Docking, MD Simulations, and DFT Calculations: Assessing W254’s Function and Sartan Binding in Furin
Abstract
1. Introduction
2. Materials and Methods
2.1. Quantum Mechanics
2.2. Induced Fit Docking
2.3. Molecular Dynamics
3. Results and Discussion
3.1. Quantum Mechanics Calculations
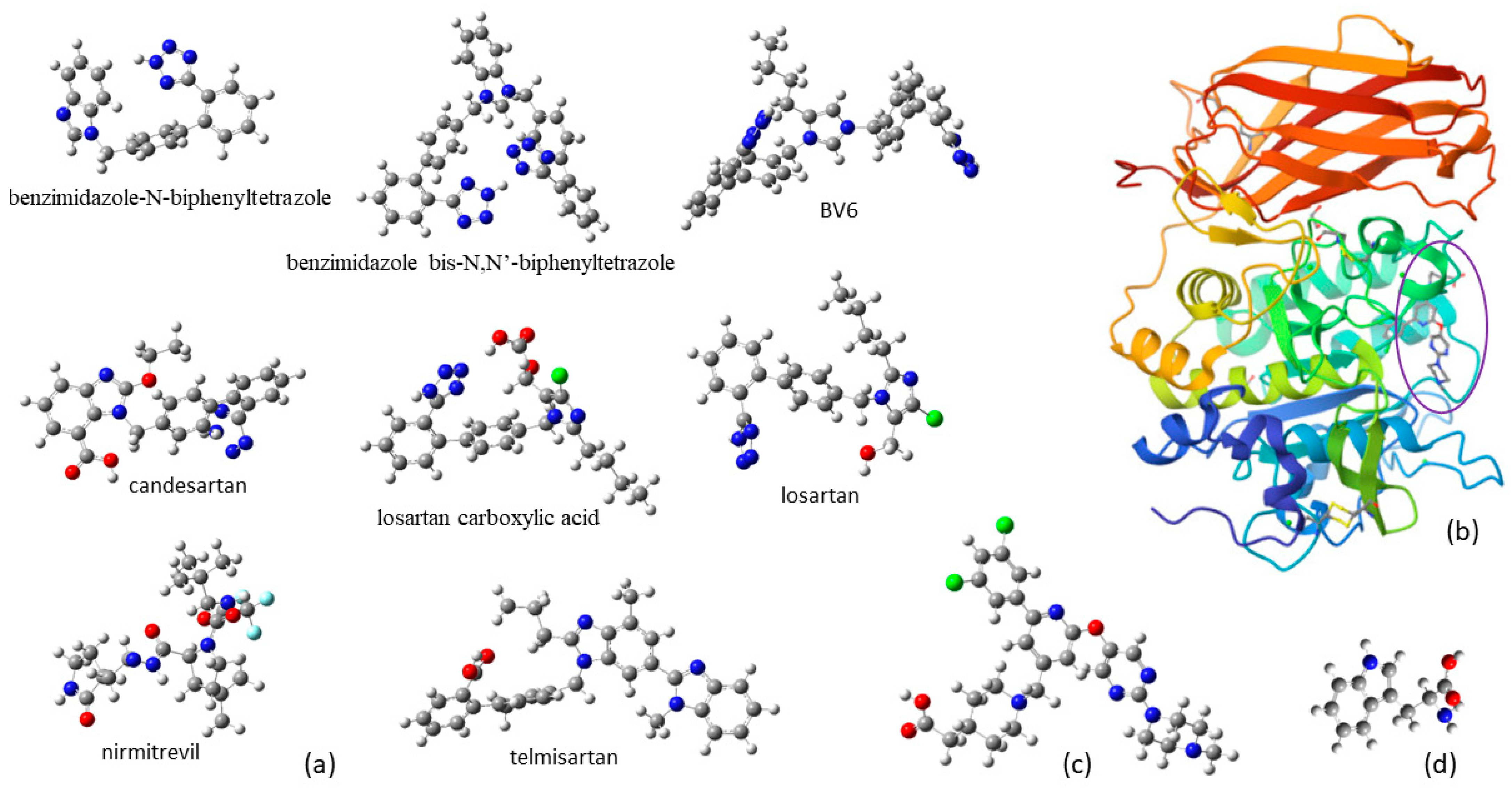
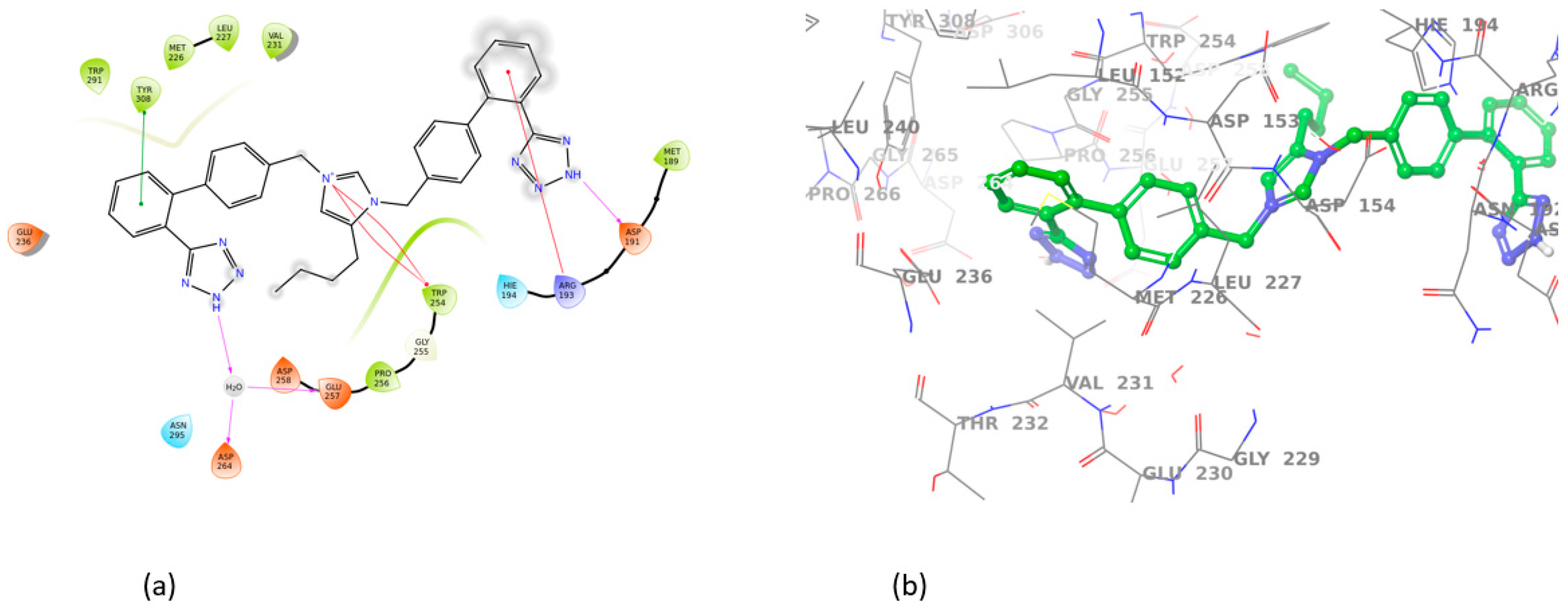
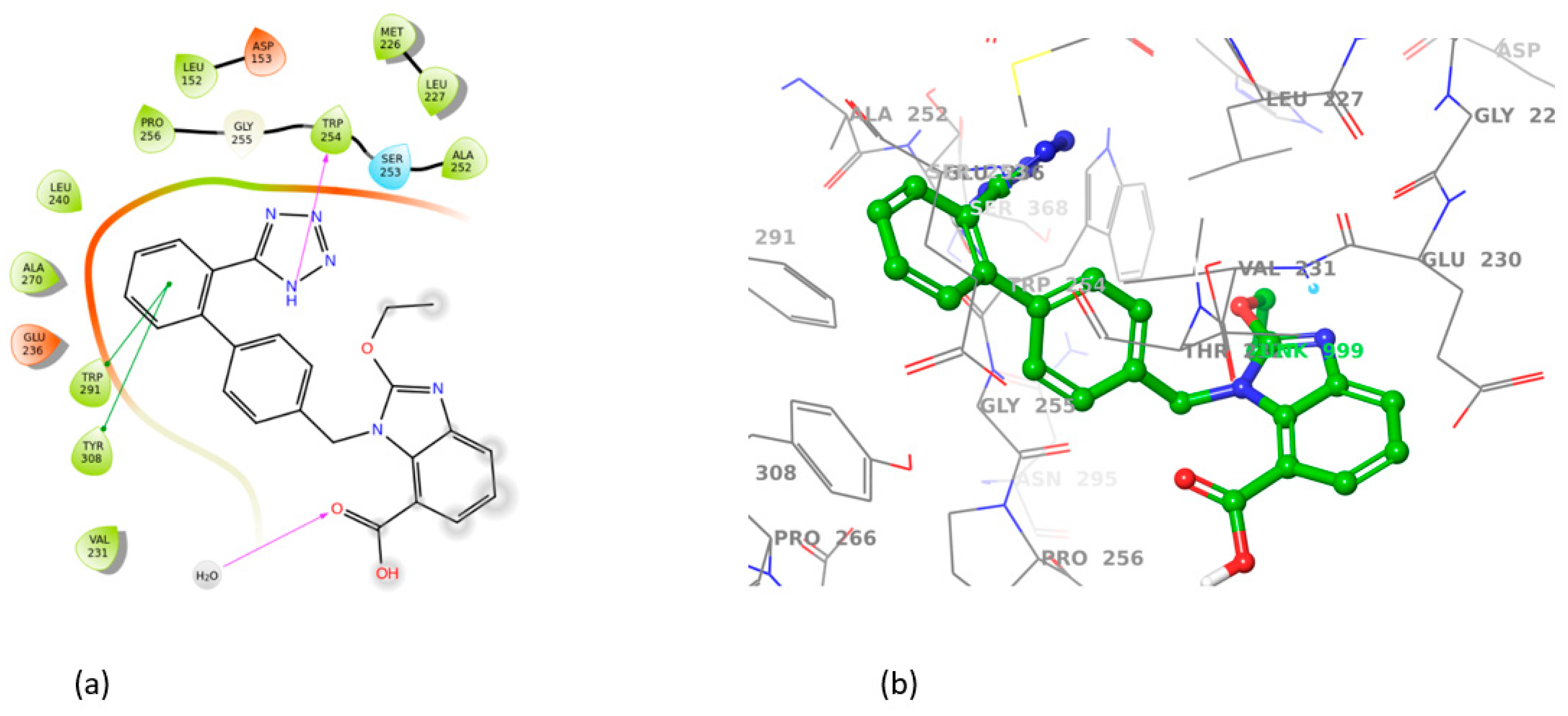
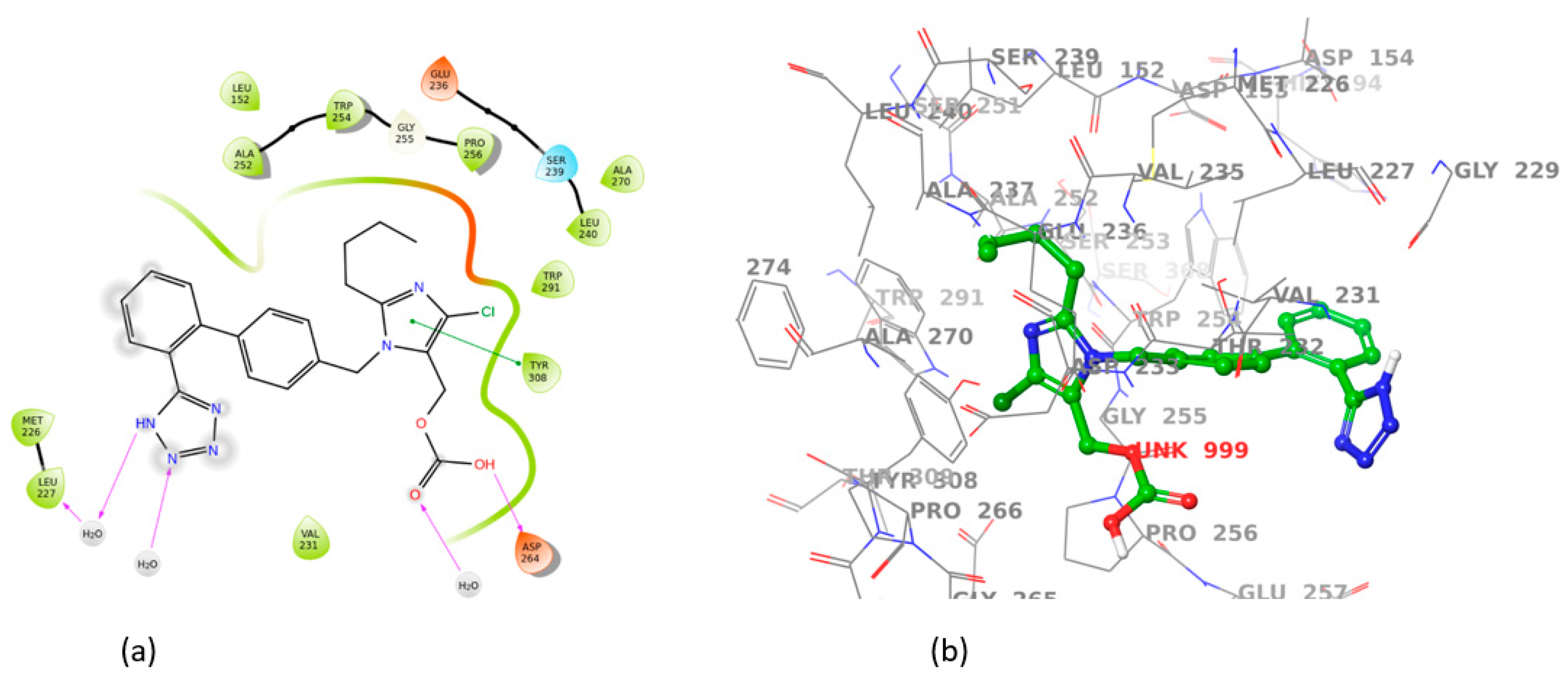
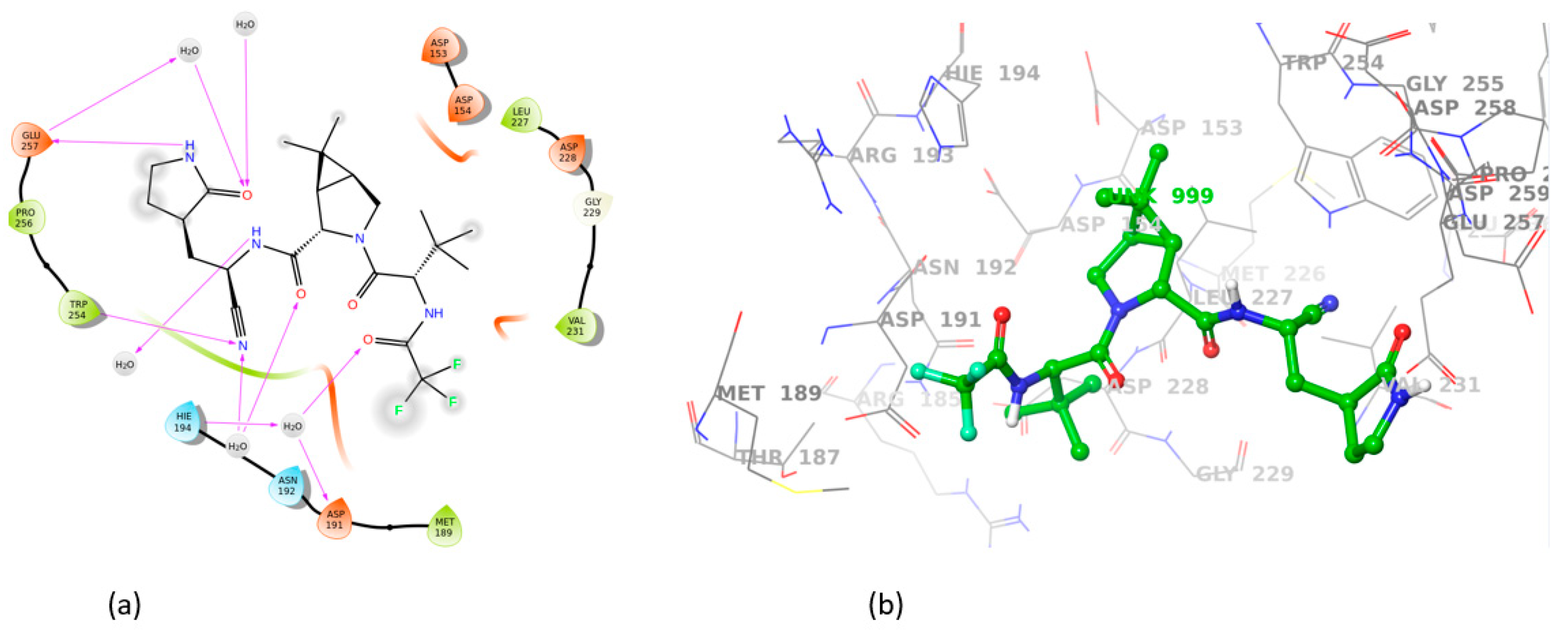
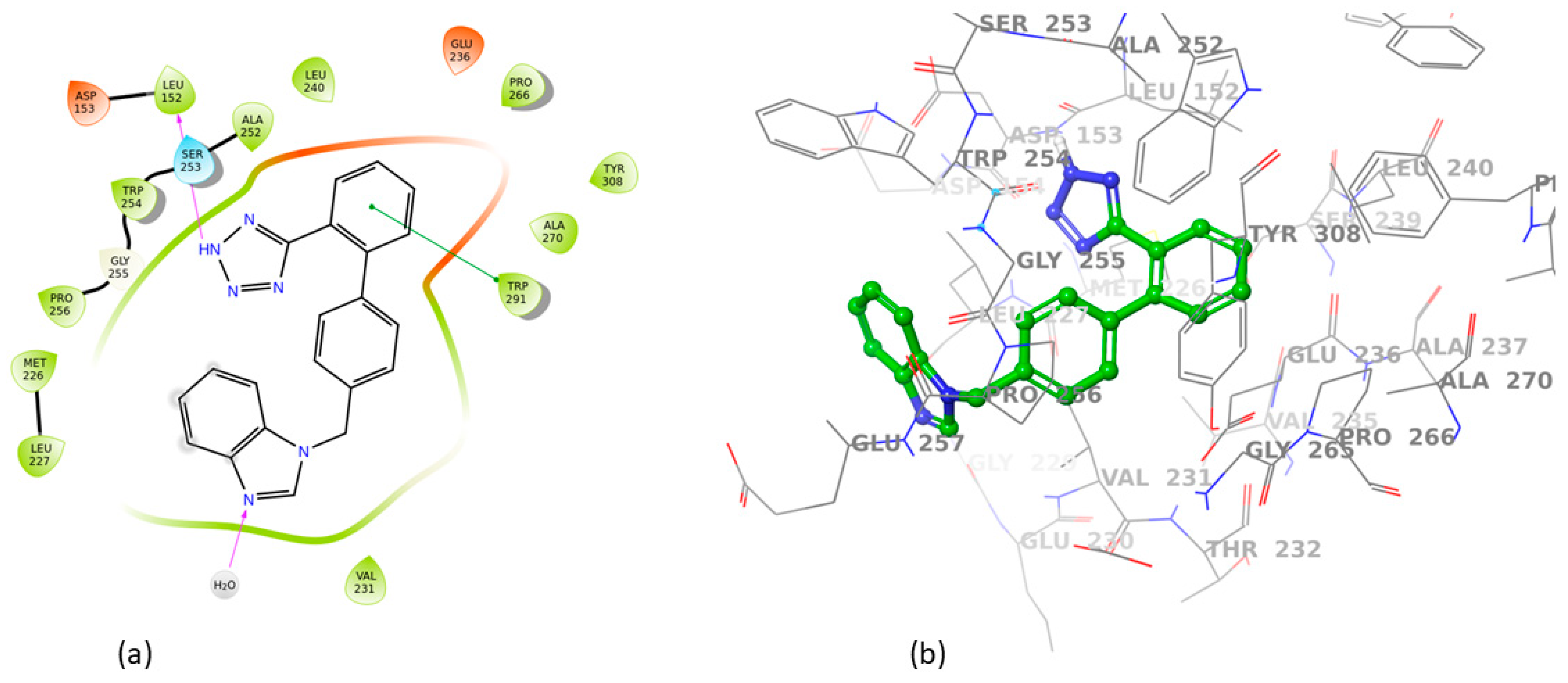
3.2. Induced Fit Docking
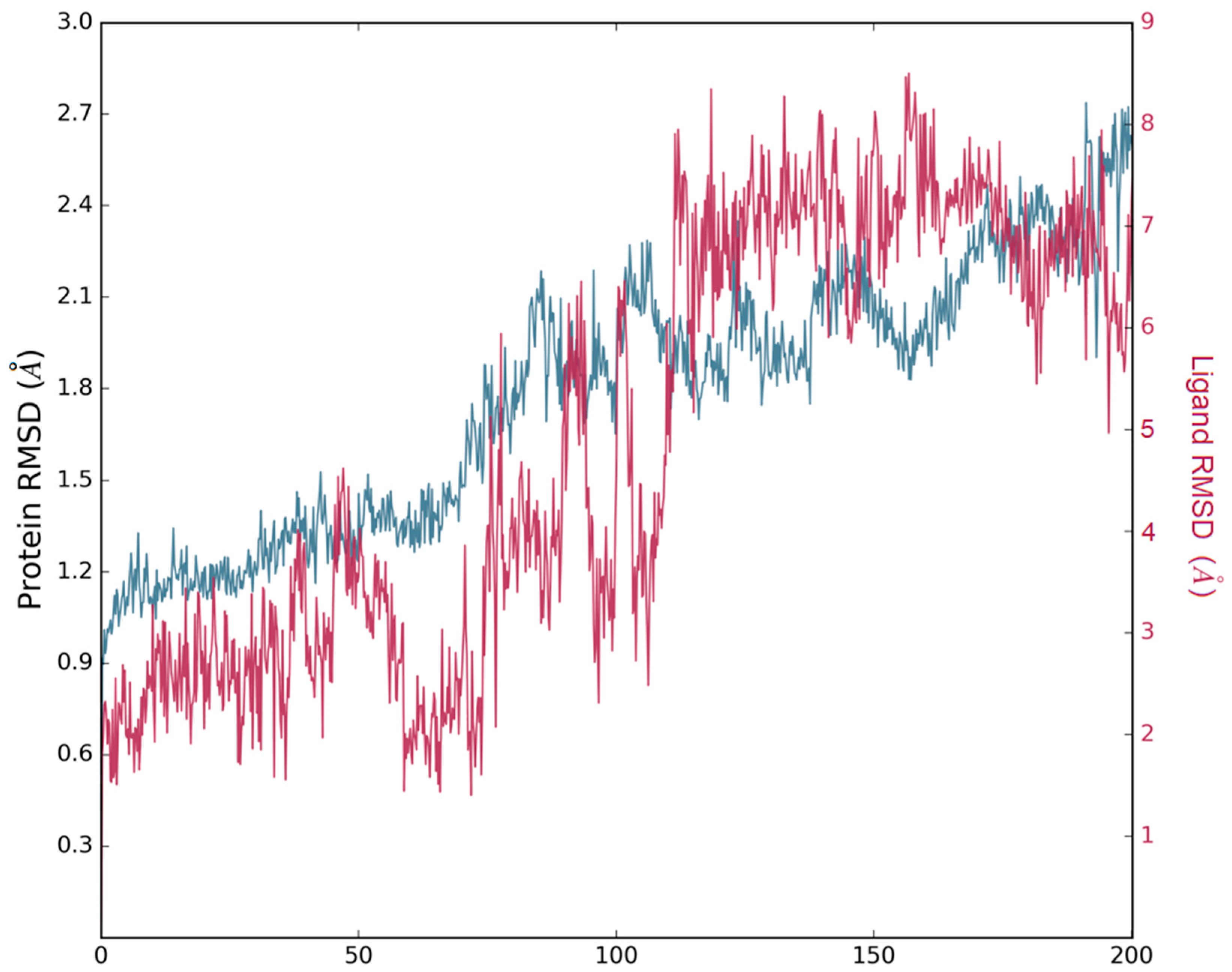
3.3. Molecular Dynamics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dahms, S.O.; Schnapp, G.; Winter, M.; Büttner, F.H.; Schlepütz, M.; Gnamm, C.; Pautsch, A.; Brandstetter, H. Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism. ACS Chem. Biol. 2022, 17, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G. Furin at the Cutting Edge: From Protein Traffic to Embryogenesis and Disease. Nat. Rev. Mol. Cell Biol. 2002, 3, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Than, M.E.; Henrich, S.; Bourenkov, G.P.; Bartunik, H.D.; Huber, R.; Bode, W. The Endoproteinase Furin Contains Two Essential Ca2+ Ions Stabilizing Its N-Terminus and the Unique S1 Specificity Pocket. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 61, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.; Sauter, D. Furin-mediated Protein Processing in Infectious Diseases and Cancer. Clin. Transl. Immunol. 2019, 8, e1073. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.; Rysä, J.; Almgren, P.; Nilsson, J.; Engström, G.; Orho-Melander, M.; Ruskoaho, H.; Melander, O. Plasma Levels of the Proprotein Convertase Furin and Incidence of Diabetes and Mortality. J. Intern. Med. 2018, 284, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Swärd, P.; Rosengren, B.E.; Jehpsson, L.; Karlsson, M.K. Association between Circulating Furin Levels, Obesity and Pro-inflammatory Markers in Children. Acta Paediatr. 2021, 110, 1863–1868. [Google Scholar] [CrossRef]
- Ren, K.; Jiang, T.; Zheng, X.-L.; Zhao, G.-J. Proprotein Convertase Furin/PCSK3 and Atherosclerosis: New Insights and Potential Therapeutic Targets. Atherosclerosis 2017, 262, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K. Furin Protease: From SARS CoV-2 to Anthrax, Diabetes, and Hypertension. Perm. J. 2020, 24, 20–187. [Google Scholar] [CrossRef]
- Bennett, B.D.; Denis, P.; Haniu, M.; Teplow, D.B.; Kahn, S.; Louis, J.-C.; Citron, M.; Vassar, R. A Furin-like Convertase Mediates Propeptide Cleavage of BACE, the Alzheimer’s β-Secretase. J. Biol. Chem. 2000, 275, 37712–37717. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Wu, J.; Yu, Y.; Liu, S.; Li, T.; Li, Q.; Ding, R.; Wang, H.; Nie, J.; et al. A Second Functional Furin Site in the SARS-CoV-2 Spike Protein. Emerg. Microbes Infect. 2022, 11, 182–194. [Google Scholar] [CrossRef]
- Jamison, D.A.; Anand Narayanan, S.; Trovão, N.S.; Guarnieri, J.W.; Topper, M.J.; Moraes-Vieira, P.M.; Zaksas, V.; Singh, K.K.; Wurtele, E.S.; Beheshti, A. A Comprehensive SARS-CoV-2 and COVID-19 Review, Part 1: Intracellular Overdrive for SARS-CoV-2 Infection. Eur. J. Hum. Genet. 2022, 30, 889–898. [Google Scholar] [CrossRef]
- Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 Pathogenesis. Nat. Rev. Microbiol. 2022, 20, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiang, R.; Huo, S.; Zhou, Y.; Jiang, S.; Wang, Q.; Yu, F. Molecular Mechanism of Interaction between SARS-CoV-2 and Host Cells and Interventional Therapy. Signal Transduct. Target. Ther. 2021, 6, 233. [Google Scholar] [CrossRef] [PubMed]
- Villoutreix, B.O.; Badiola, I.; Khatib, A.-M. Furin and COVID-19: Structure, Function and Chemoinformatic Analysis of Representative Active Site Inhibitors. Front. Drug Discov. 2022, 2, 899239. [Google Scholar] [CrossRef]
- Osadchuk, T.V.; Shybyryn, O.V.; Kibirev, V.K. Chemical Structure and Properties of Low-Molecular Furin Inhibitors. Ukr. Biochem. J. 2016, 88, 5–25. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zheng, M.; Yang, Y.; Gu, X.; Yang, K.; Li, M.; Liu, Y.; Zhang, Q.; Zhang, P.; Wang, Y.; et al. Furin: A Potential Therapeutic Target for COVID-19. iScience 2020, 23, 101642. [Google Scholar] [CrossRef]
- Cheng, Y.-W.; Chao, T.-L.; Li, C.-L.; Chiu, M.-F.; Kao, H.-C.; Wang, S.-H.; Pang, Y.-H.; Lin, C.-H.; Tsai, Y.-M.; Lee, W.-H.; et al. Furin Inhibitors Block SARS-CoV-2 Spike Protein Cleavage to Suppress Virus Production and Cytopathic Effects. Cell Rep. 2020, 33, 108254. [Google Scholar] [CrossRef]
- Kacprzak, M.M.; Peinado, J.R.; Than, M.E.; Appel, J.; Henrich, S.; Lipkind, G.; Houghten, R.A.; Bode, W.; Lindberg, I. Inhibition of Furin by Polyarginine-Containing Peptides. J. Biol. Chem. 2004, 279, 36788–36794. [Google Scholar] [CrossRef]
- Örd, M.; Faustova, I.; Loog, M. The Sequence at Spike S1/S2 Site Enables Cleavage by Furin and Phospho-Regulation in SARS-CoV2 but Not in SARS-CoV1 or MERS-CoV. Sci. Rep. 2020, 10, 16944. [Google Scholar] [CrossRef] [PubMed]
- Sielaff, F.; Than, M.E.; Bevec, D.; Lindberg, I.; Steinmetzer, T. New Furin Inhibitors Based on Weakly Basic Amidinohydrazones. Bioorg. Med. Chem. Lett. 2011, 21, 836–840. [Google Scholar] [CrossRef][Green Version]
- Osman, E.E.A.; Rehemtulla, A.; Neamati, N. Why All the Fury over Furin? J. Med. Chem. 2022, 65, 2747–2784. [Google Scholar] [CrossRef]
- Dahms, S.O.; Hardes, K.; Becker, G.L.; Steinmetzer, T.; Brandstetter, H.; Than, M.E. X-Ray Structures of Human Furin in Complex with Competitive Inhibitors. ACS Chem. Biol. 2014, 9, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Swiderski, J.; Gadanec, L.K.; Apostolopoulos, V.; Moore, G.J.; Kelaidonis, K.; Matsoukas, J.M.; Zulli, A. Role of Angiotensin II in Cardiovascular Diseases: Introducing Bisartans as a Novel Therapy for Coronavirus 2019. Biomolecules 2023, 13, 787. [Google Scholar] [CrossRef]
- Ridgway, H.; Moore, G.J.; Mavromoustakos, T.; Tsiodras, S.; Ligielli, I.; Kelaidonis, K.; Chasapis, C.T.; Gadanec, L.K.; Zulli, A.; Apostolopoulos, V.; et al. Discovery of a New Generation of Angiotensin Receptor Blocking Drugs: Receptor Mechanisms and in Silico Binding to Enzymes Relevant to SARS-CoV-2. Comput. Struct. Biotechnol. J. 2022, 20, 2091–2111. [Google Scholar] [CrossRef]
- Ridgway, H.; Chasapis, C.T.; Kelaidonis, K.; Ligielli, I.; Moore, G.J.; Gadanec, L.K.; Zulli, A.; Apostolopoulos, V.; Mavromoustakos, T.; Matsoukas, J.M. Understanding the Driving Forces That Trigger Mutations in SARS-CoV-2: Mutational Energetics and the Role of Arginine Blockers in COVID-19 Therapy. Viruses 2022, 14, 1029. [Google Scholar] [CrossRef] [PubMed]
- Ridgway, H.; Ntallis, C.; Chasapis, C.T.; Kelaidonis, K.; Matsoukas, M.-T.; Plotas, P.; Apostolopoulos, V.; Moore, G.; Tsiodras, S.; Paraskevis, D.; et al. Molecular Epidemiology of SARS-CoV-2: The Dominant Role of Arginine in Mutations and Infectivity. Viruses 2023, 15, 309. [Google Scholar] [CrossRef]
- Duarte, M.; Pelorosso, F.; Nicolosi, L.N.; Victoria Salgado, M.; Vetulli, H.; Aquieri, A.; Azzato, F.; Castro, M.; Coyle, J.; Davolos, I.; et al. Telmisartan for Treatment of COVID-19 Patients: An Open Multicenter Randomized Clinical Trial. EClinicalMedicine 2021, 37, 100962. [Google Scholar] [CrossRef] [PubMed]
- Elkahloun, A.G.; Saavedra, J.M. Candesartan Could Ameliorate the COVID-19 Cytokine Storm. Biomed. Pharmacother. 2020, 131, 110653. [Google Scholar] [CrossRef]
- Kelaidonis, K.; Ligielli, I.; Letsios, S.; Vidali, V.P.; Mavromoustakos, T.; Vassilaki, N.; Moore, G.J.; Hoffmann, W.; Węgrzyn, K.; Ridgway, H.; et al. Computational and Enzymatic Studies of Sartans in SARS-CoV-2 Spike RBD-ACE2 Binding: The Role of Tetrazole and Perspectives as Antihypertensive and COVID-19 Therapeutics. Int. J. Mol. Sci. 2023, 24, 8454. [Google Scholar] [CrossRef]
- Kellici, T.; Tzakos, A.; Mavromoustakos, T. Rational Drug Design and Synthesis of Molecules Targeting the Angiotensin II Type 1 and Type 2 Receptors. Molecules 2015, 20, 3868–3897. [Google Scholar] [CrossRef]
- Kellici, T.F.; Liapakis, G.; Tzakos, A.G.; Mavromoustakos, T. Pharmaceutical Compositions for Antihypertensive Treatments: A Patent Review. Expert Opin. Ther. Pat. 2015, 25, 1305–1317. [Google Scholar] [PubMed]
- Nejat, R.; Sadr, A.S. Are Losartan and Imatinib Effective against SARS-CoV2 Pathogenesis? A Pathophysiologic-Based in Silico Study. Silico Pharmacol. 2021, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Palli, V.; Leonis, G.; Zoupanou, N.; Georgiou, N.; Chountoulesi, M.; Naziris, N.; Tzeli, D.; Demetzos, C.; Valsami, G.; Marousis, K.D.; et al. Losartan Interactions with 2-Hydroxypropyl-β-CD. Molecules 2022, 27, 2421. [Google Scholar] [CrossRef] [PubMed]
- Ridgway, H.; Orbell, J.D.; Matsoukas, M.-T.; Kelaidonis, K.; Moore, G.J.; Tsiodras, S.; Gorgoulis, V.G.; Chasapis, C.T.; Apostolopoulos, V.; Matsoukas, J.M. W254 in Furin Functions as a Molecular Gate Promoting Anti-Viral Drug Binding: Elucidation of Putative Drug Tunneling and Docking by Non-Equilibrium Molecular Dynamics. Comput. Struct. Biotechnol. J. 2023, 21, 4589–4612. [Google Scholar] [CrossRef] [PubMed]
- Tzeliou, C.E.; Mermigki, M.A.; Tzeli, D. Review on the QM/MM Methodologies and Their Application to Metalloproteins. Molecules 2022, 27, 2660. [Google Scholar] [CrossRef] [PubMed]
- Kristyan, S. Theory of Variational Calculation with a Scaling Correct Moment Functional to Solve the Electronic Schrödinger Equation Directly for Ground State One-electron Density and Electronic Energy. Int. J. Quantum Chem. 2013, 113, 1479–1492. [Google Scholar] [CrossRef]
- Grimme, S.; Neese, F. Double-Hybrid Density Functional Theory for Excited Electronic States of Molecules. J. Chem. Phys. 2007, 127, 154116. [Google Scholar] [CrossRef] [PubMed]
- Vydrov, O.A.; Scuseria, G.E. Assessment of a Long-Range Corrected Hybrid Functional. J. Chem. Phys. 2006, 125, 234109. [Google Scholar] [CrossRef]
- Cohen, A.J.; Mori-Sánchez, P.; Yang, W. Challenges for Density Functional Theory. Chem. Rev. 2012, 112, 289–320. [Google Scholar] [CrossRef]
- Verma, P.; Truhlar, D.G. Status and Challenges of Density Functional Theory. Trends Chem. 2020, 2, 302–318. [Google Scholar] [CrossRef]
- Becke, A.D. Density-Functional Exchange-Energy Approximation with Correct Asymptotic Behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti Correlation-Energy Formula into a Functional of the Electron Density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 Suite of Density Functionals for Main Group Thermochemistry, Thermochemical Kinetics, Noncovalent Interactions, Excited States, and Transition Elements: Two New Functionals and Systematic Testing of Four M06-Class Functionals and 12 Other Function. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Tzeliou, C.E.; Tzeli, D. 3-Input AND Molecular Logic Gate with Enhanced Fluorescence Output: The Key Atom for the Accurate Prediction of the Spectra. J. Chem. Inf. Model. 2022, 62, 6436–6448. [Google Scholar] [CrossRef] [PubMed]
- Pagliai, M.; Osticioli, I.; Nevin, A.; Siano, S.; Cardini, G.; Schettino, V. DFT Calculations of the IR and Raman Spectra of Anthraquinone Dyes and Lakes. J. Raman Spectrosc. 2018, 49, 668–683. [Google Scholar] [CrossRef]
- Paschoal, D.; Guerra, C.F.; de Oliveira, M.A.L.; Ramalho, T.C.; Dos Santos, H.F. Predicting Pt-195 NMR Chemical Shift Using New Relativistic All-electron Basis Set. J. Comput. Chem. 2016, 37, 2360–2373. [Google Scholar] [CrossRef] [PubMed]
- Vreven, T.; Morokuma, K. Chapter 3 Hybrid Methods: ONIOM(QM:MM) and QM/MM. Annu. Rep. Comput. Chem. 2006, 2, 35–51. [Google Scholar] [CrossRef]
- Curtiss, L.A.; McGrath, M.P.; Blaudeau, J.; Davis, N.E.; Binning, R.C.; Radom, L. Extension of Gaussian-2 Theory to Molecules Containing Third-row Atoms Ga–Kr. J. Chem. Phys. 1995, 103, 6104–6113. [Google Scholar] [CrossRef]
- Bikadi, Z.; Hazai, E. Application of the PM6 Semi-Empirical Method to Modeling Proteins Enhances Docking Accuracy of AutoDock. J. Cheminform. 2009, 1, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tzeli, D.; Theodorakopoulos, G.; Petsalakis, I.D.; Ajami, D.; Rebek, J. Jr Theoretical study of hydrogen bonding in homodimers and heterodimers of amide, boronic acid and carboxylic acid, free and in encapsulation complexes. J. Am. Chem. Soc. 2011, 133, 16977–16985. [Google Scholar] [CrossRef]
- Cossi, M.; Scalmani, G.; Rega, N.; Barone, V. New Developments in the Polarizable Continuum Model for Quantum Mechanical and Classical Calculations on Molecules in Solution. J. Chem. Phys. 2002, 117, 43–54. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision B.01; Gaussian, Inc.: Wallingford CT, USA, 2016. [Google Scholar]
- Douglas, L.E.J.; Reihill, J.A.; Ho, M.W.Y.; Axten, J.M.; Campobasso, N.; Schneck, J.L.; Rendina, A.R.; Wilcoxen, K.M.; Martin, S.L. A Highly Selective, Cell-Permeable Furin Inhibitor BOS-318 Rescues Key Features of Cystic Fibrosis Airway Disease. Cell Chem. Biol. 2022, 29, 947–957.e8. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L.L.C. MacroModel, Version 10.2; Schrödinger, LLC: New York, NY, USA, 2013. [Google Scholar]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Georgiou, N.; Gouleni, N.; Chontzopoulou, E.; Skoufas, G.S.; Gkionis, A.; Tzeli, D.; Vassiliou, S.; Mavromoustakos, T. Structure Assignment, Conformational Properties and Discovery of Potential Targets of the Ugi Cinnamic Adduct NGI25. J. Biomol. Struct. Dyn. 2021, 41, 1253–1266. [Google Scholar] [CrossRef]
- Imtiaz, S.; Muzaffar, S.; Ali, S.M. Demonstrating Accuracy of the Already Proposed Protocol for Structure Elucidation of Cyclodextrin Inclusion Complexes by Validation Using Quantitative ROESY Analysis. J. Incl. Phenom. Macrocycl. Chem. 2021, 100, 71–87. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Humphreys, D.D.; Friesner, R.A.; Berne, B.J. A Multiple-Time-Step Molecular Dynamics Algorithm for Macromolecules. J. Phys. Chem. 1994, 98, 6885–6892. [Google Scholar] [CrossRef]
- Lyman, E.; Zuckerman, D.M. Ensemble-Based Convergence Analysis of Biomolecular Trajectories. Biophys. J. 2006, 91, 164–172. [Google Scholar] [CrossRef]
- Bütikofer, A.; Løken, K.V.; Salvanes, K.G. Infant Health Care and Long-Term Outcomes. Rev. Econ. Stat. 2019, 101, 341–354. [Google Scholar] [CrossRef]
- Sheybani, Z.; Heydari Dokoohaki, M.; Negahdaripour, M.; Dehdashti, M.; Zolghadr, H.; Moghadami, M.; Masoompour, S.M.; Zolghadr, A.R. The Interactions of Folate with the Enzyme Furin: A Computational Study. RSC Adv. 2021, 11, 23815–23824. [Google Scholar] [CrossRef] [PubMed]
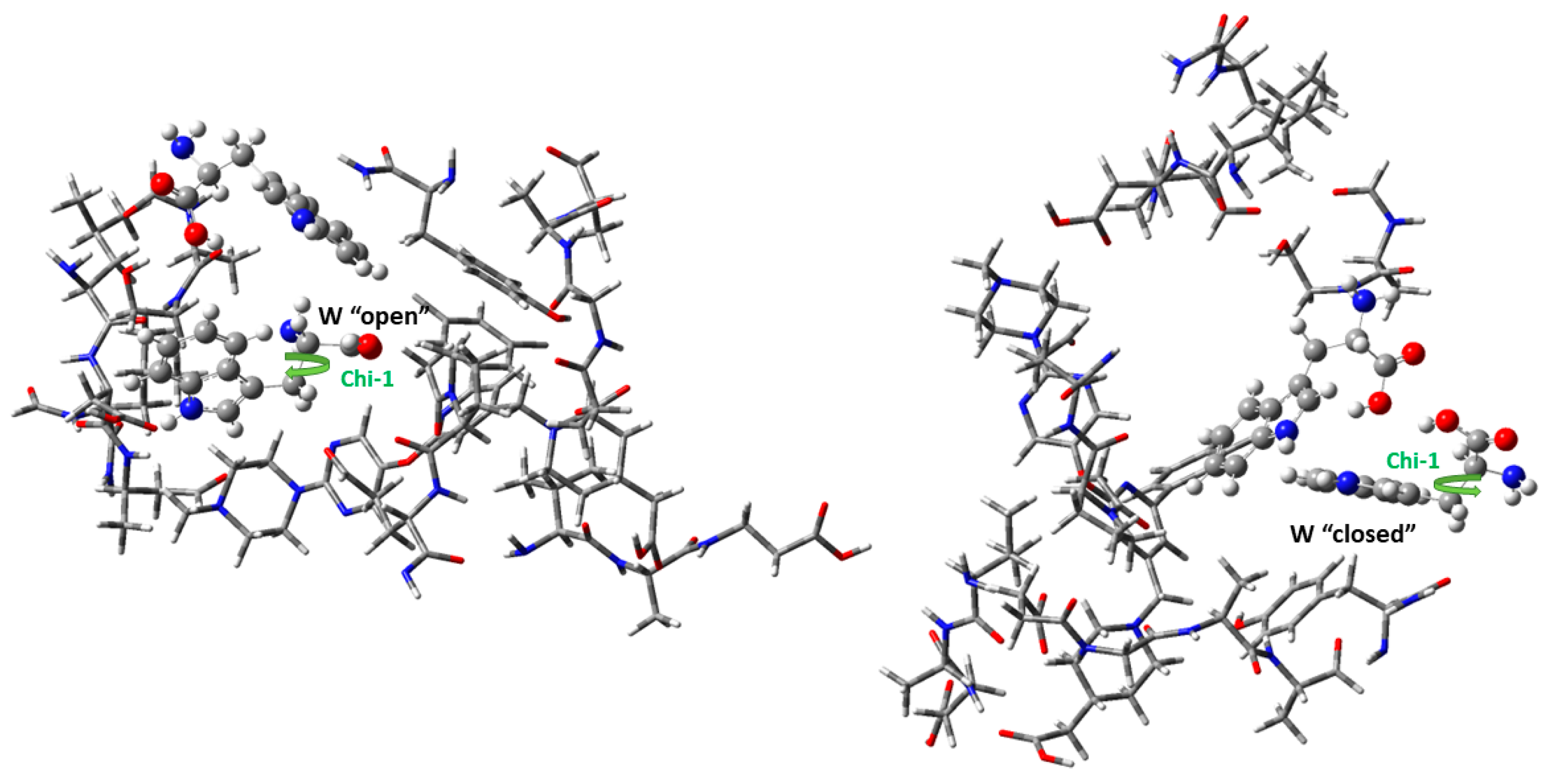



| “Open” Conformation | “Closed” Conformation | |
|---|---|---|
| CCCC (Chi-1) | 174.8 | 75.3 |
| CCCC | 63.3 | 78.9 |
| BE | −32.6 | −19.3 |
| ΔH a | −32.8 | −18.8 |
| Def | 16.0 | 18.0 |
| Rel | 0.0 | 30.5 |
| Compound | Enzyme | Hydrogen Bonds | π-π Stacking | IFD Score a |
|---|---|---|---|---|
| Losartan | 7LCU | ALA252, ASP264 | TRP254, HIE194 | −8.98 |
| Benzimidazole-N-biphenyltetrazole | 7LCU | LEU152 | TRP291 | −10.48 |
| Benzimidazole bis-N,N′-biphenyltetrazole | 7LCU | ASP191, TRP254, HIE194 | TRP291 | −12.06 |
| BV6 | 7LCU | ASP191, GLU257 | ARG193, TRP254 | −10.02 |
| Candesartan | 7LCU | TRP254 | TRP291, TYR308 | −9.38 |
| Losartan carboxylic acid | 7LCU | ASP264 | TYR308 | −9.41 |
| Nirmitrevil | 7LCU | ASP191, HIE194, TRP254, GLU257 | −7.30 | |
| Telmisartan | 7LCU | ASP264, GLU257, GLY255 | TRP254, TYR308 | −11.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgiou, N.; Mavromoustakos, T.; Tzeli, D. Docking, MD Simulations, and DFT Calculations: Assessing W254’s Function and Sartan Binding in Furin. Curr. Issues Mol. Biol. 2024, 46, 8226-8238. https://doi.org/10.3390/cimb46080486
Georgiou N, Mavromoustakos T, Tzeli D. Docking, MD Simulations, and DFT Calculations: Assessing W254’s Function and Sartan Binding in Furin. Current Issues in Molecular Biology. 2024; 46(8):8226-8238. https://doi.org/10.3390/cimb46080486
Chicago/Turabian StyleGeorgiou, Nikitas, Thomas Mavromoustakos, and Demeter Tzeli. 2024. "Docking, MD Simulations, and DFT Calculations: Assessing W254’s Function and Sartan Binding in Furin" Current Issues in Molecular Biology 46, no. 8: 8226-8238. https://doi.org/10.3390/cimb46080486
APA StyleGeorgiou, N., Mavromoustakos, T., & Tzeli, D. (2024). Docking, MD Simulations, and DFT Calculations: Assessing W254’s Function and Sartan Binding in Furin. Current Issues in Molecular Biology, 46(8), 8226-8238. https://doi.org/10.3390/cimb46080486








