Nemati, F.; Ghanati, F.; Gavlighi, H.A.; Sharifi, M. Comparison of sucrose metabolism in wheat seedlings during drought stress and subsequent recovery. Biol. Plant. 2018, 62, 595–599. [Google Scholar] [CrossRef]
- Winter, H.; Huber, S.C. Regulation of sucrose metabolism in higher plants: Localization and regulation of activity of key enzymes. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 253–289. [Google Scholar] [CrossRef]
- Huber, S.C.; Huber, J.L. Activation of sucrose-phosphate synthase from darkened spinach leaves by an endogenous protein phosphatase. Arch. Biochem. Biophys. 1990, 282, 421–426. [Google Scholar] [CrossRef] [PubMed]
- McMichael, R.W., Jr.; Bachmann, M.; Huber, S.C. Spinach Leaf Sucrose-Phosphate Synthase and Nitrate Reductase Are Phosphorylated/Inactivated by Multiple Protein Kinases in Vitro. Plant Physiol. 1995, 108, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.N.; Sklodowski, K.; Encke, B.; Schulze, W.X. A kinase-phosphatase signaling module with BSK8 and BSL2 involved in regulation of sucrose-phosphate synthase. J. Proteome Res. 2014, 13, 3397–3409. [Google Scholar] [CrossRef]
- Lutfiyya, L.L.; Xu, N.; D’Ordine, R.L.; Morrell, J.A.; Miller, P.W.; Duff, S.M. Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J. Plant Physiol. 2007, 164, 923–933. [Google Scholar] [CrossRef]
- Okamura, M.; Aoki, N.; Hirose, T.; Yonekura, M.; Ohto, C.; Ohsugi, R. Tissue specificity and diurnal change in gene expression of the sucrose phosphate synthase gene family in rice. Plant Sci. 2011, 181, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, J.; Hu, B.; Li, J.; Qin, Y.; Chen, L.; Qin, Y.; Hu, G. Identification and expression profile analysis of the sucrose phosphate synthase gene family in Litchi chinensis Sonn. PeerJ 2018, 6, e4379. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, T.; Sanada, T.; Yamaki, S. Seasonal Fluctuations of Some Enzymes Relating to Sucrose and Sorbitol Metabolism in Peach Fruit. J. Am. Soc. Hortic. Sci. 1990, 115, 278–281. [Google Scholar] [CrossRef]
- Zhang, L.H.; Zhu, L.C.; Xu, Y.; LÜ, L.; Li, X.G.; Li, W.H.; Liu, W.D.; Ma, F.W.; Li, M.J.; Han, D.G. Genome-wide identification and function analysis of the sucrose phosphate synthase MdSPS gene family in apple. J. Integr. Agric. 2023, 22, 2080–2093. [Google Scholar] [CrossRef]



| Treatment | Fruit Weight (g) | Soluble Solid (%) | Soluble Sugar (%) | Titratable Acid (‰) | Soluble Protein (mg/g) | Vitamin C (mg/100 g) |
|---|---|---|---|---|---|---|
| Control | 86.89 ± 2.26 b | 15.85 ± 0.29 b | 10.18 ± 0.30 b | 3.48 ± 0.04 a | 6.03 ± 0.18b | 4.20 ± 0.31ab |
| ALA | 98.79 ± 3.01 a | 17.06 ± 0.45 a | 11.64 ± 0.13 a | 3.29 ± 0.04 b | 7.25 ± 0.24 a | 4.67 ± 0.16 a |
| Gene Name | Gene ID | Genome Location | CDS Length (bp) | Protein Length (aa) | Mw (KDa) | Subcellular Localization |
|---|---|---|---|---|---|---|
| PpSPS1 | Prupe.7G249900 | Chr07: 21151882-21157785 (−) | 3174 | 1057 | 118.17 | Cytoplasmic |
| PpSPS2 | Prupe.1G159700 | Chr01:12702147-12709381 (−) | 3207 | 1068 | 119.69 | Cytoplasmic; Nuclear |
| PpSPS3 | Prupe.1G483200 | Chr01:40288494-40295210 (−) | 3180 | 1059 | 118.26 | Nuclear |
| PpSPS4 | Prupe.8G003700 | Chr08: 302873-308259 (−) | 3078 | 1025 | 115.41 | Cytoplasmic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Guo, X.; Du, J.; Yu, M. ALA Promotes Sucrose Accumulation in Early Peach Fruit by Regulating SPS Activity. Curr. Issues Mol. Biol. 2024, 46, 7944-7954. https://doi.org/10.3390/cimb46080469
Chen Z, Guo X, Du J, Yu M. ALA Promotes Sucrose Accumulation in Early Peach Fruit by Regulating SPS Activity. Current Issues in Molecular Biology. 2024; 46(8):7944-7954. https://doi.org/10.3390/cimb46080469
Chicago/Turabian StyleChen, Zheng, Xin Guo, Jinhua Du, and Mingliang Yu. 2024. "ALA Promotes Sucrose Accumulation in Early Peach Fruit by Regulating SPS Activity" Current Issues in Molecular Biology 46, no. 8: 7944-7954. https://doi.org/10.3390/cimb46080469




