Ontogeny of Skin Stem Cells and Molecular Underpinnings
Abstract
:1. Introduction
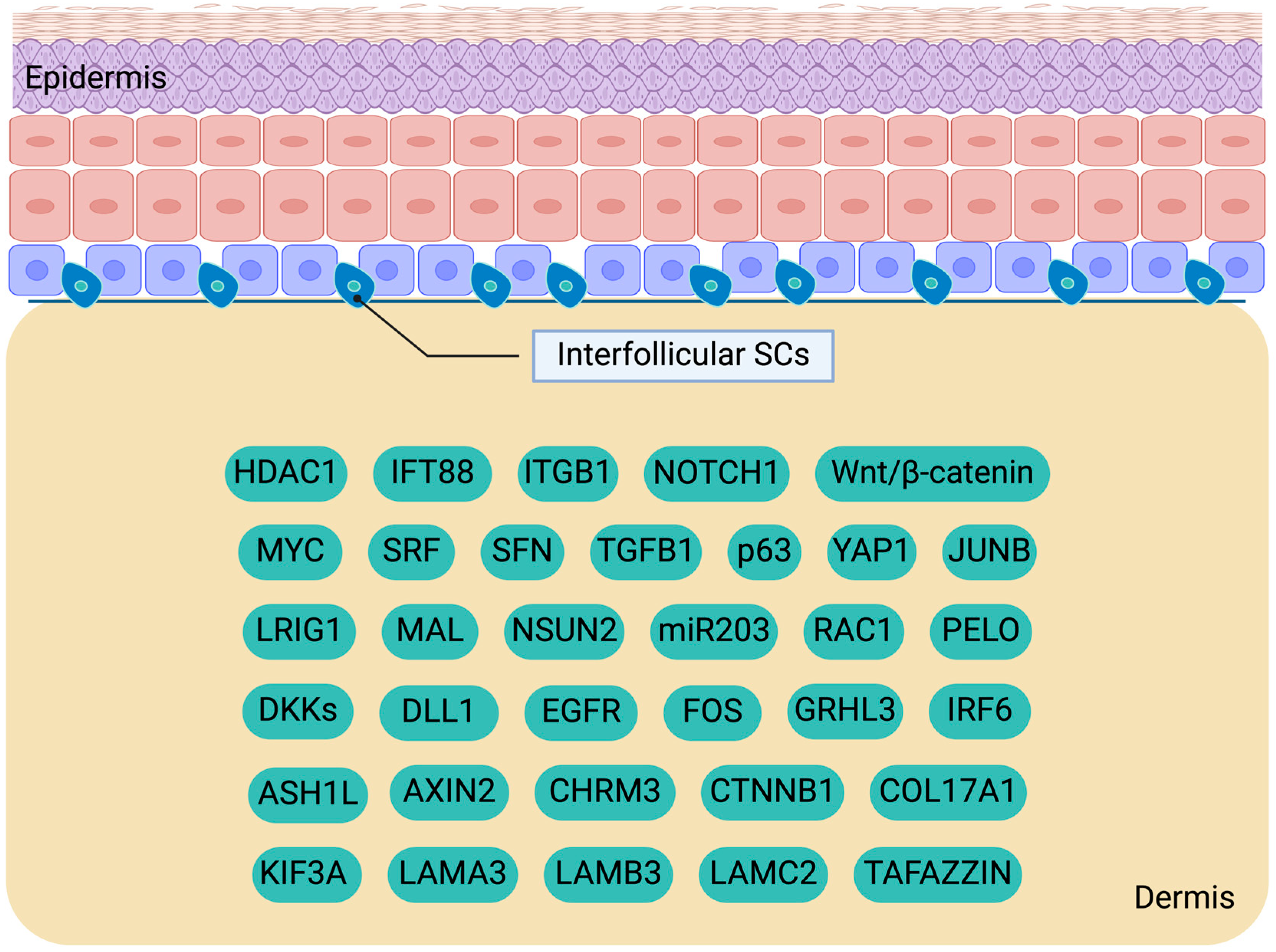
2. Investigating Interfollicular Epidermal SCs: Key Players in Skin Homeostasis
2.1. Interfollicular Epidermal SC Proliferation and Differentiation
2.2. Molecular Cues Regulating IFESCs
3. Insights into Hair Follicle SCs: Bulge and Hair Germ Dynamics
3.1. Identification and Characteristics of Hair Follicle SCs
3.2. Molecular Mechanisms Governing Activation and Quiescence of HFSCs
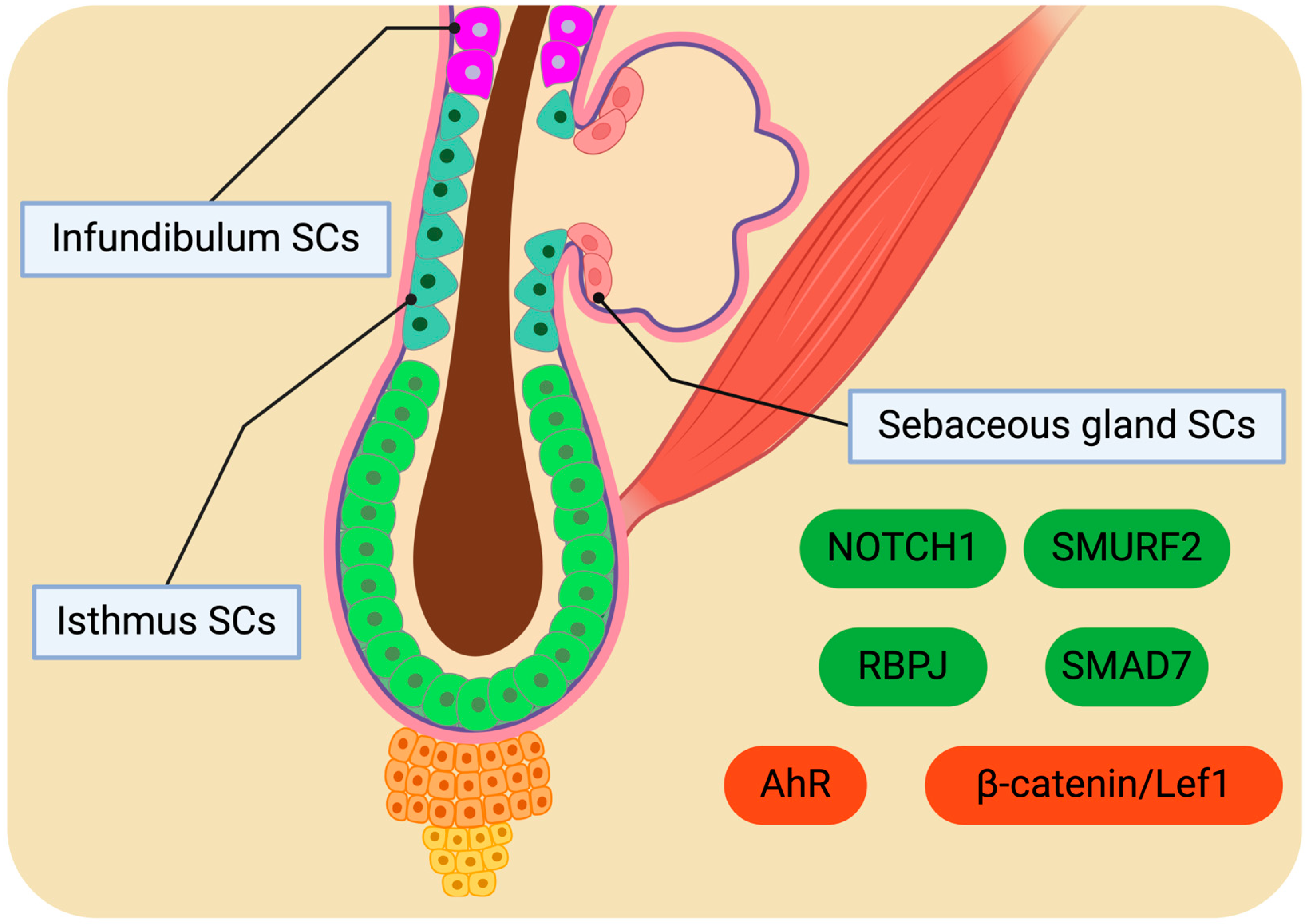
4. Potential of Isthmus, Infundibulum, and Sebaceous Gland SCs for Homeostatic Regulation
4.1. Distinctive SC Populations
4.2. Molecular Drivers Orchestrating SC Behaviour
5. Molecular Profile and Modulation of Sweat Gland SCs
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 14-3-3σ | 14-3-3 protein sigma |
| 3D | Three-dimensional |
| ACDs | Asymmetric cell divisions |
| AhR | Aryl hydrocarbon receptor |
| aPKC | Atypical protein kinase C |
| ASH1L | ASH1-like histone lysine methyltransferase |
| BLIMP1 | B lymphocyte-induced maturation protein 1 |
| BMI-1 | B cell-specific Moloney murine leukemia virus integration site 1 |
| BMP | Bone morphogenetic protein |
| BMPR1A | Bone morphogenetic protein receptor type 1A |
| BuSCs | Bulge stem cells |
| CD29 | Cluster of differentiation 29 |
| CD34 | Cluster of differentiation 34 |
| CERS4 | Ceramide synthase 4 |
| CHRM3 | Cholinergic receptor muscarinic 3 |
| COL17 | Type XVII collagen |
| CTHRC1 | Collagen triple helix repeat containing 1 |
| CYP1A1 | Cytochrome P450 family 1 subfamily A member 1 |
| DKKs | Dickkopf Wnt signaling pathway inhibitors |
| DNA | Deoxyribonucleic acid |
| DP | Dermal papilla |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial-to-mesenchymal transition |
| ENG | Endoglin |
| EPUs | Epidermal proliferative units |
| EzH2 | Enhancer of zeste homolog 2 |
| FGF | Fibroblast growth factor |
| FOS | Fos proto-oncogene, AP-1 transcription factor subunit |
| FOXC1 | Forkhead box protein C1 |
| FOXI3 | Forkead box I3 |
| FOXP1 | Forkhead box protein P1 |
| GATA6 | GATA binding protein 6 |
| GRHL3 | Grainyhead-like transcription factor 3 |
| GTP | Guanosine triphosphate |
| H2B-GFP | Histone 2B-green fluorescent protein |
| HAP | Hair follicle-associated pluripotent |
| HES1 | Hairy/enhancer of split-1 |
| HF | Hair follicle |
| HFSCs | Hair follicle stem cells |
| HG | Hair germ |
| HGSCs | Hair germ stem cells |
| HMOX | Heme oxygenase |
| IFE | Interfollicular epidermis |
| IFT88 | Intraflagellar transport protein 88 homolog |
| IRF6 | Interferon regulatory factor 6 |
| IRS | Inner root sheath |
| Iα6 | Integrin alpha-5 subunit |
| JUNB | Transcription factor jun-B |
| K | Keratin |
| K15-GFP | K15 promoter-driven green fluorescent protein |
| KIF3A | Kinesin family member 3A |
| LEF1 | Lymphoid enhancer-binding factor 1 |
| LGR | Leucine-rich repeat containing G protein-coupled receptor |
| LHX2 | LIM homeobox 2 |
| lncRNAs | Long non-coding RNA |
| LR | Label-retaining |
| LRIG1 | Leucine-rich repeats and immunoglobulin-like domains 1 |
| LRP6 | Low-density lipoprotein receptor-related protein 6 |
| MAL | Myelin and lymphocyte protein |
| miRNA | Micro ribonucleic acid |
| mRNA | Messenger ribonucleic acid |
| MSI2 | Musashi RNA binding protein 2 |
| mTOR | Mammalian target of rapamycin |
| NFATC1 | Nuclear factor of activated T cells 1 |
| non-LR | Non-label-retaining |
| NOTCH1 | Neurogenic locus notch homolog protein 1 |
| ORS | Outer root sheath |
| OVOL2 | Ovo like zinc finger 2 |
| P450 | Cytochrome P450 |
| PAR | Protease-activated receptor |
| PCR | Polymerase chain reaction |
| PELO | Pelota mRNA surveillance and ribosome rescue factor |
| PI3K/Akt | Phosphoinositide-3-kinase–protein kinase B/Akt |
| PLET1 | Placenta expressed transcript 1 |
| PlncRNA-1 | Prostate cancer-upregulated long non-coding RNA 1 |
| PORCN | Porcupine O-acyltransferase |
| PRC1 | Protein regulator of cytokinesis 1 |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 |
| RBPJ | Recombination signal binding protein for immunoglobulin kappa J region |
| RNA | Ribonucleic acid |
| rRNA | Ribosomal ribonucleic acid |
| RUNX1 | Runt-related transcription factor 1 |
| SCA-1 | Stem cell antigen 1 |
| SCD1 | Stearoyl-CoA desaturase 1 |
| SCDS | Symmetric cell divisions |
| SCs | Stem cells |
| SFN | Stratifin |
| SFRP1 | Secreted frizzled-related protein 1 |
| SG | Sebaceous gland |
| SGSCs | Sebaceous gland stem cells |
| SHH | Sonic hedgehog |
| SIRT7 | Sirtuin 7 |
| SLC1A3 | Solute carrier family 1 member 3 |
| SMAD1 | Mothers against decapentaplegic homolog 1 |
| SMAD4 | Mothers against decapentaplegic homolog 4 |
| SMAD7 | Mothers against decapentaplegic homolog 7 |
| SMURF2 | SMAD ubiquitination regulatory factor 2 |
| SOAT1 | Sterol O-acyltransferase |
| SOX9 | SRY-box transcription factor 9 |
| sPLA2-IIA | Secreted phospholipase A2 type IIA |
| SRF | Serum response factor |
| SwD | Sweat duct |
| SwGs | Sweat glands |
| SwGSCs | Sweat gland stem cells |
| TACs | Transit-amplifying cells |
| TARBP2 | TAR (HIV) RNA-binding protein 2 |
| TAZ | Tafazzin |
| TCDD | 2,3,7,8-Tetrachlorodibenzo-p-dioxin |
| TCF3 | Transcription factor 3 |
| TCF4 | Transcription factor 4 |
| Tet-o-H2B-GFP | “Tet On” regulated expression of the H2B-GFP fusion protein |
| TGFβ | Transforming growth factor β |
| YAP | Yes-associated protein |
| ΔΝp63α | Delta Np63 alpha |
References
- Wysocki, A.B. Skin Anatomy, Physiology, and Pathophysiology. Nurs. Clin. N. Am. 1999, 34, 777–797. [Google Scholar] [CrossRef]
- Gilaberte, Y.; Prieto-Torres, L.; Pastushenko, I.; Juarranz, Á. Chapter 1—Anatomy and Function of the Skin. In Nanoscience in Dermatology; Hamblin, M.R., Avci, P., Prow, T.W., Eds.; Academic Press: Boston, FL, USA, 2016; pp. 1–14. ISBN 978-0-12-802926-8. [Google Scholar]
- Castellano-Pellicena, I.; Morrison, C.G.; Bell, M.; O’Connor, C.; Tobin, D.J. Melanin Distribution in Human Skin: Influence of Cytoskeletal, Polarity, and Centrosome-Related Machinery of Stratum Basale Keratinocytes. Int. J. Mol. Sci. 2021, 22, 3143. [Google Scholar] [CrossRef] [PubMed]
- Cichorek, M.; Wachulska, M.; Stasiewicz, A.; Tymińska, A. Skin Melanocytes: Biology and Development. Adv. Dermatol. Allergol. Dermatol. Alergol. 2013, 30, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.; Mathew, S. Merkel Cells: A Collective Review of Current Concepts. Int. J. Appl. Basic Med. Res. 2019, 9, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ojeda, W.; Pandey, A.; Alhajj, M.; Oakley, A.M. Anatomy, Skin (Integument). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Brown, T.M.; Krishnamurthy, K. Histology, Dermis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Ibrahim, A.A.E.; Bagherani, N.; Smoller, B.; Bagherani, N.; Reyes-Barron, C. Anatomy and Organization of Human Skin. In Atlas of Dermatology, Dermatopathology and Venereology: Cutaneous Anatomy, Biology and Inherited Disorders and General Dermatologic Concepts; Smoller, B., Bagherani, N., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 109–132. ISBN 978-3-319-53811-2. [Google Scholar]
- Valle, M.; Zamorani, M.P. Skin and Subcutaneous Tissue. In Ultrasound of the Musculoskeletal System; Bianchi, S., Martinoli, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 19–43. ISBN 978-3-540-28163-4. [Google Scholar]
- Jin, R.; Luo, L.; Zheng, J. The Trinity of Skin: Skin Homeostasis as a Neuro–Endocrine–Immune Organ. Life 2022, 12, 725. [Google Scholar] [CrossRef]
- Jiao, Q.; Zhi, L.; You, B.; Wang, G.; Wu, N.; Jia, Y. Skin Homeostasis: Mechanism and Influencing Factors. J. Cosmet. Dermatol. 2024, 23, 1518–1526. [Google Scholar] [CrossRef] [PubMed]
- Dekoninck, S.; Blanpain, C. Stem Cell Dynamics, Migration and Plasticity during Wound Healing. Nat. Cell Biol. 2019, 21, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Mokry, J.; Pisal, R. Development and Maintenance of Epidermal Stem Cells in Skin Adnexa. Int. J. Mol. Sci. 2020, 21, 9736. [Google Scholar] [CrossRef]
- Morita, R.; Sanzen, N.; Sasaki, H.; Hayashi, T.; Umeda, M.; Yoshimura, M.; Yamamoto, T.; Shibata, T.; Abe, T.; Kiyonari, H.; et al. Tracing the Origin of Hair Follicle Stem Cells. Nature 2021, 594, 547–552. [Google Scholar] [CrossRef]
- Kaur, P. Interfollicular Epidermal Stem Cells: Identification, Challenges, Potential. J. Investig. Dermatol. 2006, 126, 1450–1458. [Google Scholar] [CrossRef]
- Morris, R.J.; Liu, Y.; Marles, L.; Yang, Z.; Trempus, C.; Li, S.; Lin, J.S.; Sawicki, J.A.; Cotsarelis, G. Capturing and Profiling Adult Hair Follicle Stem Cells. Nat. Biotechnol. 2004, 22, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Benitah, S.A.; Frye, M. Stem Cells in Ectodermal Development. J. Mol. Med. 2012, 90, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Dermitzakis, I.; Manthou, M.E.; Meditskou, S.; Miliaras, D.; Kesidou, E.; Boziki, M.; Petratos, S.; Grigoriadis, N.; Theotokis, P. Developmental Cues and Molecular Drivers in Myelinogenesis: Revisiting Early Life to Re-Evaluate the Integrity of CNS Myelin. Curr. Issues Mol. Biol. 2022, 44, 3208–3237. [Google Scholar] [CrossRef] [PubMed]
- Dermitzakis, I.; Manthou, M.E.; Meditskou, S.; Tremblay, M.-È.; Petratos, S.; Zoupi, L.; Boziki, M.; Kesidou, E.; Simeonidou, C.; Theotokis, P. Origin and Emergence of Microglia in the CNS—An Interesting (Hi)Story of an Eccentric Cell. Curr. Issues Mol. Biol. 2023, 45, 2609–2628. [Google Scholar] [CrossRef]
- Dermitzakis, I.; Theotokis, P.; Evangelidis, P.; Delilampou, E.; Evangelidis, N.; Chatzisavvidou, A.; Avramidou, E.; Manthou, M.E. CNS Border-Associated Macrophages: Ontogeny and Potential Implication in Disease. Curr. Issues Mol. Biol. 2023, 45, 4285–4300. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, H.; Duan, E. Epidermal Development in Mammals: Key Regulators, Signals from beneath, and Stem Cells. Int. J. Mol. Sci. 2013, 14, 10869–10895. [Google Scholar] [CrossRef] [PubMed]
- Sotiropoulou, P.A.; Blanpain, C. Development and Homeostasis of the Skin Epidermis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008383. [Google Scholar] [CrossRef]
- Chen, M.; Przyborowski, M.; Berthiaume, F. Stem Cells for Skin Tissue Engineering and Wound Healing. Crit. Rev. Biomed. Eng. 2009, 37, 399–421. [Google Scholar] [CrossRef]
- Jin, Y.; Li, S.; Yu, Q.; Chen, T.; Liu, D. Application of Stem Cells in Regeneration Medicine. MedComm 2023, 4, e291. [Google Scholar] [CrossRef]
- Piccione, M.; Di Liddo, R. Skin Stem Cells in Cancer. In Cancer Stem Cells: New Horizons in Cancer Therapies; Pathak, S., Banerjee, A., Eds.; Springer: Singapore, 2020; pp. 111–124. ISBN 9789811551208. [Google Scholar]
- Yang, R.; Wang, J.; Chen, X.; Shi, Y.; Xie, J. Epidermal Stem Cells in Wound Healing and Regeneration. Stem Cells Int. 2020, 2020, 9148310. [Google Scholar] [CrossRef]
- Ogliari, K.S.; Marinowic, D.; Brum, D.E.; Loth, F. Stem Cells in Dermatology. An. Bras. Dermatol. 2014, 89, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Khandpur, S.; Gupta, S.; Gunaabalaji, D.R. Stem Cell Therapy in Dermatology. Indian J. Dermatol. Venereol. Leprol. 2021, 87, 753–767. [Google Scholar] [CrossRef]
- Blanpain, C.; Fuchs, E. Epidermal Homeostasis: A Balancing Act of Stem Cells in the Skin. Nat. Rev. Mol. Cell Biol. 2009, 10, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.L.; Patel, D.M.; Green, K.J. Deconstructing the Skin: Cytoarchitectural Determinants of Epidermal Morphogenesis. Nat. Rev. Mol. Cell Biol. 2011, 12, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.H.; Harper, S.; Watt, F.M. Stem Cell Patterning and Fate in Human Epidermis. Cell 1995, 80, 83–93. [Google Scholar] [CrossRef]
- Jensen, U.B.; Lowell, S.; Watt, F.M. The Spatial Relationship between Stem Cells and Their Progeny in the Basal Layer of Human Epidermis: A New View Based on Whole-Mount Labelling and Lineage Analysis. Dev. Camb. Engl. 1999, 126, 2409–2418. [Google Scholar] [CrossRef] [PubMed]
- Koster, M.I. Making an Epidermis. Ann. N. Y. Acad. Sci. 2009, 1170, 7–10. [Google Scholar] [CrossRef]
- Mackenzie, I.C. Retroviral Transduction of Murine Epidermal Stem Cells Demonstrates Clonal Units of Epidermal Structure. J. Investig. Dermatol. 1997, 109, 377–383. [Google Scholar] [CrossRef]
- Clayton, E.; Doupé, D.P.; Klein, A.M.; Winton, D.J.; Simons, B.D.; Jones, P.H. A Single Type of Progenitor Cell Maintains Normal Epidermis. Nature 2007, 446, 185–189. [Google Scholar] [CrossRef]
- Mascré, G.; Dekoninck, S.; Drogat, B.; Youssef, K.K.; Broheé, S.; Sotiropoulou, P.A.; Simons, B.D.; Blanpain, C. Distinct Contribution of Stem and Progenitor Cells to Epidermal Maintenance. Nature 2012, 489, 257–262. [Google Scholar] [CrossRef]
- Potten, C.S. The Epidermal Proliferative Unit: The Possible Role of the Central Basal Cell. Cell Tissue Kinet. 1974, 7, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Barrandon, Y.; Green, H. Three Clonal Types of Keratinocyte with Different Capacities for Multiplication. Proc. Natl. Acad. Sci. USA 1987, 84, 2302–2306. [Google Scholar] [CrossRef]
- Alonso, L.; Fuchs, E. Stem Cells of the Skin Epithelium. Proc. Natl. Acad. Sci. USA 2003, 100 (Suppl. S1), 11830–11835. [Google Scholar] [CrossRef] [PubMed]
- Ro, S.; Rannala, B. A Stop-EGFP Transgenic Mouse to Detect Clonal Cell Lineages Generated by Mutation. EMBO Rep. 2004, 5, 914–920. [Google Scholar] [CrossRef]
- Piedrafita, G.; Kostiou, V.; Wabik, A.; Colom, B.; Fernandez-Antoran, D.; Herms, A.; Murai, K.; Hall, B.A.; Jones, P.H. A Single-Progenitor Model as the Unifying Paradigm of Epidermal and Esophageal Epithelial Maintenance in Mice. Nat. Commun. 2020, 11, 1429. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.J.; Fischer, S.M.; Slaga, T.J. Evidence That the Centrally and Peripherally Located Cells in the Murine Epidermal Proliferative Unit Are Two Distinct Cell Populations. J. Investig. Dermatol. 1985, 84, 277–281. [Google Scholar] [CrossRef]
- Cotsarelis, G.; Cheng, S.Z.; Dong, G.; Sun, T.T.; Lavker, R.M. Existence of Slow-Cycling Limbal Epithelial Basal Cells That Can Be Preferentially Stimulated to Proliferate: Implications on Epithelial Stem Cells. Cell 1989, 57, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Cotsarelis, G.; Sun, T.T.; Lavker, R.M. Label-Retaining Cells Reside in the Bulge Area of Pilosebaceous Unit: Implications for Follicular Stem Cells, Hair Cycle, and Skin Carcinogenesis. Cell 1990, 61, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Laurenti, E.; Oser, G.; van der Wath, R.C.; Blanco-Bose, W.; Jaworski, M.; Offner, S.; Dunant, C.F.; Eshkind, L.; Bockamp, E.; et al. Hematopoietic Stem Cells Reversibly Switch from Dormancy to Self-Renewal during Homeostasis and Repair. Cell 2008, 135, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Bonaguidi, M.A.; Wheeler, M.A.; Shapiro, J.S.; Stadel, R.P.; Sun, G.J.; Ming, G.; Song, H. In Vivo Clonal Analysis Reveals Self-Renewing and Multipotent Adult Neural Stem Cell Characteristics. Cell 2011, 145, 1142–1155. [Google Scholar] [CrossRef]
- Rocheteau, P.; Gayraud-Morel, B.; Siegl-Cachedenier, I.; Blasco, M.A.; Tajbakhsh, S. A Subpopulation of Adult Skeletal Muscle Stem Cells Retains All Template DNA Strands after Cell Division. Cell 2012, 148, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Sada, A.; Jacob, F.; Leung, E.; Wang, S.; White, B.S.; Shalloway, D.; Tumbar, T. Defining the Cellular Lineage Hierarchy in the Interfollicular Epidermis of Adult Skin. Nat. Cell Biol. 2016, 18, 619–631. [Google Scholar] [CrossRef]
- Paquet-Fifield, S.; Schlüter, H.; Li, A.; Aitken, T.; Gangatirkar, P.; Blashki, D.; Koelmeyer, R.; Pouliot, N.; Palatsides, M.; Ellis, S.; et al. A Role for Pericytes as Microenvironmental Regulators of Human Skin Tissue Regeneration. J. Clin. Investig. 2009, 119, 2795–2806. [Google Scholar] [CrossRef] [PubMed]
- Lechler, T.; Fuchs, E. Asymmetric Cell Divisions Promote Stratification and Differentiation of Mammalian Skin. Nature 2005, 437, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Rompolas, P.; Mesa, K.R.; Kawaguchi, K.; Park, S.; Gonzalez, D.; Brown, S.; Boucher, J.; Klein, A.M.; Greco, V. Spatiotemporal Coordination of Stem Cell Commitment during Epidermal Homeostasis. Science 2016, 352, 1471–1474. [Google Scholar] [CrossRef]
- Jones, P.H.; Watt, F.M. Separation of Human Epidermal Stem Cells from Transit Amplifying Cells on the Basis of Differences in Integrin Function and Expression. Cell 1993, 73, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Stone, M.G.; Simpson, C.; Reynolds, L.E.; Marshall, J.F.; Hart, I.R.; Hodivala-Dilke, K.M.; Eady, R.A.J. Desmosomal Proteins, Including Desmoglein 3, Serve as Novel Negative Markers for Epidermal Stem Cell-Containing Population of Keratinocytes. J. Cell Sci. 2003, 116, 4239–4248. [Google Scholar] [CrossRef]
- Brakebusch, C.; Grose, R.; Quondamatteo, F.; Ramirez, A.; Jorcano, J.L.; Pirro, A.; Svensson, M.; Herken, R.; Sasaki, T.; Timpl, R.; et al. Skin and Hair Follicle Integrity Is Crucially Dependent on Β1 Integrin Expression on Keratinocytes. EMBO J. 2000, 19, 3990–4003. [Google Scholar] [CrossRef] [PubMed]
- Piwko-Czuchra, A.; Koegel, H.; Meyer, H.; Bauer, M.; Werner, S.; Brakebusch, C.; Fässler, R. Beta1 Integrin-Mediated Adhesion Signalling Is Essential for Epidermal Progenitor Cell Expansion. PLoS ONE 2009, 4, e5488. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Liu, Y.; Yang, Y.; Yan, Y.; Kim, A.J.; Guo, C.; Fisher, G.J.; Quan, T. Reduced Expression of Collagen 17A1 in Naturally Aged, Photoaged, and UV-Irradiated Human Skin In Vivo: Potential Links to Epidermal Aging. J. Cell Commun. Signal. 2022, 16, 421–432. [Google Scholar] [CrossRef]
- Watanabe, M.; Natsuga, K.; Nishie, W.; Kobayashi, Y.; Donati, G.; Suzuki, S.; Fujimura, Y.; Tsukiyama, T.; Ujiie, H.; Shinkuma, S.; et al. Type XVII Collagen Coordinates Proliferation in the Interfollicular Epidermis. eLife 2017, 6, e26635. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Matsumura, H.; Kato, T.; Ichinose, S.; Takada, A.; Namiki, T.; Asakawa, K.; Morinaga, H.; Mohri, Y.; De Arcangelis, A.; et al. Stem Cell Competition Orchestrates Skin Homeostasis and Ageing. Nature 2019, 568, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Connelly, J.T.; Gautrot, J.E.; Trappmann, B.; Tan, D.W.-M.; Donati, G.; Huck, W.T.S.; Watt, F.M. Actin and Serum Response Factor Transduce Physical Cues from the Microenvironment to Regulate Epidermal Stem Cell Fate Decisions. Nat. Cell Biol. 2010, 12, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Nanba, D.; Toki, F.; Matsushita, N.; Matsushita, S.; Higashiyama, S.; Barrandon, Y. Actin Filament Dynamics Impacts Keratinocyte Stem Cell Maintenance. EMBO Mol. Med. 2013, 5, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Anvarian, Z.; Mykytyn, K.; Mukhopadhyay, S.; Pedersen, L.B.; Christensen, S.T. Cellular Signalling by Primary Cilia in Development, Organ Function and Disease. Nat. Rev. Nephrol. 2019, 15, 199–219. [Google Scholar] [CrossRef] [PubMed]
- Croyle, M.J.; Lehman, J.M.; O’Connor, A.K.; Wong, S.Y.; Malarkey, E.B.; Iribarne, D.; Dowdle, W.E.; Schoeb, T.R.; Verney, Z.M.; Athar, M.; et al. Role of Epidermal Primary Cilia in the Homeostasis of Skin and Hair Follicles. Dev. Camb. Engl. 2011, 138, 1675–1685. [Google Scholar] [CrossRef]
- Lowell, S.; Jones, P.; Le Roux, I.; Dunne, J.; Watt, F.M. Stimulation of Human Epidermal Differentiation by Delta-Notch Signalling at the Boundaries of Stem-Cell Clusters. Curr. Biol. 2000, 10, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Lim, X.; Tan, S.H.; Koh, W.L.C.; Chau, R.M.W.; Yan, K.S.; Kuo, C.J.; van Amerongen, R.; Klein, A.M.; Nusse, R. Interfollicular Epidermal Stem Cells Self-Renew via Autocrine Wnt Signaling. Science 2013, 342, 1226–1230. [Google Scholar] [CrossRef]
- Zhu, A.J.; Watt, F.M. Beta-Catenin Signalling Modulates Proliferative Potential of Human Epidermal Keratinocytes Independently of Intercellular Adhesion. Dev. Camb. Engl. 1999, 126, 2285–2298. [Google Scholar] [CrossRef]
- Lin, Z.; Jin, S.; Chen, J.; Li, Z.; Lin, Z.; Tang, L.; Nie, Q.; Andersen, B. Murine Interfollicular Epidermal Differentiation Is Gradualistic with GRHL3 Controlling Progression from Stem to Transition Cell States. Nat. Commun. 2020, 11, 5434. [Google Scholar] [CrossRef]
- Huelsken, J.; Vogel, R.; Erdmann, B.; Cotsarelis, G.; Birchmeier, W. Beta-Catenin Controls Hair Follicle Morphogenesis and Stem Cell Differentiation in the Skin. Cell 2001, 105, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Merrill, B.J.; Gat, U.; DasGupta, R.; Fuchs, E. Tcf3 and Lef1 Regulate Lineage Differentiation of Multipotent Stem Cells in Skin. Genes Dev. 2001, 15, 1688–1705. [Google Scholar] [CrossRef] [PubMed]
- Barrott, J.J.; Cash, G.M.; Smith, A.P.; Barrow, J.R.; Murtaugh, L.C. Deletion of Mouse Porcn Blocks Wnt Ligand Secretion and Reveals an Ectodermal Etiology of Human Focal Dermal Hypoplasia/Goltz Syndrome. Proc. Natl. Acad. Sci. USA 2011, 108, 12752–12757. [Google Scholar] [CrossRef] [PubMed]
- Lim, X.; Nusse, R. Wnt Signaling in Skin Development, Homeostasis, and Disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a008029. [Google Scholar] [CrossRef] [PubMed]
- Niemann, C.; Watt, F.M. Designer Skin: Lineage Commitment in Postnatal Epidermis. Trends Cell Biol. 2002, 12, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Niemann, C.; Owens, D.M.; Hülsken, J.; Birchmeier, W.; Watt, F.M. Expression of DeltaNLef1 in Mouse Epidermis Results in Differentiation of Hair Follicles into Squamous Epidermal Cysts and Formation of Skin Tumours. Dev. Camb. Engl. 2002, 129, 95–109. [Google Scholar] [CrossRef]
- Silva-Vargas, V.; Lo Celso, C.; Giangreco, A.; Ofstad, T.; Prowse, D.M.; Braun, K.M.; Watt, F.M. Beta-Catenin and Hedgehog Signal Strength Can Specify Number and Location of Hair Follicles in Adult Epidermis without Recruitment of Bulge Stem Cells. Dev. Cell 2005, 9, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Yang, Z.; Andl, T.; Cui, C.; Kim, N.; Millar, S.E.; Cotsarelis, G. Wnt-Dependent de Novo Hair Follicle Regeneration in Adult Mouse Skin after Wounding. Nature 2007, 447, 316–320. [Google Scholar] [CrossRef]
- Gandarillas, A.; Watt, F.M. C-Myc Promotes Differentiation of Human Epidermal Stem Cells. Genes Dev. 1997, 11, 2869–2882. [Google Scholar] [CrossRef]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of C-MYC as a Target of the APC Pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef]
- Jensen, K.B.; Watt, F.M. Single-Cell Expression Profiling of Human Epidermal Stem and Transit-Amplifying Cells: Lrig1 Is a Regulator of Stem Cell Quiescence. Proc. Natl. Acad. Sci. USA 2006, 103, 11958–11963. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Watt, F.M. The RNA Methyltransferase Misu (NSun2) Mediates Myc-Induced Proliferation and Is Upregulated in Tumors. Curr. Biol. 2006, 16, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Watt, F.M.; Frye, M.; Benitah, S.A. Myc in Mammalian Epidermis. Nat. Rev. Cancer 2008, 8, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Waikel, R.L.; Kawachi, Y.; Waikel, P.A.; Wang, X.J.; Roop, D.R. Deregulated Expression of C-Myc Depletes Epidermal Stem Cells. Nat. Genet. 2001, 28, 165–168. [Google Scholar] [CrossRef]
- Frye, M.; Gardner, C.; Li, E.R.; Arnold, I.; Watt, F.M. Evidence That Myc Activation Depletes the Epidermal Stem Cell Compartment by Modulating Adhesive Interactions with the Local Microenvironment. Dev. Camb. Engl. 2003, 130, 2793–2808. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Ye, Z.; Shi, C.; Sun, L.; Han, M.; Zhuang, Y.; Xu, T.; Zhao, S.; Wu, X. The Histone Methyltransferase Ash1l Is Required for Epidermal Homeostasis in Mice. Sci. Rep. 2017, 7, 45401. [Google Scholar] [CrossRef]
- Shin, J.-W.; Choi, H.-R.; Nam, K.-M.; Lee, H.-S.; Kim, S.-A.; Joe, H.-J.; Kazumi, T.; Park, K.-C. The Co-Expression Pattern of P63 and HDAC1: A Potential Way to Disclose Stem Cells in Interfollicular Epidermis. Int. J. Mol. Sci. 2017, 18, 1360. [Google Scholar] [CrossRef]
- Koster, M.I.; Roop, D.R. The Role of P63 in Development and Differentiation of the Epidermis. J. Dermatol. Sci. 2004, 34, 3–9. [Google Scholar] [CrossRef]
- Lena, A.M.; Shalom-Feuerstein, R.; Rivetti di Val Cervo, P.; Aberdam, D.; Knight, R.A.; Melino, G.; Candi, E. MiR-203 Represses “stemness” by Repressing DeltaNp63. Cell Death Differ. 2008, 15, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Koster, M.I.; Kim, S.; Roop, D.R. P63 Deficiency: A Failure of Lineage Commitment or Stem Cell Maintenance? J. Investig. Dermatol. Symp. Proc. 2005, 10, 118–123. [Google Scholar] [CrossRef]
- Hammond, N.L.; Headon, D.J.; Dixon, M.J. The Cell-Cycle Regulator Protein 14-3-3σ Is Essential for Hair Follicle Integrity and Epidermal Homeostasis. J. Investig. Dermatol. 2012, 132, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Elbediwy, A.; Vincent-Mistiaen, Z.I.; Spencer-Dene, B.; Stone, R.K.; Boeing, S.; Wculek, S.K.; Cordero, J.; Tan, E.H.; Ridgway, R.; Brunton, V.G.; et al. Integrin Signalling Regulates YAP and TAZ to Control Skin Homeostasis. Dev. Camb. Engl. 2016, 143, 1674–1687. [Google Scholar] [CrossRef]
- De Rosa, L.; Secone Seconetti, A.; De Santis, G.; Pellacani, G.; Hirsch, T.; Rothoeft, T.; Teig, N.; Pellegrini, G.; Bauer, J.W.; De Luca, M. Laminin 332-Dependent YAP Dysregulation Depletes Epidermal Stem Cells in Junctional Epidermolysis Bullosa. Cell Rep. 2019, 27, 2036–2049.e6. [Google Scholar] [CrossRef]
- Richardson, R.J.; Dixon, J.; Malhotra, S.; Hardman, M.J.; Knowles, L.; Boot-Handford, R.P.; Shore, P.; Whitmarsh, A.; Dixon, M.J. Irf6 Is a Key Determinant of the Keratinocyte Proliferation-Differentiation Switch. Nat. Genet. 2006, 38, 1329–1334. [Google Scholar] [CrossRef] [PubMed]
- Liakath-Ali, K.; Mills, E.W.; Sequeira, I.; Lichtenberger, B.M.; Pisco, A.O.; Sipilä, K.H.; Mishra, A.; Yoshikawa, H.; Wu, C.C.-C.; Ly, T.; et al. An Evolutionarily Conserved Ribosome-Rescue Pathway Maintains Epidermal Homeostasis. Nature 2018, 556, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Miura, H.; Tanemura, A.; Kobayashi, K.; Kondoh, G.; Sano, S.; Ozawa, K.; Inui, S.; Nakata, A.; Takagi, T.; et al. Targeted Disruption of LIG-1 Gene Results in Psoriasiform Epidermal Hyperplasia. FEBS Lett. 2002, 521, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.B.; Collins, C.A.; Nascimento, E.; Tan, D.W.; Frye, M.; Itami, S.; Watt, F.M. Lrig1 Expression Defines a Distinct Multipotent Stem Cell Population in Mammalian Epidermis. Cell Stem Cell 2009, 4, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.K.; Tomić-Canić, M.; Lucas, D.J.; Simon, M.; Blumenberg, M. TGF Beta Promotes the Basal Phenotype of Epidermal Keratinocytes: Transcriptional Induction of K#5 and K#14 Keratin Genes. Growth Factors 1995, 12, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Grando, C.; Liu, S.; Chernyavsky, A.; Chen, J.K.; Andersen, B.; Grando, S.A. The M3 Muscarinic Acetylcholine Receptor Promotes Epidermal Differentiation. J. Investig. Dermatol. 2022, 142, 3211–3221.e2. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 14 June 2024).
- Garcin, C.L.; Ansell, D.M. The Battle of the Bulge: Re-Evaluating Hair Follicle Stem Cells in Wound Repair. Exp. Dermatol. 2017, 26, 101–104. [Google Scholar] [CrossRef]
- Topouzi, H.; Logan, N.J.; Williams, G.; Higgins, C.A. Methods for the Isolation and 3D Culture of Dermal Papilla Cells from Human Hair Follicles. Exp. Dermatol. 2017, 26, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Greco, V.; Chen, T.; Rendl, M.; Schober, M.; Pasolli, H.A.; Stokes, N.; Dela Cruz-Racelis, J.; Fuchs, E. A Two-Step Mechanism for Stem Cell Activation during Hair Regeneration. Cell Stem Cell 2009, 4, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Bickenbach, J.R. Identification and Behavior of Label-Retaining Cells in Oral Mucosa and Skin. J. Dent. Res. 1981, 60, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Blanpain, C.; Lowry, W.E.; Geoghegan, A.; Polak, L.; Fuchs, E. Self-Renewal, Multipotency, and the Existence of Two Cell Populations within an Epithelial Stem Cell Niche. Cell 2004, 118, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Trempus, C.S.; Morris, R.J.; Bortner, C.D.; Cotsarelis, G.; Faircloth, R.S.; Reece, J.M.; Tennant, R.W. Enrichment for Living Murine Keratinocytes from the Hair Follicle Bulge with the Cell Surface Marker CD34. J. Investig. Dermatol. 2003, 120, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Oshima, H.; Rochat, A.; Kedzia, C.; Kobayashi, K.; Barrandon, Y. Morphogenesis and Renewal of Hair Follicles from Adult Multipotent Stem Cells. Cell 2001, 104, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Rochat, A.; Kobayashi, K.; Barrandon, Y. Location of Stem Cells of Human Hair Follicles by Clonal Analysis. Cell 1994, 76, 1063–1073. [Google Scholar] [CrossRef]
- Claudinot, S.; Nicolas, M.; Oshima, H.; Rochat, A.; Barrandon, Y. Long-Term Renewal of Hair Follicles from Clonogenic Multipotent Stem Cells. Proc. Natl. Acad. Sci. USA 2005, 102, 14677–14682. [Google Scholar] [CrossRef] [PubMed]
- Youssef, K.K.; Van Keymeulen, A.; Lapouge, G.; Beck, B.; Michaux, C.; Achouri, Y.; Sotiropoulou, P.A.; Blanpain, C. Identification of the Cell Lineage at the Origin of Basal Cell Carcinoma. Nat. Cell Biol. 2010, 12, 299–305. [Google Scholar] [CrossRef]
- Jaks, V.; Barker, N.; Kasper, M.; van Es, J.H.; Snippert, H.J.; Clevers, H.; Toftgård, R. Lgr5 Marks Cycling, yet Long-Lived, Hair Follicle Stem Cells. Nat. Genet. 2008, 40, 1291–1299. [Google Scholar] [CrossRef]
- Tumbar, T.; Guasch, G.; Greco, V.; Blanpain, C.; Lowry, W.E.; Rendl, M.; Fuchs, E. Defining the Epithelial Stem Cell Niche in Skin. Science 2004, 303, 359–363. [Google Scholar] [CrossRef]
- Ito, M.; Liu, Y.; Yang, Z.; Nguyen, J.; Liang, F.; Morris, R.J.; Cotsarelis, G. Stem Cells in the Hair Follicle Bulge Contribute to Wound Repair but Not to Homeostasis of the Epidermis. Nat. Med. 2005, 11, 1351–1354. [Google Scholar] [CrossRef] [PubMed]
- Levy, V.; Lindon, C.; Zheng, Y.; Harfe, B.D.; Morgan, B.A. Epidermal Stem Cells Arise from the Hair Follicle after Wounding. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2007, 21, 1358–1366. [Google Scholar] [CrossRef] [PubMed]
- Levy, V.; Lindon, C.; Harfe, B.D.; Morgan, B.A. Distinct Stem Cell Populations Regenerate the Follicle and Interfollicular Epidermis. Dev. Cell 2005, 9, 855–861. [Google Scholar] [CrossRef]
- Nowak, J.A.; Polak, L.; Pasolli, H.A.; Fuchs, E. Hair Follicle Stem Cells Are Specified and Function in Early Skin Morphogenesis. Cell Stem Cell 2008, 3, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Waghmare, S.K.; Bansal, R.; Lee, J.; Zhang, Y.V.; McDermitt, D.J.; Tumbar, T. Quantitative Proliferation Dynamics and Random Chromosome Segregation of Hair Follicle Stem Cells. EMBO J. 2008, 27, 1309–1320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.V.; Cheong, J.; Ciapurin, N.; McDermitt, D.J.; Tumbar, T. Distinct Self-Renewal and Differentiation Phases in the Niche of Infrequently Dividing Hair Follicle Stem Cells. Cell Stem Cell 2009, 5, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-C.; Pasolli, H.A.; Fuchs, E. Dynamics Between Stem Cells, Niche and Progeny in the Hair Follicle. Cell 2011, 144, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Kizawa, K.; Hamada, K.; Cotsarelis, G. Hair Follicle Stem Cells in the Lower Bulge Form the Secondary Germ, a Biochemically Distinct but Functionally Equivalent Progenitor Cell Population, at the Termination of Catagen. Differ. Res. Biol. Divers. 2004, 72, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-C.; Li, L.; Fuchs, E. Transit-Amplifying Cells Orchestrate Stem Cell Activity and Tissue Regeneration. Cell 2014, 157, 935–949. [Google Scholar] [CrossRef]
- Lien, W.-H.; Guo, X.; Polak, L.; Lawton, L.N.; Young, R.A.; Zheng, D.; Fuchs, E. Genome-Wide Maps of Histone Modifications Unwind In Vivo Chromatin States of the Hair Follicle Lineage. Cell Stem Cell 2011, 9, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Rompolas, P.; Mesa, K.R.; Greco, V. Spatial Organization within a Niche as a Determinant of Stem-Cell Fate. Nature 2013, 502, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Kobielak, K.; Stokes, N.; de la Cruz, J.; Polak, L.; Fuchs, E. Loss of a Quiescent Niche but Not Follicle Stem Cells in the Absence of Bone Morphogenetic Protein Signaling. Proc. Natl. Acad. Sci. USA 2007, 104, 10063–10068. [Google Scholar] [CrossRef]
- Plikus, M.V.; Mayer, J.; de la Cruz, D.; Baker, R.E.; Maini, P.K.; Maxson, R.; Chuong, C.-M. Cyclic Dermal BMP Signaling Regulates Stem Cell Activation during Hair Regeneration. Nature 2008, 451, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Botchkarev, V.A.; Botchkareva, N.V.; Nakamura, M.; Huber, O.; Funa, K.; Lauster, R.; Paus, R.; Gilchrest, B.A. Noggin Is Required for Induction of the Hair Follicle Growth Phase in Postnatal Skin. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2001, 15, 2205–2214. [Google Scholar] [CrossRef]
- Zhang, J.; He, X.C.; Tong, W.-G.; Johnson, T.; Wiedemann, L.M.; Mishina, Y.; Feng, J.Q.; Li, L. Bone Morphogenetic Protein Signaling Inhibits Hair Follicle Anagen Induction by Restricting Epithelial Stem/Progenitor Cell Activation and Expansion. Stem Cells 2006, 24, 2826–2839. [Google Scholar] [CrossRef] [PubMed]
- Osada, S.-I.; Minematsu, N.; Oda, F.; Akimoto, K.; Kawana, S.; Ohno, S. Atypical Protein Kinase C Isoform, APKCλ, Is Essential for Maintaining Hair Follicle Stem Cell Quiescence. J. Investig. Dermatol. 2015, 135, 2584–2592. [Google Scholar] [CrossRef]
- Leishman, E.; Howard, J.M.; Garcia, G.E.; Miao, Q.; Ku, A.T.; Dekker, J.D.; Tucker, H.; Nguyen, H. Foxp1 Maintains Hair Follicle Stem Cell Quiescence through Regulation of Fgf18. Dev. Camb. Engl. 2013, 140, 3809–3818. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Siegenthaler, J.A.; Dowell, R.D.; Yi, R. Foxc1 Reinforces Quiescence in Self-Renewing Hair Follicle Stem Cells. Science 2016, 351, 613–617. [Google Scholar] [CrossRef]
- Lay, K.; Kume, T.; Fuchs, E. FOXC1 Maintains the Hair Follicle Stem Cell Niche and Governs Stem Cell Quiescence to Preserve Long-Term Tissue-Regenerating Potential. Proc. Natl. Acad. Sci. USA 2016, 113, E1506–E1515. [Google Scholar] [CrossRef]
- Suen, W.-J.; Li, S.-T.; Yang, L.-T. Hes1 Regulates Anagen Initiation and Hair Follicle Regeneration through Modulation of Hedgehog Signaling. Stem Cells 2020, 38, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Tanigaki, K.; Han, H.; Hiai, H.; Honjo, T. Notch/RBP-J Signaling Regulates Epidermis/Hair Fate Determination of Hair Follicular Stem Cells. Curr. Biol. 2003, 13, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Cadigan, K.M.; Waterman, M.L. TCF/LEFs and Wnt Signaling in the Nucleus. Cold Spring Harb. Perspect. Biol. 2012, 4, a007906. [Google Scholar] [CrossRef] [PubMed]
- Lien, W.-H.; Polak, L.; Lin, M.; Lay, K.; Zheng, D.; Fuchs, E. In Vivo Transcriptional Governance of Hair Follicle Stem Cells by Canonical Wnt Regulators. Nat. Cell Biol. 2014, 16, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, J.; Shi, C.; Huang, Y.; Wang, Y.; Yang, T.; Yang, J. Lef1 Contributes to the Differentiation of Bulge Stem Cells by Nuclear Translocation and Cross-Talk with the Notch Signaling Pathway. Int. J. Med. Sci. 2013, 10, 738–746. [Google Scholar] [CrossRef]
- Adam, R.C.; Yang, H.; Ge, Y.; Lien, W.-H.; Wang, P.; Zhao, Y.; Polak, L.; Levorse, J.; Baksh, S.C.; Zheng, D.; et al. Temporal Layering of Signaling Effectors Drives Chromatin Remodeling during Hair Follicle Stem Cell Lineage Progression. Cell Stem Cell 2018, 22, 398–413.e7. [Google Scholar] [CrossRef] [PubMed]
- Shirokova, V.; Biggs, L.C.; Jussila, M.; Ohyama, T.; Groves, A.K.; Mikkola, M.L. Foxi3 Deficiency Compromises Hair Follicle Stem Cell Specification and Activation. Stem Cells 2016, 34, 1896–1908. [Google Scholar] [CrossRef] [PubMed]
- Lim, X.; Tan, S.H.; Yu, K.L.; Lim, S.B.H.; Nusse, R. Axin2 Marks Quiescent Hair Follicle Bulge Stem Cells That Are Maintained by Autocrine Wnt/β-Catenin Signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E1498–E1505. [Google Scholar] [CrossRef]
- Joost, S.; Jacob, T.; Sun, X.; Annusver, K.; La Manno, G.; Sur, I.; Kasper, M. Single-Cell Transcriptomics of Traced Epidermal and Hair Follicle Stem Cells Reveals Rapid Adaptations during Wound Healing. Cell Rep. 2018, 25, 585–597.e7. [Google Scholar] [CrossRef]
- de Lau, W.; Peng, W.C.; Gros, P.; Clevers, H. The R-Spondin/Lgr5/Rnf43 Module: Regulator of Wnt Signal Strength. Genes Dev. 2014, 28, 305–316. [Google Scholar] [CrossRef]
- Hoeck, J.D.; Biehs, B.; Kurtova, A.V.; Kljavin, N.M.; de Sousa E Melo, F.; Alicke, B.; Koeppen, H.; Modrusan, Z.; Piskol, R.; de Sauvage, F.J. Stem Cell Plasticity Enables Hair Regeneration Following Lgr5+ Cell Loss. Nat. Cell Biol. 2017, 19, 666–676. [Google Scholar] [CrossRef]
- Ren, X.; Xia, W.; Xu, P.; Shen, H.; Dai, X.; Liu, M.; Shi, Y.; Ye, X.; Dang, Y. Lgr4 Deletion Delays the Hair Cycle and Inhibits the Activation of Hair Follicle Stem Cells. J. Investig. Dermatol. 2020, 140, 1706–1712.e4. [Google Scholar] [CrossRef] [PubMed]
- Osorio, K.M.; Lilja, K.C.; Tumbar, T. Runx1 Modulates Adult Hair Follicle Stem Cell Emergence and Maintenance from Distinct Embryonic Skin Compartments. J. Cell Biol. 2011, 193, 235–250. [Google Scholar] [CrossRef]
- Lee, J.; Hoi, C.S.L.; Lilja, K.C.; White, B.S.; Lee, S.E.; Shalloway, D.; Tumbar, T. Runx1 and P21 Synergistically Limit the Extent of Hair Follicle Stem Cell Quiescence In Vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 4634–4639. [Google Scholar] [CrossRef]
- Hoi, C.S.L.; Lee, S.E.; Lu, S.-Y.; McDermitt, D.J.; Osorio, K.M.; Piskun, C.M.; Peters, R.M.; Paus, R.; Tumbar, T. Runx1 Directly Promotes Proliferation of Hair Follicle Stem Cells and Epithelial Tumor Formation in Mouse Skin. Mol. Cell. Biol. 2010, 30, 2518–2536. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Nattakom, M.; Holowka, D.; Wang, D.H.; Thomas Brenna, J.; Ku, A.T.; Nguyen, H.; Ibrahim, S.F.; Tumbar, T. Runx1 Role in Epithelial and Cancer Cell Proliferation Implicates Lipid Metabolism and Scd1 and Soat1 Activity. Stem Cells 2018, 36, 1603–1616. [Google Scholar] [CrossRef]
- Calvo-Sánchez, M.I.; Fernández-Martos, S.; Carrasco, E.; Moreno-Bueno, G.; Bernabéu, C.; Quintanilla, M.; Espada, J. A Role for the Tgf-β/Bmp Co-Receptor Endoglin in the Molecular Oscillator That Regulates the Hair Follicle Cycle. J. Mol. Cell Biol. 2019, 11, 39–52. [Google Scholar] [CrossRef]
- Wang, A.B.; Zhang, Y.V.; Tumbar, T. Gata6 Promotes Hair Follicle Progenitor Cell Renewal by Genome Maintenance during Proliferation. EMBO J. 2017, 36, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Veselá, B.; Svandová, E.; Smarda, J.; Matalová, E. Mybs in Mouse Hair Follicle Development. Tissue Cell 2014, 46, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Haensel, D.; Sun, P.; MacLean, A.L.; Ma, X.; Zhou, Y.; Stemmler, M.P.; Brabletz, S.; Berx, G.; Plikus, M.V.; Nie, Q.; et al. An Ovol2-Zeb1 Transcriptional Circuit Regulates Epithelial Directional Migration and Proliferation. EMBO Rep. 2019, 20, e46273. [Google Scholar] [CrossRef]
- Li, G.; Tang, X.; Zhang, S.; Jin, M.; Wang, M.; Deng, Z.; Liu, Z.; Qian, M.; Shi, W.; Wang, Z.; et al. SIRT7 Activates Quiescent Hair Follicle Stem Cells to Ensure Hair Growth in Mice. EMBO J. 2020, 39, e104365. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Tian, Y.; Song, Y.; Shi, J.; Xu, J.; Xiong, K.; Li, J.; Xu, W.; Zhao, Y.; Shuai, J.; et al. Msi2 Maintains Quiescent State of Hair Follicle Stem Cells by Directly Repressing the Hh Signaling Pathway. J. Investig. Dermatol. 2017, 137, 1015–1024. [Google Scholar] [CrossRef]
- Adam, R.C.; Yang, H.; Rockowitz, S.; Larsen, S.B.; Nikolova, M.; Oristian, D.S.; Polak, L.; Kadaja, M.; Asare, A.; Zheng, D.; et al. Pioneer Factors Govern Super-Enhancer Dynamics in Stem Cell Plasticity and Lineage Choice. Nature 2015, 521, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Rhee, H.; Polak, L.; Fuchs, E. Lhx2 Maintains Stem Cell Character in Hair Follicles. Science 2006, 312, 1946–1949. [Google Scholar] [CrossRef] [PubMed]
- Horsley, V.; Aliprantis, A.O.; Polak, L.; Glimcher, L.H.; Fuchs, E. NFATc1 Balances Quiescence and Proliferation of Skin Stem Cells. Cell 2008, 132, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Pivetti, S.; Fernandez-Perez, D.; D’Ambrosio, A.; Barbieri, C.M.; Manganaro, D.; Rossi, A.; Barnabei, L.; Zanotti, M.; Scelfo, A.; Chiacchiera, F.; et al. Loss of PRC1 Activity in Different Stem Cell Compartments Activates a Common Transcriptional Program with Cell Type-Dependent Outcomes. Sci. Adv. 2019, 5, eaav1594. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; He, J.; Wang, J.; Chen, X.; Yang, R. Regulation of Signaling Pathways in Hair Follicle Stem Cells. Burn. Trauma 2022, 10, tkac022. [Google Scholar] [CrossRef]
- Yuan, S.; Li, F.; Meng, Q.; Zhao, Y.; Chen, L.; Zhang, H.; Xue, L.; Zhang, X.; Lengner, C.; Yu, Z. Post-Transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by MiR-22. PLoS Genet. 2015, 11, e1005253. [Google Scholar] [CrossRef]
- Zhang, L.; Stokes, N.; Polak, L.; Fuchs, E. Specific MicroRNAs Are Preferentially Expressed by Skin Stem Cells to Balance Self-Renewal and Early Lineage Commitment. Cell Stem Cell 2011, 8, 294–308. [Google Scholar] [CrossRef]
- Zhang, L.; Ge, Y.; Fuchs, E. MiR-125b Can Enhance Skin Tumor Initiation and Promote Malignant Progression by Repressing Differentiation and Prolonging Cell Survival. Genes Dev. 2014, 28, 2532–2546. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.; Liu, C.; Li, L.; Lan, M.; Yu, Y.; Gu, L.; Su, Y.; Zhang, K.; Zhang, Y.; Wang, T.; et al. MiR-29a/B1 Inhibits Hair Follicle Stem Cell Lineage Progression by Spatiotemporally Suppressing WNT and BMP Signaling. Cell Rep. 2019, 29, 2489–2504.e4. [Google Scholar] [CrossRef]
- Du, K.-T.; Deng, J.-Q.; He, X.-G.; Liu, Z.-P.; Peng, C.; Zhang, M.-S. MiR-214 Regulates the Human Hair Follicle Stem Cell Proliferation and Differentiation by Targeting EZH2 and Wnt/β-Catenin Signaling Way In Vitro. Tissue Eng. Regen. Med. 2018, 15, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.I.; Alam, M.; Emelianov, V.U.; Poterlowicz, K.; Patel, A.; Sharov, A.A.; Mardaryev, A.N.; Botchkareva, N.V. MicroRNA-214 Controls Skin and Hair Follicle Development by Modulating the Activity of the Wnt Pathway. J. Cell Biol. 2014, 207, 549–567. [Google Scholar] [CrossRef] [PubMed]
- Si, Y.; Bai, J.; Wu, J.; Li, Q.; Mo, Y.; Fang, R.; Lai, W. LncRNA PlncRNA-1 Regulates Proliferation and Differentiation of Hair Follicle Stem Cells through TGF-β1-mediated Wnt/Β-catenin Signal Pathway. Mol. Med. Rep. 2018, 17, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Zheng, Y.; Ma, S.; Xing, Q.; Wang, X.; Yang, B.; Yin, G.; Guan, F. Long Non-coding RNA Regulates Hair Follicle Stem Cell Proliferation and Differentiation through PI3K/AKT Signal Pathway. Mol. Med. Rep. 2018, 17, 5477–5483. [Google Scholar] [CrossRef]
- Vishlaghi, N.; Lisse, T.S. Dicer- and Bulge Stem Cell-Dependent MicroRNAs during Induced Anagen Hair Follicle Development. Front. Cell Dev. Biol. 2020, 8, 338. [Google Scholar] [CrossRef] [PubMed]
- Reichenbach, B.; Classon, J.; Aida, T.; Tanaka, K.; Genander, M.; Göritz, C. Glutamate Transporter Slc1a3 Mediates Inter-Niche Stem Cell Activation during Skin Growth. EMBO J. 2018, 37, e98280. [Google Scholar] [CrossRef]
- Tang, Y.; Luo, B.; Deng, Z.; Wang, B.; Liu, F.; Li, J.; Shi, W.; Xie, H.; Hu, X.; Li, J. Mitochondrial Aerobic Respiration Is Activated during Hair Follicle Stem Cell Differentiation, and Its Dysfunction Retards Hair Regeneration. PeerJ 2016, 4, e1821. [Google Scholar] [CrossRef] [PubMed]
- Flores, A.; Schell, J.; Krall, A.S.; Jelinek, D.; Miranda, M.; Grigorian, M.; Braas, D.; White, A.C.; Zhou, J.L.; Graham, N.A.; et al. Lactate Dehydrogenase Activity Drives Hair Follicle Stem Cell Activation. Nat. Cell Biol. 2017, 19, 1017–1026. [Google Scholar] [CrossRef]
- Sarate, R.M.; Chovatiya, G.L.; Ravi, V.; Khade, B.; Gupta, S.; Waghmare, S.K. SPLA2 -IIA Overexpression in Mice Epidermis Depletes Hair Follicle Stem Cells and Induces Differentiation Mediated through Enhanced JNK/c-Jun Activation. Stem Cells 2016, 34, 2407–2417. [Google Scholar] [CrossRef]
- Peters, F.; Vorhagen, S.; Brodesser, S.; Jakobshagen, K.; Brüning, J.C.; Niessen, C.M.; Krönke, M. Ceramide Synthase 4 Regulates Stem Cell Homeostasis and Hair Follicle Cycling. J. Investig. Dermatol. 2015, 135, 1501–1509. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-L.; Xu, R.; Yi, J.K.; Li, F.; Chen, J.; Jones, E.C.; Slutsky, J.B.; Huang, L.; Rigas, B.; Cao, J.; et al. Alkaline Ceramidase 1 Protects Mice from Premature Hair Loss by Maintaining the Homeostasis of Hair Follicle Stem Cells. Stem Cell Rep. 2017, 9, 1488–1500. [Google Scholar] [CrossRef]
- Liakath-Ali, K.; Vancollie, V.E.; Lelliott, C.J.; Speak, A.O.; Lafont, D.; Protheroe, H.J.; Ingvorsen, C.; Galli, A.; Green, A.; Gleeson, D.; et al. Alkaline Ceramidase 1 Is Essential for Mammalian Skin Homeostasis and Regulating Whole-Body Energy Expenditure. J. Pathol. 2016, 239, 374–383. [Google Scholar] [CrossRef]
- Martel, J.L.; Miao, J.H.; Badri, T. Anatomy, Hair Follicle. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Thody, A.J.; Shuster, S. Control and Function of Sebaceous Glands. Physiol. Rev. 1989, 69, 383–416. [Google Scholar] [CrossRef]
- Schneider, M.R.; Paus, R. Sebocytes, Multifaceted Epithelial Cells: Lipid Production and Holocrine Secretion. Int. J. Biochem. Cell Biol. 2010, 42, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Page, M.E.; Lombard, P.; Ng, F.; Göttgens, B.; Jensen, K.B. The Epidermis Comprises Autonomous Compartments Maintained by Distinct Stem Cell Populations. Cell Stem Cell 2013, 13, 471–482. [Google Scholar] [CrossRef]
- Nijhof, J.G.W.; Braun, K.M.; Giangreco, A.; van Pelt, C.; Kawamoto, H.; Boyd, R.L.; Willemze, R.; Mullenders, L.H.F.; Watt, F.M.; de Gruijl, F.R.; et al. The Cell-Surface Marker MTS24 Identifies a Novel Population of Follicular Keratinocytes with Characteristics of Progenitor Cells. Dev. Camb. Engl. 2006, 133, 3027–3037. [Google Scholar] [CrossRef] [PubMed]
- Depreter, M.G.L.; Blair, N.F.; Gaskell, T.L.; Nowell, C.S.; Davern, K.; Pagliocca, A.; Stenhouse, F.H.; Farley, A.M.; Fraser, A.; Vrana, J.; et al. Identification of Plet-1 as a Specific Marker of Early Thymic Epithelial Progenitor Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 961–966. [Google Scholar] [CrossRef]
- Raymond, K.; Richter, A.; Kreft, M.; Frijns, E.; Janssen, H.; Slijper, M.; Praetzel-Wunder, S.; Langbein, L.; Sonnenberg, A. Expression of the Orphan Protein Plet-1 during Trichilemmal Differentiation of Anagen Hair Follicles. J. Investig. Dermatol. 2010, 130, 1500–1513. [Google Scholar] [CrossRef] [PubMed]
- Jensen, U.B.; Yan, X.; Triel, C.; Woo, S.-H.; Christensen, R.; Owens, D.M. A Distinct Population of Clonogenic and Multipotent Murine Follicular Keratinocytes Residing in the Upper Isthmus. J. Cell Sci. 2008, 121, 609–617. [Google Scholar] [CrossRef]
- Snippert, H.J.; Haegebarth, A.; Kasper, M.; Jaks, V.; van Es, J.H.; Barker, N.; van de Wetering, M.; van den Born, M.; Begthel, H.; Vries, R.G.; et al. Lgr6 Marks Stem Cells in the Hair Follicle That Generate All Cell Lineages of the Skin. Science 2010, 327, 1385–1389. [Google Scholar] [CrossRef]
- Frances, D.; Niemann, C. Stem Cell Dynamics in Sebaceous Gland Morphogenesis in Mouse Skin. Dev. Biol. 2012, 363, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Horsley, V.; O’Carroll, D.; Tooze, R.; Ohinata, Y.; Saitou, M.; Obukhanych, T.; Nussenzweig, M.; Tarakhovsky, A.; Fuchs, E. Blimp1 Defines a Progenitor Population That Governs Cellular Input to the Sebaceous Gland. Cell 2006, 126, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, K.; Cottle, D.L.; Donati, G.; Chiang, M.-F.; Quist, S.R.; Gollnick, H.P.; Natsuga, K.; Lin, K.-I.; Watt, F.M. BLIMP1 Is Required for Postnatal Epidermal Homeostasis but Does Not Define a Sebaceous Gland Progenitor under Steady-State Conditions. Stem Cell Rep. 2014, 3, 620–633. [Google Scholar] [CrossRef] [PubMed]
- Sellheyer, K.; Krahl, D. Blimp-1: A Marker of Terminal Differentiation but Not of Sebocytic Progenitor Cells. J. Cutan. Pathol. 2010, 37, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Ghazizadeh, S.; Taichman, L.B. Multiple Classes of Stem Cells in Cutaneous Epithelium: A Lineage Analysis of Adult Mouse Skin. EMBO J. 2001, 20, 1215–1222. [Google Scholar] [CrossRef]
- Veniaminova, N.A.; Grachtchouk, M.; Doane, O.J.; Peterson, J.K.; Quigley, D.A.; Lull, M.V.; Pyrozhenko, D.V.; Nair, R.R.; Patrick, M.T.; Balmain, A.; et al. Niche-Specific Factors Dynamically Regulate Sebaceous Gland Stem Cells in the Skin. Dev. Cell 2019, 51, 326–340.e4. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.; Lehrer, M.S.; Jensen, P.J.; Sun, T.T.; Lavker, R.M. Involvement of Follicular Stem Cells in Forming Not Only the Follicle But Also the Epidermis. Cell 2000, 102, 451–461. [Google Scholar] [CrossRef]
- Panteleyev, A.A.; Rosenbach, T.; Paus, R.; Christiano, A.M. The Bulge Is the Source of Cellular Renewal in the Sebaceous Gland of Mouse Skin. Arch. Dermatol. Res. 2000, 292, 573–576. [Google Scholar] [CrossRef]
- Petersson, M.; Brylka, H.; Kraus, A.; John, S.; Rappl, G.; Schettina, P.; Niemann, C. TCF/Lef1 Activity Controls Establishment of Diverse Stem and Progenitor Cell Compartments in Mouse Epidermis. EMBO J. 2011, 30, 3004–3018. [Google Scholar] [CrossRef]
- Han, J.; Lin, K.; Choo, H.Q.; Chen, Y.; Zhang, X.; Xu, R.H.; Wang, X.; Wu, Y. Distinct Bulge Stem Cell Populations Maintain the Pilosebaceous Unit in a β-Catenin-Dependent Manner. iScience 2023, 26, 105805. [Google Scholar] [CrossRef] [PubMed]
- Krieger, T.; Simons, B.D. Dynamic Stem Cell Heterogeneity. Dev. Camb. Engl. 2015, 142, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mignone, J.; Yang, M.; Matic, M.; Penman, S.; Enikolopov, G.; Hoffman, R.M. Nestin Expression in Hair Follicle Sheath Progenitor Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 9958–9961. [Google Scholar] [CrossRef] [PubMed]
- Amoh, Y.; Hoffman, R.M. Hair Follicle-Associated-Pluripotent (HAP) Stem Cells. Cell Cycle Georget. Tex 2017, 16, 2169–2175. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M. The Pluripotency of Hair Follicle Stem Cells. Cell Cycle Georget. Tex 2006, 5, 232–233. [Google Scholar] [CrossRef] [PubMed]
- Amoh, Y.; Li, L.; Campillo, R.; Kawahara, K.; Katsuoka, K.; Penman, S.; Hoffman, R.M. Implanted Hair Follicle Stem Cells Form Schwann Cells That Support Repair of Severed Peripheral Nerves. Proc. Natl. Acad. Sci. USA 2005, 102, 17734–17738. [Google Scholar] [CrossRef] [PubMed]
- Amoh, Y.; Li, L.; Katsuoka, K.; Penman, S.; Hoffman, R.M. Multipotent Nestin-Positive, Keratin-Negative Hair-Follicle Bulge Stem Cells Can Form Neurons. Proc. Natl. Acad. Sci. USA 2005, 102, 5530–5534. [Google Scholar] [CrossRef]
- Han, G.; Li, A.G.; Liang, Y.-Y.; Owens, P.; He, W.; Lu, S.; Yoshimatsu, Y.; Wang, D.; Ten Dijke, P.; Lin, X.; et al. Smad7-Induced Beta-Catenin Degradation Alters Epidermal Appendage Development. Dev. Cell 2006, 11, 301–312. [Google Scholar] [CrossRef]
- Ikuta, T.; Ohba, M.; Zouboulis, C.C.; Fujii-Kuriyama, Y.; Kawajiri, K. B Lymphocyte-Induced Maturation Protein 1 Is a Novel Target Gene of Aryl Hydrocarbon Receptor. J. Dermatol. Sci. 2010, 58, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Ju, Q.; Fimmel, S.; Hinz, N.; Stahlmann, R.; Xia, L.; Zouboulis, C.C. 2,3,7,8-Tetrachlorodibenzo-p-Dioxin Alters Sebaceous Gland Cell Differentiation In Vitro. Exp. Dermatol. 2011, 20, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Clayton, R.W.; Göbel, K.; Niessen, C.M.; Paus, R.; van Steensel, M.a.M.; Lim, X. Homeostasis of the Sebaceous Gland and Mechanisms of Acne Pathogenesis. Br. J. Dermatol. 2019, 181, 677–690. [Google Scholar] [CrossRef]
- Baker, L.B. Physiology of Sweat Gland Function: The Roles of Sweating and Sweat Composition in Human Health. Temp. Multidiscip. Biomed. J. 2019, 6, 211–259. [Google Scholar] [CrossRef] [PubMed]
- Hodge, B.D.; Sanvictores, T.; Brodell, R.T. Anatomy, Skin Sweat Glands. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Song, W.; Yao, B.; Zhu, D.; Zhang, Y.; Li, Z.; Huang, S.; Fu, X. 3D-Bioprinted Microenvironments for Sweat Gland Regeneration. Burn. Trauma 2022, 10, tkab044. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.P.; Polak, L.; Rocha, A.S.; Pasolli, H.A.; Chen, S.-C.; Sharma, N.; Blanpain, C.; Fuchs, E. Identification of Stem Cell Populations in Sweat Glands and Ducts Reveals Roles in Homeostasis and Wound Repair. Cell 2012, 150, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Ohe, S.; Tanaka, T.; Yanai, H.; Komai, Y.; Omachi, T.; Kanno, S.; Tanaka, K.; Ishigaki, K.; Saiga, K.; Nakamura, N.; et al. Maintenance of Sweat Glands by Stem Cells Located in the Acral Epithelium. Biochem. Biophys. Res. Commun. 2015, 466, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Petschnik, A.E.; Klatte, J.E.; Evers, L.H.; Kruse, C.; Paus, R.; Danner, S. Phenotypic Indications That Human Sweat Glands Are a Rich Source of Nestin-Positive Stem Cell Populations. Br. J. Dermatol. 2010, 162, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Tiede, S.; Kloepper, J.E.; Ernst, N.; Poeggeler, B.; Kruse, C.; Paus, R. Nestin in Human Skin: Exclusive Expression in Intramesenchymal Skin Compartments and Regulation by Leptin. J. Investig. Dermatol. 2009, 129, 2711–2720. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yao, B.; Duan, X.; Li, J.; Song, W.; Enhejirigala; Li, Z.; Yuan, X.; Kong, Y.; Zhang, Y.; et al. Notch1 Down-Regulation in Lineage-Restricted Niches Is Involved in the Development of Mouse Eccrine Sweat Glands. J. Mol. Histol. 2022, 53, 857–867. [Google Scholar] [CrossRef]
- Ma, K.; Tan, Z.; Zhang, C.; Fu, X. Mesenchymal Stem Cells for Sweat Gland Regeneration after Burns: From Possibility to Reality. Burn. J. Int. Soc. Burn Inj. 2016, 42, 492–499. [Google Scholar] [CrossRef]
- Shikiji, T.; Minami, M.; Inoue, T.; Hirose, K.; Oura, H.; Arase, S. Keratinocytes Can Differentiate into Eccrine Sweat Ducts in Vitro: Involvement of Epidermal Growth Factor and Fetal Bovine Serum. J. Dermatol. Sci. 2003, 33, 141–150. [Google Scholar] [CrossRef]
- Xu, Y.; Hong, Y.; Xu, M.; Ma, K.; Fu, X.; Zhang, M.; Wang, G. Role of Keratinocyte Growth Factor in the Differentiation of Sweat Gland-Like Cells from Human Umbilical Cord-Derived Mesenchymal Stem Cells. Stem Cells Transl. Med. 2016, 5, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, S.; Ma, K.; Fu, X.; Han, W.; Sheng, Z. Promising New Potential for Mesenchymal Stem Cells Derived from Human Umbilical Cord Wharton’s Jelly: Sweat Gland Cell-like Differentiative Capacity. J. Tissue Eng. Regen. Med. 2012, 6, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Sun, Q.; Zhen, Y.; Li, F.; Xu, Y.; Liu, Y.; Zhang, X.; Qin, M. The Differentiation of Amniotic Fluid Stem Cells into Sweat Glandlike Cells Is Enhanced by the Presence of Sonic Hedgehog in the Conditioned Medium. Exp. Dermatol. 2016, 25, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, Y.; Fu, X. Sweat Gland Regeneration after Burn Injury: Is Stem Cell Therapy a New Hope? Cytotherapy 2015, 17, 526–535. [Google Scholar] [CrossRef]
- Nakamura, M.; Tokura, Y. The Localization of Label-Retaining Cells in Eccrine Glands. J. Investig. Dermatol. 2009, 129, 2077–2078. [Google Scholar] [CrossRef] [PubMed]
- Danner, S.; Kremer, M.; Petschnik, A.E.; Nagel, S.; Zhang, Z.; Hopfner, U.; Reckhenrich, A.K.; Weber, C.; Schenck, T.L.; Becker, T.; et al. The Use of Human Sweat Gland-Derived Stem Cells for Enhancing Vascularization during Dermal Regeneration. J. Investig. Dermatol. 2012, 132, 1707–1716. [Google Scholar] [CrossRef] [PubMed]
- Leung, Y.; Kandyba, E.; Chen, Y.-B.; Ruffins, S.; Kobielak, K. Label Retaining Cells (LRCs) with Myoepithelial Characteristic from the Proximal Acinar Region Define Stem Cells in the Sweat Gland. PLoS ONE 2013, 8, e74174. [Google Scholar] [CrossRef] [PubMed]
- Rittié, L.; Sachs, D.L.; Orringer, J.S.; Voorhees, J.J.; Fisher, G.J. Eccrine Sweat Glands Are Major Contributors to Reepithelialization of Human Wounds. Am. J. Pathol. 2013, 182, 163–171. [Google Scholar] [CrossRef]
- Pontiggia, L.; Biedermann, T.; Böttcher-Haberzeth, S.; Oliveira, C.; Braziulis, E.; Klar, A.S.; Meuli-Simmen, C.; Meuli, M.; Reichmann, E. De Novo Epidermal Regeneration Using Human Eccrine Sweat Gland Cells: Higher Competence of Secretory over Absorptive Cells. J. Investig. Dermatol. 2014, 134, 1735–1742. [Google Scholar] [CrossRef]
- Scadden, D.T. Nice Neighborhood: Emerging Concepts of the Stem Cell Niche. Cell 2014, 157, 41–50. [Google Scholar] [CrossRef]
- Renner, M.; Lancaster, M.A.; Bian, S.; Choi, H.; Ku, T.; Peer, A.; Chung, K.; Knoblich, J.A. Self-Organized Developmental Patterning and Differentiation in Cerebral Organoids. EMBO J. 2017, 36, 1316–1329. [Google Scholar] [CrossRef]
- Bagley, J.A.; Reumann, D.; Bian, S.; Lévi-Strauss, J.; Knoblich, J.A. Fused Cerebral Organoids Model Interactions between Brain Regions. Nat. Methods 2017, 14, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.S.; Janda, C.Y.; Chang, J.; Zheng, G.X.Y.; Larkin, K.A.; Luca, V.C.; Chia, L.A.; Mah, A.T.; Han, A.; Terry, J.M.; et al. Non-Equivalence of Wnt and R-Spondin Ligands during Lgr5+ Intestinal Stem-Cell Self-Renewal. Nature 2017, 545, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Date, S.; Sato, T. Mini-Gut Organoids: Reconstitution of the Stem Cell Niche. Annu. Rev. Cell Dev. Biol. 2015, 31, 269–289. [Google Scholar] [CrossRef]
- Lindemans, C.A.; Calafiore, M.; Mertelsmann, A.M.; O’Connor, M.H.; Dudakov, J.A.; Jenq, R.R.; Velardi, E.; Young, L.F.; Smith, O.M.; Lawrence, G.; et al. Interleukin-22 Promotes Intestinal-Stem-Cell-Mediated Epithelial Regeneration. Nature 2015, 528, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Huch, M.; Dorrell, C.; Boj, S.F.; van Es, J.H.; Li, V.S.W.; van de Wetering, M.; Sato, T.; Hamer, K.; Sasaki, N.; Finegold, M.J.; et al. In Vitro Expansion of Single Lgr5+ Liver Stem Cells Induced by Wnt-Driven Regeneration. Nature 2013, 494, 247–250. [Google Scholar] [CrossRef]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.A.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Boj, S.F.; Hwang, C.-I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef]
- Roe, J.-S.; Hwang, C.-I.; Somerville, T.D.D.; Milazzo, J.P.; Lee, E.J.; Da Silva, B.; Maiorino, L.; Tiriac, H.; Young, C.M.; Miyabayashi, K.; et al. Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell 2017, 170, 875–888.e20. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Clevers, H. SnapShot: Growing Organoids from Stem Cells. Cell 2015, 161, 1700–1700.e1. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Wang, R.; Wang, Y.; Zhang, Y.; Hu, T.; Song, W.; Li, Z.; Huang, S.; Fu, X. Biochemical and Structural Cues of 3D-Printed Matrix Synergistically Direct MSC Differentiation for Functional Sweat Gland Regeneration. Sci. Adv. 2020, 6, eaaz1094. [Google Scholar] [CrossRef] [PubMed]


| Gene | Chromosomal Locus | Protein | Reference |
|---|---|---|---|
| ASH1L | 1q22 | ASH1-like histone lysine methyltransferase | [82] |
| AXIN2 | 17q24.1 | Axin 2 | [64] |
| CHRM3 | 1q43 | Cholinergic receptor muscarinic 3 | [95] |
| COL17A1 | 10q25.1 | Collagen type XVII alpha 1 chain | [56,57,58] |
| CTNNB1 | 3p22.1 | Catenin beta 1 | [65] |
| DLL1 | 6q27 | Delta-like canonical Notch ligand 1 | [63] |
| EGFR | 7p11.2 | Epidermal growth factor receptor | [77] |
| FOS | 14q24.3 | Fos proto-oncogene | [59] |
| GRHL3 | 1p36.11 | Grainyhead-like transcription factor 3 | [66] |
| HDAC1 | 1p35.2–p35.1 | Histone deacetylase 1 | [83] |
| IFT88 | 13q12.11 | Intraflagellar transport 88 | [62] |
| IRF6 | 1q32.2 | Interferon regulatory factor 6 | [90] |
| ITGB1 | 10p11.22 | Integrin subunit beta 1 | [52,54,55,88] |
| JUNB | 19p13.13 | JunB proto-oncogene | [59] |
| KIF3A | 5q31.1 | Kinesin family member 3A | [62] |
| LAMA3 | 18q11.2 | Laminin subunit alpha 3 | [89] |
| LAMB3 | 1q32.2 | Laminin subunit beta 3 | [89] |
| LAMC2 | 1q25.3 | Laminin subunit gamma 2 | [89] |
| LRIG1 | 3p14.1 | Leucine-rich repeats and immunoglobulin-like domains 1 | [77,92,93] |
| MAL | 2q11.1 | Mal | [59] |
| MYC | 8q24.21 | MYC proto-oncogene | [75,77,78,79,80,81] |
| NOTCH1 | 9q34.3 | Notch receptor 1 | [63] |
| NSUN2 | 5p15.31 | NOP2/Sun RNA methyltransferase 2 | [78] |
| PELO | 5q11.2 | Pelota mRNA surveillance and ribosome rescue factor | [91] |
| RAC1 | 7p22.1 | Rac family small GTPase 1 | [60] |
| SFN | 1p36.11 | Stratifin | [87] |
| SRF | 6p21.1 | Serum response factor | [59] |
| TAFAZZIN | Xq28 | Tafazzin | [88] |
| TGFB1 | 19q13.2 | Transforming growth factor beta 1 | [94] |
| TP63 | 3q28 | Tumor protein p63 | [83,84,85,86,87] |
| WNT10A | 2q35 | Wnt family member 10A | [64] |
| WNT4 | 1p36.12 | Wnt family member 4 | [64] |
| YAP1 | 11q22.1 | Yes1 associated transcriptional regulator | [87,88,89] |
| Gene | Chromosomal Locus | Protein | Reference |
|---|---|---|---|
| AXIN2 | 17q24.1 | Axin 2 | [135] |
| BMP2 | 20p12.3 | Bone morphogenetic protein 2 | [126] |
| BMP5 | 6p12.1 | Bone morphogenetic protein 5 | [126] |
| BMP6 | 6p24.3 | Bone morphogenetic protein 6 | [124] |
| BMPR1A | 10q23.2 | Bone morphogenetic protein receptor type 1A | [158] |
| CDH1 | 16q22.1 | Cadherin 1 | [127] |
| CDKN1A | 6p21.2 | Cyclin-dependent kinase inhibitor 1A | [141] |
| CDKN2B | 9p21.3 | Cyclin-dependent kinase inhibitor 2B | [141] |
| CERS4 | 19p13.2 | Ceramide synthase 4 | [168] |
| CTNNB1 | 3p22.1 | Catenin beta 1 | [131,132,144] |
| DICER1 | 14q32.13 | Dicer 1 | [163] |
| ENG | 9q34.11 | Endoglin | [144] |
| FGF18 | 5q35.1 | Fibroblast growth factor 18 | [124,125,126] |
| FOSB | 19q13.32 | FosB proto-oncogene | [167] |
| FOXC1 | 6p25.3 | Forkhead box C1 | [126,127] |
| FOXI3 | 2p11.2 | Forkhead box I3 | [134] |
| FOXP1 | 3p13 | Forkhead box P1 | [125] |
| GAΤA6 | 18q11.2 | GATA binding protein 6 | [145] |
| HES1 | 3q29 | Hes family bHLH transcription factor 1 | [128] |
| JAG1 | 20p12.2 | Jagged canonical Notch ligand 1 | [128,132] |
| JUN | 1p32.1 | Jun proto-oncogene | [167] |
| LEF1 | 4q25 | Lymphoid enhancer binding factor 1 | [132,133] |
| LGR4 | 11p14.1 | Leucine-rich repeat containing G protein-coupled receptor 4 | [139] |
| LGR5 | 12q21.1 | Leucine-rich repeat containing G protein-coupled receptor 5 | [107,137,138] |
| LHX2 | 9q33.3 | LIM homeobox 2 | [150,151] |
| LRP6 | 12p13.2 | LDL receptor-related protein 6 | [158] |
| MSI2 | 17q22 | Musashi RNA binding protein 2 | [149] |
| MTOR | 1p36.22 | Mechanistic target of rapamycin kinase | [139] |
| MYB | 6q23.3 | MYB proto-oncogene, transcription factor | [146] |
| MYC | 8q24.21 | MYC proto-oncogene | [132] |
| NFATC1 | 18q23 | Nuclear factor of activated T cells 1 | [126,148,150,152] |
| NOTCH1 | 9q34.3 | Notch receptor 1 | [132] |
| OVOL2 | 20p11.23 | Ovo like zinc finger 2 | [147] |
| PLA2G2A | 1p36.13 | Phospholipase A2 group IIA | [167] |
| PRC1 | 15q26.1 | Protein regulator of cytokinesis 1 | [153] |
| PRKCI | 3q26.2 | Protein kinase C iota | [124] |
| RBPJ | 4p15.2 | Recombination signal binding protein for immunoglobulin kappa J region | [133] |
| RUNX1 | 21q22.12 | RUNX family transcription factor 1 | [140,141,142,143] |
| SCD | 10q24.31 | Stearoyl-CoA desaturase | [143] |
| SFRP1 | 8p11.21 | Secreted frizzled related protein 1 | [135] |
| SHH | 7q36.3 | Sonic hedgehog signaling molecule | [117,153,154] |
| SIRT7 | 17q25.3 | Sirtuin 7 | [148] |
| SLC1A3 | 5p13.2 | Solute carrier family 1 member 3 | [48,164] |
| SMAD1 | 4q31.21 | SMAD family member 1 | [133] |
| SMAD4 | 18q21.2 | SMAD family member 4 | [144] |
| SOAT1 | 1q25.2 | Sterol O-acyltransferase 1 | [143] |
| SOX9 | 17q24.3 | SRY-box transcription factor 9 | [150] |
| TCF3 | 19p13.3 | Transcription factor 3 | [131,133] |
| TCF4 | 18q21.2 | Transcription factor 4 | [131,133] |
| TGFB1 | 19q13.2 | Transforming growth factor beta 1 | [144,161] |
| ZEB1 | 10p11.22 | Zinc finger E-box binding homeobox 1 | [147] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dermitzakis, I.; Kampitsi, D.D.; Manthou, M.E.; Evangelidis, P.; Vakirlis, E.; Meditskou, S.; Theotokis, P. Ontogeny of Skin Stem Cells and Molecular Underpinnings. Curr. Issues Mol. Biol. 2024, 46, 8118-8147. https://doi.org/10.3390/cimb46080481
Dermitzakis I, Kampitsi DD, Manthou ME, Evangelidis P, Vakirlis E, Meditskou S, Theotokis P. Ontogeny of Skin Stem Cells and Molecular Underpinnings. Current Issues in Molecular Biology. 2024; 46(8):8118-8147. https://doi.org/10.3390/cimb46080481
Chicago/Turabian StyleDermitzakis, Iasonas, Despoina Dimitria Kampitsi, Maria Eleni Manthou, Paschalis Evangelidis, Efstratios Vakirlis, Soultana Meditskou, and Paschalis Theotokis. 2024. "Ontogeny of Skin Stem Cells and Molecular Underpinnings" Current Issues in Molecular Biology 46, no. 8: 8118-8147. https://doi.org/10.3390/cimb46080481









