The Role of WNT3A Protein and Gene Variants in Allergic Rhinitis: A Case-Control Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Determination of Serum WNT3A Protein Levels by ELISA
2.3. Genomic DNA Extraction and Genoytping
2.4. Statistical Analysis
3. Results
3.1. Serum WNT3A Levels
3.2. Distribution of Genotypes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meng, Y.; Wang, C.; Zhang, L. Advances and novel developments in allergic rhinitis. Allergy 2020, 75, 3069–3076. [Google Scholar] [CrossRef] [PubMed]
- Testera-Montes, A.; Jurado, R.; Salas, M.; Eguiluz-Gracia, I.; Mayorga, C. Diagnostic Tools in Allergic Rhinitis. Front. Allergy 2021, 2, 721851. [Google Scholar] [CrossRef] [PubMed]
- Zoabi, Y.; Levi-Schaffer, F.; Eliashar, R. Allergic Rhinitis: Pathophysiology and Treatment Focusing on Mast Cells. Biomedicines 2022, 10, 2486. [Google Scholar] [CrossRef] [PubMed]
- Brozek, J.L.; Bousquet, J.; Agache, I.; Agarwal, A.; Bachert, C.; Bosnic-Anticevich, S.; Brignardello-Petersen, R.; Canonica, G.W.; Casale, T.; Chavannes, N.H.; et al. Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines-2016 revision. J. Allergy Clin. Immunol. 2017, 140, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/beta-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Reuter, S.; Beckert, H.; Taube, C. Take the Wnt out of the inflammatory sails: Modulatory effects of Wnt in airway diseases. Lab. Investig. 2016, 96, 177–185. [Google Scholar] [CrossRef]
- Tebroke, J.; Lieverse, J.E.; Safholm, J.; Schulte, G.; Nilsson, G.; Ronnberg, E. Wnt-3a Induces Cytokine Release in Human Mast Cells. Cells 2019, 8, 1372. [Google Scholar] [CrossRef]
- Li, J.; Xue, K.; Zheng, Y.; Wang, Y.; Xu, C. RORA Overexpression Alleviates Nasal Mucosal Injury and Enhances Red Blood Cell Immune Adhesion Function in a Mouse Model of Allergic Rhinitis via Inactivation of the Wnt/beta-Catenin Signaling Pathway. Int. Arch. Allergy Immunol. 2019, 180, 79–90. [Google Scholar] [CrossRef]
- Greiner, A.N.; Hellings, P.W.; Rotiroti, G.; Scadding, G.K. Allergic rhinitis. Lancet 2011, 378, 2112–2122. [Google Scholar] [CrossRef]
- Huang, S.X.; Green, M.D.; de Carvalho, A.T.; Mumau, M.; Chen, Y.W.; D’Souza, S.L.; Snoeck, H.W. The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat. Protoc. 2015, 10, 413–425. [Google Scholar] [CrossRef]
- McCauley, K.B.; Hawkins, F.; Serra, M.; Thomas, D.C.; Jacob, A.; Kotton, D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 2017, 20, 844–857.E6. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Luo, J.Q.; Ouyang, F.; Cheng, L.; Chen, X.P.; Zhou, H.H.; Huang, W.H.; Zhang, W. WNT3A rs752107(C > T) Polymorphism Is Associated With an Increased Risk of Essential Hypertension and Related Cardiovascular Diseases. Front. Cardiovasc. Med. 2021, 8, 675222. [Google Scholar] [CrossRef] [PubMed]
- Hernando, B.; Pena-Chilet, M.; Ibarrola-Villava, M.; Martin-Gonzalez, M.; Gomez-Fernandez, C.; Ribas, G.; Martinez-Cadenas, C. Genetic 3′UTR variation is associated with human pigmentation characteristics and sensitivity to sunlight. Exp. Dermatol. 2017, 26, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Yang, L.; Li, P.Q.; Wu, H.; Xie, H.B.; Shen, X.; Xie, X.D. Association of Wnt3A gene variants with non-syndromic cleft lip with or without cleft palate in Chinese population. Arch. Oral Biol. 2011, 56, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Cruz, R.; Garcia-Ortiz, H.; Castillejos-Lopez, M.; Quiterio, M.; Valdes-Flores, M.; Orozco, L.; Villarreal-Molina, T.; Salmeron, J. WNT3A gene polymorphisms are associated with bone mineral density variation in postmenopausal mestizo women of an urban Mexican population: Findings of a pathway-based high-density single nucleotide screening. Age 2014, 36, 9635. [Google Scholar] [CrossRef] [PubMed]
- Messoudi, S.; Al-Sulaiti, M.A.; Al-Busaidi, A.S.; Dendana, M.; Nsiri, B.; Almawi, W.Y.; Mahjoub, T. Contribution of JAK2 and STAT3 variants to the genetic susceptibility of recurrent miscarriage among Bahraini and Tunisian Arabs. Mol. Biol. Rep. 2013, 40, 585–589. [Google Scholar] [CrossRef]
- Vine, A.E.; Curtis, D. Markers typed in genome-wide analysis identify regions showing deviation from Hardy-Weinberg equilibrium. BMC Res. Notes 2009, 2, 29. [Google Scholar] [CrossRef]
- Maan Hasan, S.; Adnan, F.A.-A.; Akeel Hussain Ali, A.-A. Intronic SNPs and Genetic Diseases: A Review. Int. J. Res. Appl. Sci. Biotechnol. 2021, 8, 267–274. [Google Scholar] [CrossRef]
- Reuter, S.; Martin, H.; Beckert, H.; Bros, M.; Montermann, E.; Belz, C.; Heinz, A.; Ohngemach, S.; Sahin, U.; Stassen, M.; et al. The Wnt/beta-catenin pathway attenuates experimental allergic airway disease. J. Immunol. 2014, 193, 485–495. [Google Scholar] [CrossRef]
- Qiu, C.Y.; Cui, X.Y.; Lu, M.P.; Yin, M.; Xu, W.Y.; Zhu, X.J.; Yang, Q.; Cheng, L. CircRNA expression profiles and circRNA-miRNA-mRNA crosstalk in allergic rhinitis. World Allergy Organ. J. 2021, 14, 100548. [Google Scholar] [CrossRef]
- Wu, G.; Yang, G.; Zhang, R.; Xu, G.; Zhang, L.; Wen, W.; Lu, J.; Liu, J.; Yu, Y. Altered microRNA Expression Profiles of Extracellular Vesicles in Nasal Mucus From Patients with Allergic Rhinitis. Allergy Asthma Immunol. Res. 2015, 7, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Z.Q.; Zhang, R.X.; Zhao, R.W.; Dong, W.Y.; Wang, H.; Deng, C.R.; Zhuang, G.S. Effect of PM2.5 on MicroRNA Expression and Function in Nasal Mucosa of Rats with Allergic Rhinitis. Am. J. Rhinol. Allergy 2020, 34, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Bryborn, M.; Adner, M.; Cardell, L.O. Psoriasin, one of several new proteins identified in nasal lavage fluid from allergic and non-allergic individuals using 2-dimensional gel electrophoresis and mass spectrometry. Respir. Res. 2005, 6, 118. [Google Scholar] [CrossRef]
- Howarth, P.H.; Salagean, M.; Dokic, D. Allergic rhinitis: Not purely a histamine-related disease. Allergy 2000, 55 (Suppl. S64), 7–16. [Google Scholar] [CrossRef]
- Mizuguchi, H.; Kitamura, Y.; Takeda, N.; Fukui, H. Molecular Signaling and Transcriptional Regulation of Histamine H1 Receptor Gene. Curr. Top. Behav. Neurosci. 2022, 59, 91–110. [Google Scholar] [CrossRef]
- Nappi, E.; Paoletti, G.; Malvezzi, L.; Ferri, S.; Racca, F.; Messina, M.R.; Puggioni, F.; Heffler, E.; Canonica, G.W. Comorbid allergic rhinitis and asthma: Important clinical considerations. Expert Rev. Clin. Immunol. 2022, 18, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, T.; Gosens, R. Revisiting asthma therapeutics: Focus on WNT signal transduction. Drug Discov. Today 2018, 23, 49–62. [Google Scholar] [CrossRef]
- Lewis, A.; Sanchez, S.; Berti, G.; Pan-Castillo, B.; Nijhuis, A.; Mehta, S.; Eleid, L.; Gordon, H.; Gadhok, R.; Kimberley, C.; et al. Small-molecule Wnt inhibitors are a potential novel therapy for intestinal fibrosis in Crohns disease. Clin Sci. 2022, 136, 1405–1423. [Google Scholar] [CrossRef]
- Singla, A.; Reuter, S.; Taube, C.; Peters, M.; Peters, K. The molecular mechanisms of remodeling in asthma, COPD and IPF with a special emphasis on the complex role of Wnt5A. Inflamm. Res. 2023, 72, 577–588. [Google Scholar] [CrossRef]
- Liu, J.; Ren, X.; Zhang, M.; Lei, Y.; Chen, Y.; He, H. Roles of Wnt3a and Dkk1 in experimental periodontitis. J. Dent. Sci. 2017, 12, 220–225. [Google Scholar] [CrossRef]
- Xu, G.; Emmons, R.; Hernández-Saavedra, D.; Kriska, A.; Pan, Y.-X.; Chen, H. Regulation of Gene Expression of Wnt signaling Pathway by Dietary High Fat and Effects on Colon Epithelia of Male Mice. FASEB J. 2018, 31, 643.22. [Google Scholar] [CrossRef]
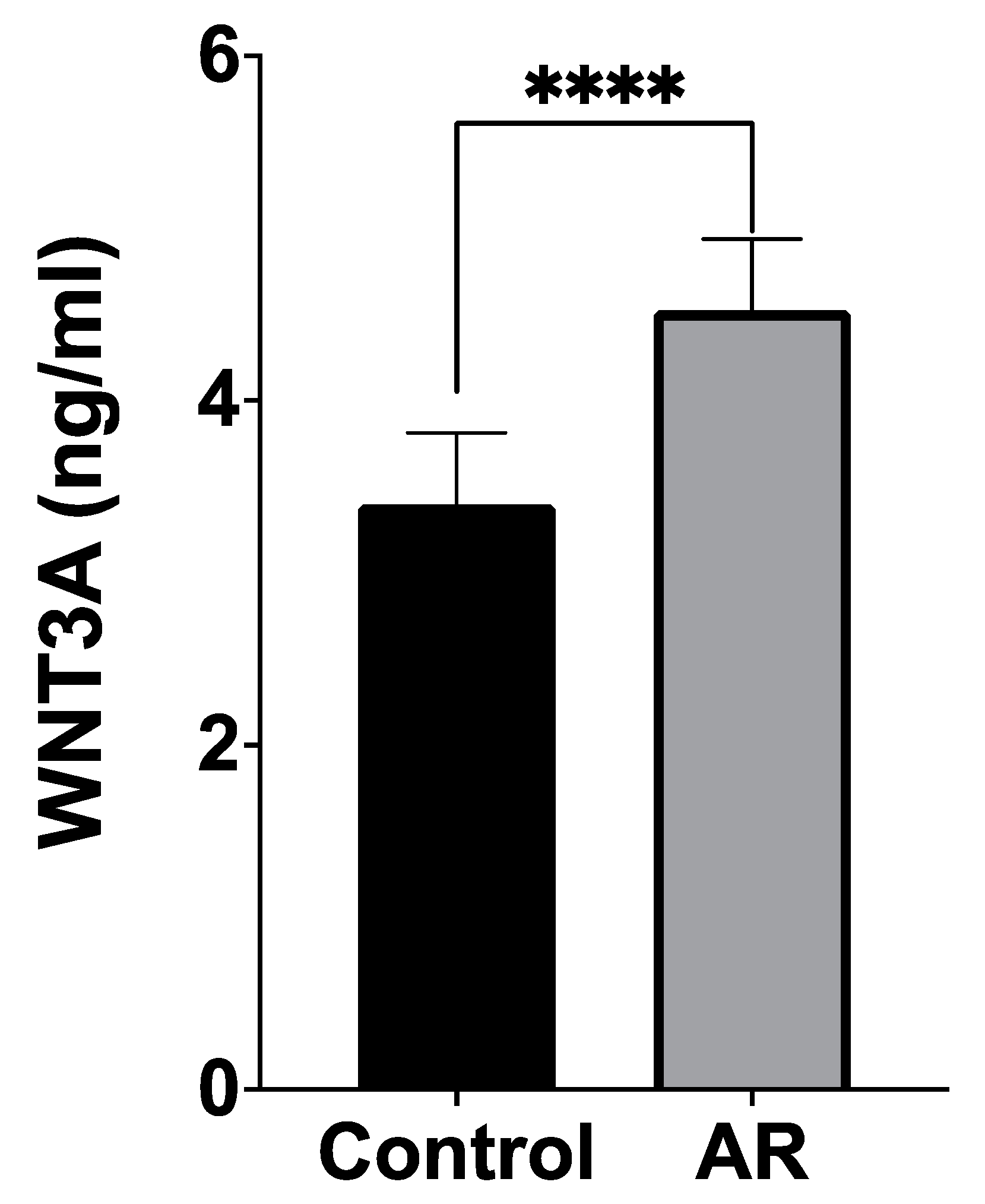


| SNP | Oligonucleotide Primer Sequences | Annealing Temperature (°C) | Fragment Sizes (bp) |
|---|---|---|---|
| rs752107 (C>T) | F: 5′-AGCAGGACTCCCACCTAAAC-3′ R: 5′-GCCTCATCCACCATAAAACC-3′ | 60.5 | CC: 302 + 165 CT: 302 + 165 + 118 + 47 TT: 302 + 118 + 47 |
| rs3121310 (G>A) | F: 5′-ATGCTCGGTGCCCTCTAAC-3′ R: 5′-GGCTTACTGACATGTGGTGC-3′ | 60.5 | GG: 267 + 100 + 43 GA: 367 + 267 + 100 + 43 AA: 367 + 43 |
| Characteristics | AR (n = 92) | Control (n = 86) | p | |
|---|---|---|---|---|
| Male (%)/ Female (%) | 32 (34.8%)/ 60 (65.2%) | 21 (24.4%)/ 65 (75.6%) | 0.1429 * | |
| Mean age (years) ± SD | 37.04 ± 12.47 | 33.16 ± 11.67 | 0.0550 ** | |
| Duration of AR diagnosis (months) (min–max) | 28.46 (18–72) | |||
| Severity of AR | Mild | 64 (69.6%) | ||
| Moderate/Severe | 28 (30.4%) | |||
| Symptom duration | Intermittent | 75 (81.5%) | ||
| Persistent | 17 (18.5%) | |||
| Symptom Duration | Severity of AR | |||||
|---|---|---|---|---|---|---|
| Intermittent (n = 75) | Persistent (n = 17) | p * | Mild (n = 64) | Moderate/Severe (n = 28) | p * | |
| WNT3A levels ng/mL (mean± SD) | 4.133 ± 3.733 | 5.064 ± 3.851 | 0.0984 | 4.828 ± 4.054 | 3.109 ± 2.636 | 0.0532 |
| SNP | AR n (%) | Control n (%) | OR ** (95% CI) | p * | |
|---|---|---|---|---|---|
| rs752107 | Genotype | n = 92 | n = 86 | ||
| CC | 71 (77.2%) | 71 (82.6%) | Ref | Ref | |
| CT | 16 (17.4%) | 8 (9.3%) | 0.5 (0.2073–1.234) | 0.184 | |
| TT | 5 (5.4%) | 7 (8.1%) | 1.4 (0.4574–4.075) | 0.765 | |
| CT + TT | 21 (22.8%) | 15(%) | 0.6818 (0.3205–1.452) | 0.356 | |
| Allele | n = 184 | n = 172 | |||
| C | 158 (85.9%) | 150 (87.2%) | Ref | Ref | |
| T | 26 (14.1%) | 22 (12.8%) | 0.8913 (0.486–1.616) | 0.758 | |
| rs3121310 | Genotype | ||||
| GG | 50 (54.4%) | 34 (39.5%) | Ref | Ref | |
| GA | 35 (38%) | 46 (53.5%) | 0.5174 (0.280–0.978) | 0.0433 | |
| AA | 7 (7.6%) | 6 (7%) | 0.7933 (0.2607–2.731) | 0.767 | |
| GA + AA | 42 (45.6%) | 52 (60.5%) | 1.821 1.008–3.371 | 0.0523 | |
| Allele | n = 184 | n = 172 | |||
| G | 135 (73.4%) | 114 (66.3%) | Ref | Ref | |
| A | 49 (26.6%) | 58 (33.7%) | 0.7134 (0.4482–1.125) | 0.165 |
| SNP | Rare Allele Frequency | p (HWE) | ||
|---|---|---|---|---|
| AR | Control | AR | Control | |
| rs752107 | 0.141 | 0.128 | 0.0147 | 0.00001 |
| rs3121310 | 0.266 | 0.337 | 0.7917 | 0.0929 |
| rs3121310 | WNT3A Levels (ng/mL) | IgE Levels (UI/mL) | Symptom Duration | Severity of AR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | p * | Mean ± SD | p * | Intermittent n (%) | Persistent n (%) | OR (95% CI) | p ** | Mild n (%) | Moderate/Severe n (%) | OR (95% CI) | p ** | |
| GG | 4.410 ± 3.975 | Ref. | 170.9 ± 147.5 | Ref. | 42 (45.6%) | 8 (8.7%) | Ref. | Ref. | 36 (39.1%) | 14 (15.2%) | Ref. | Ref. |
| GA | 4.157 ± 3.484 | >0.999 | 172.8 ± 206.6 | >0.999 | 26 (28.3%) | 9 (9.8%) | 1.817 (0.6203–5.663) | 0.2858 | 23 (25%) | 12 (13.1%) | 1.342 (0.5269–3.331) | 0.6341 |
| AA | 4.300 ± 3.927 | >0.999 | 166.9 ± 137.1 | >0.999 | 7 (7.6%) | 0 (0%) | 0.000 (0.000–2.857) | 0.5769 | 5 (5.4%) | 2 (2.2%) | 1.003 (0.8223–1.389) | >0.999 |
| GA + AA | 4.181 ± 3.511 | >0.999 | 171.9 ± 195.3 | >0.999 | 33 (35.9%) | 9 (9.8%) | 1.432 (0.5025–4.335) | 0.5936 | 28 (30.4%) | 14 (15.3%) | 1.286 (0.5509–2.971) | 0.6520 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demir Eksi, D.; Gunizi, H. The Role of WNT3A Protein and Gene Variants in Allergic Rhinitis: A Case-Control Study. Curr. Issues Mol. Biol. 2024, 46, 9523-9533. https://doi.org/10.3390/cimb46090565
Demir Eksi D, Gunizi H. The Role of WNT3A Protein and Gene Variants in Allergic Rhinitis: A Case-Control Study. Current Issues in Molecular Biology. 2024; 46(9):9523-9533. https://doi.org/10.3390/cimb46090565
Chicago/Turabian StyleDemir Eksi, Durkadin, and Huseyin Gunizi. 2024. "The Role of WNT3A Protein and Gene Variants in Allergic Rhinitis: A Case-Control Study" Current Issues in Molecular Biology 46, no. 9: 9523-9533. https://doi.org/10.3390/cimb46090565





