Molecular Integrative Study on Inhibitory Effects of Pentapeptides on Polymerization and Cell Toxicity of Amyloid-β Peptide (1–42)
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Molecular Docking
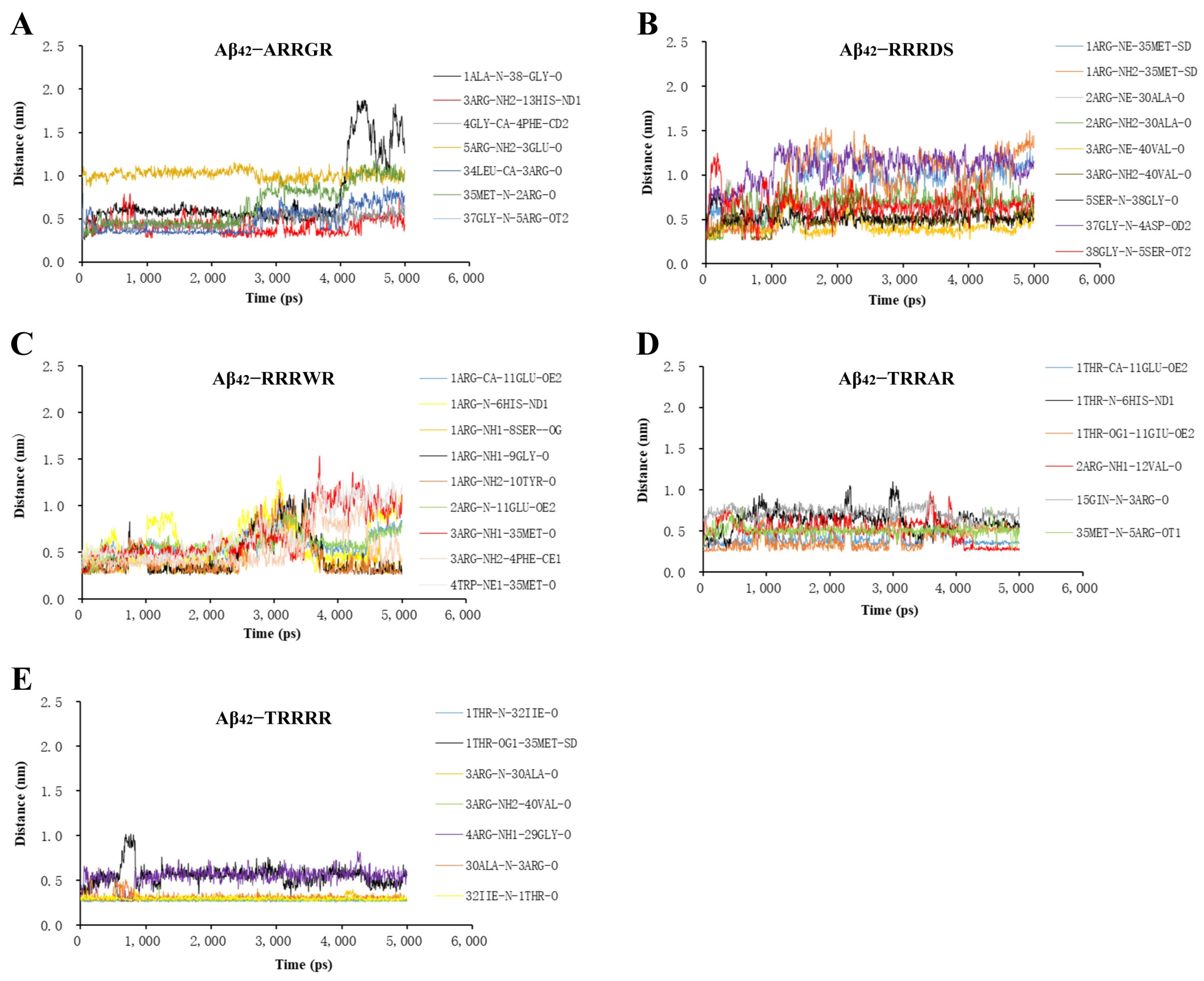
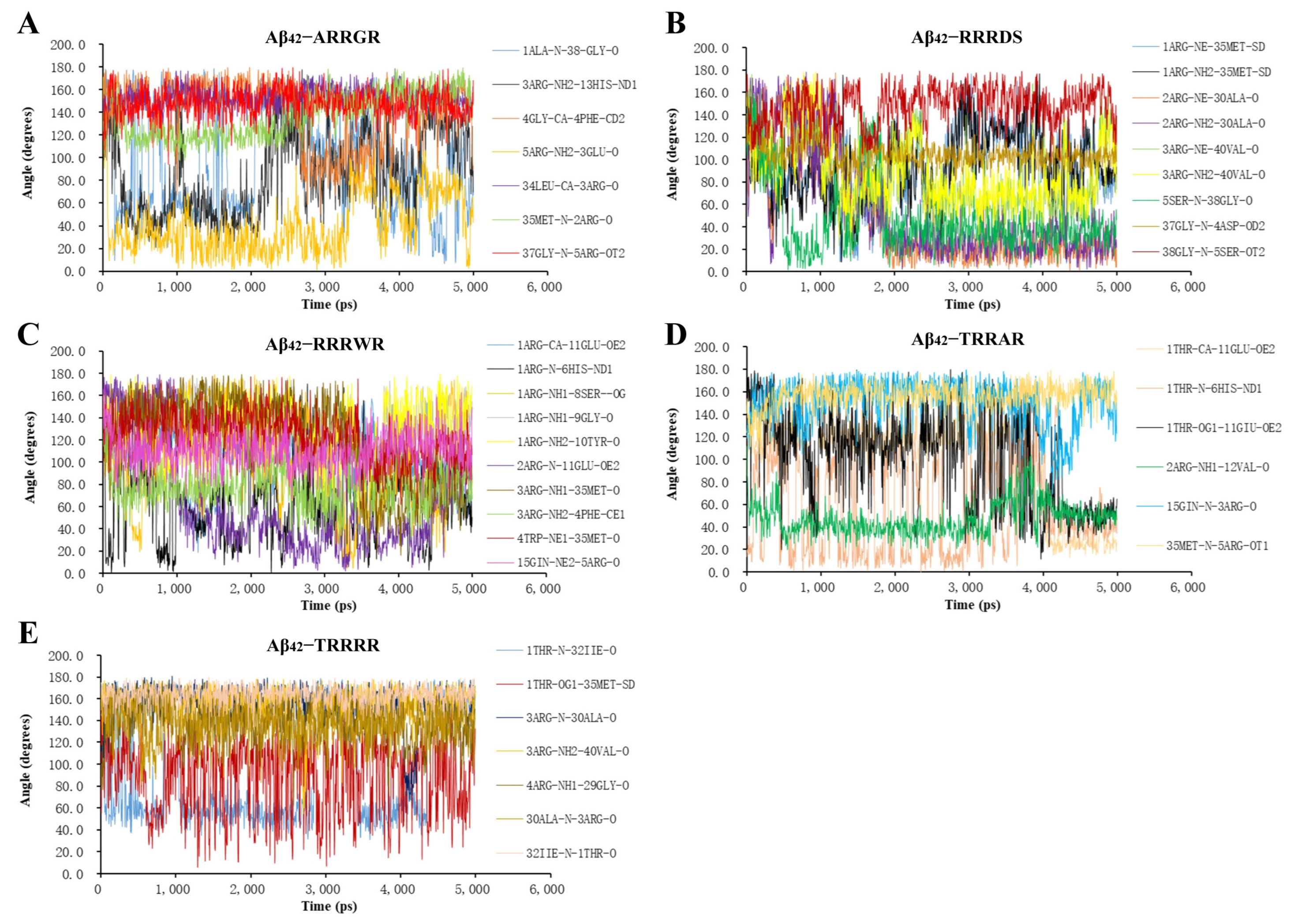
2.3. Molecular Dynamics Analysis of Binding Stability
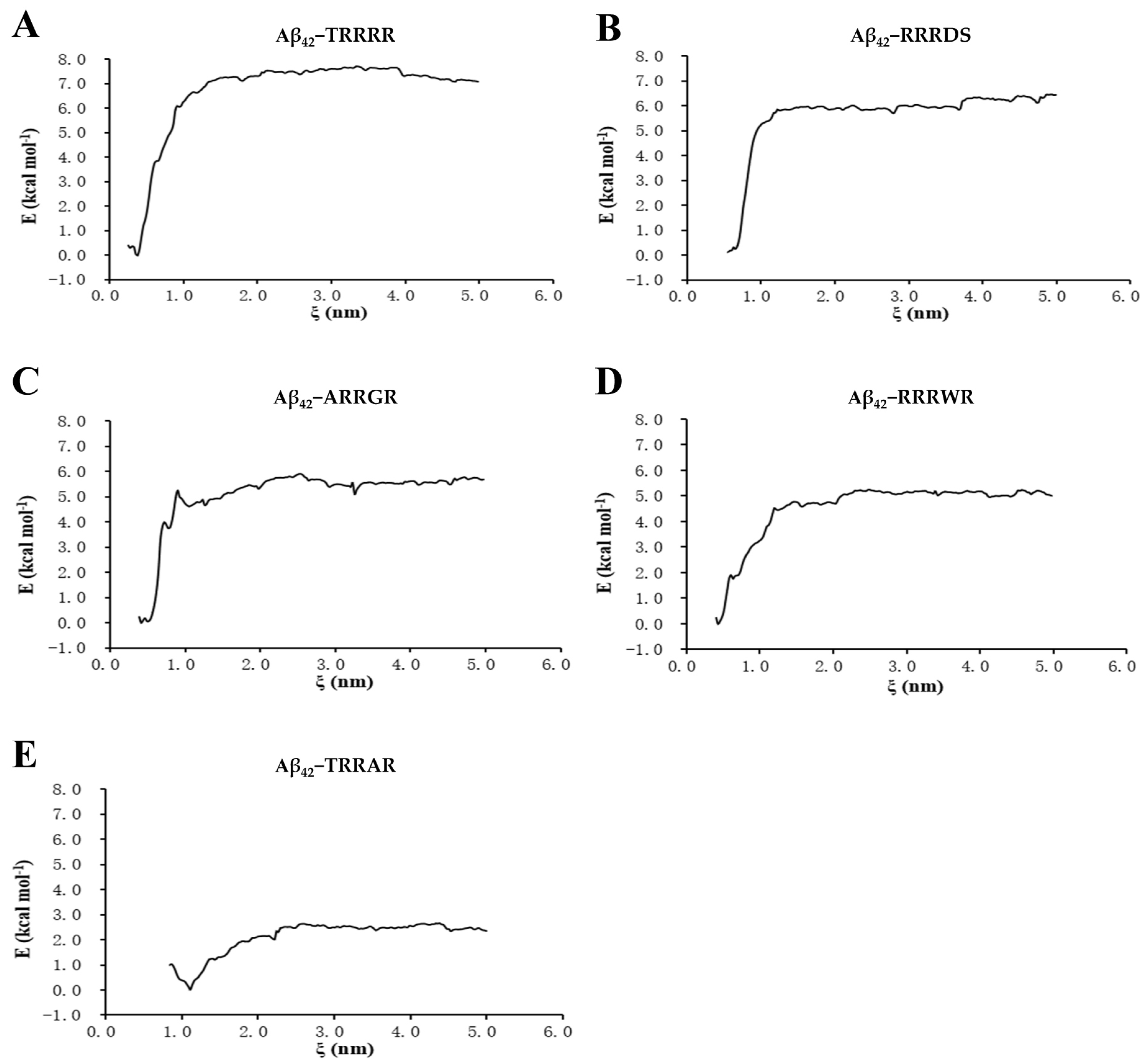
2.4. Umbrella Sampling of Binding Free Energy
2.5. ThT Fluorescent Detection of Aggregation of Aβ42
2.6. Transmission Electron Microscope Observation of Aggregated Aβ42
2.7. Cell Culture
2.8. Transfection and Expression of Secretable Aβ42
2.9. ELISA Experiment
2.10. Microscopic Observation of Cell Death
2.11. Flow Cytometry Methods to Detect Reactive Oxygen Species and Apoptosis
2.12. Statistical Analysis
3. Results
3.1. Molecular Docking of Pentapeptides
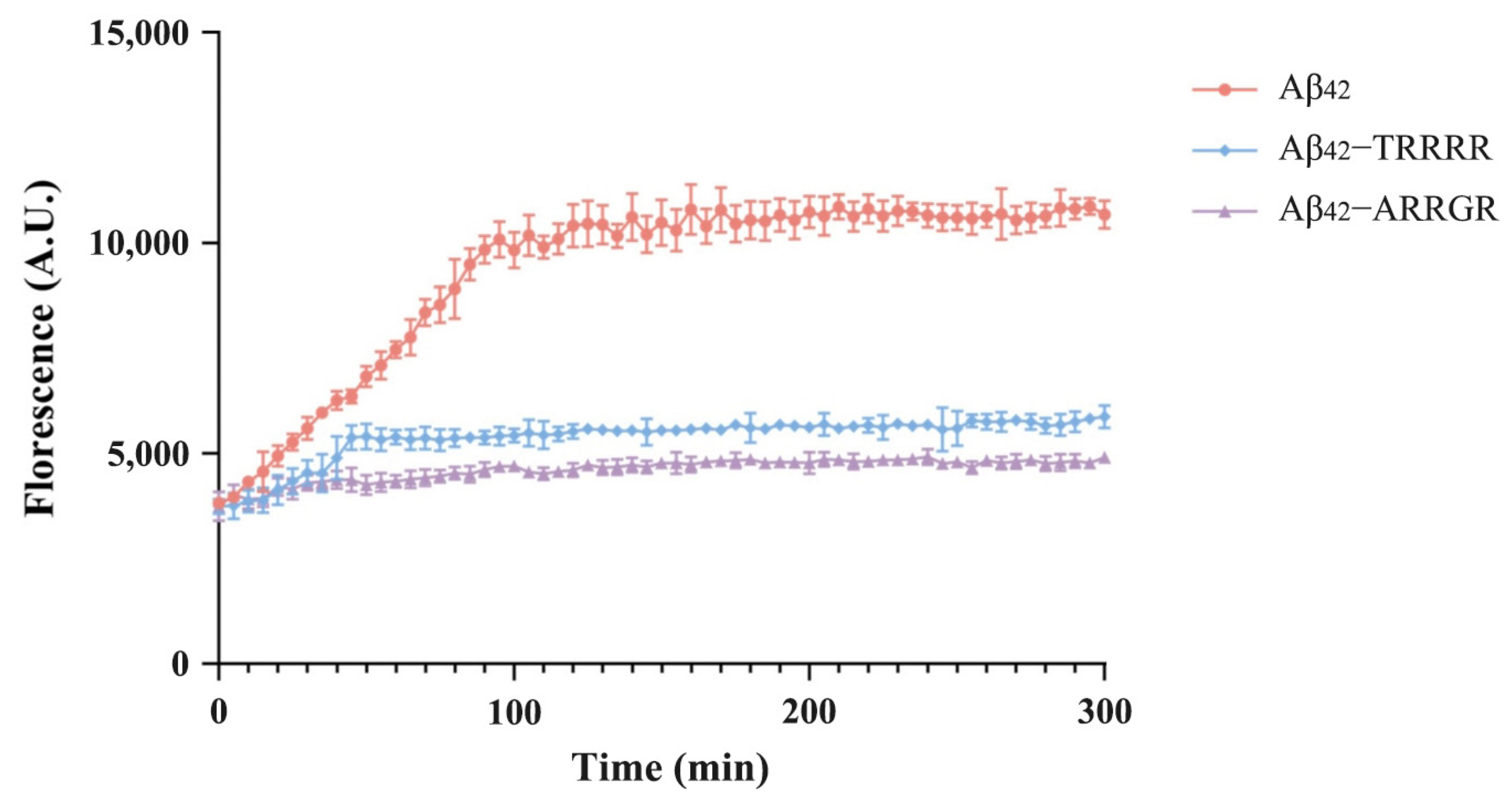
3.2. Effects of Pentapeptides on Aggregation of Aβ42 Detected by ThT Fluorescence Assay
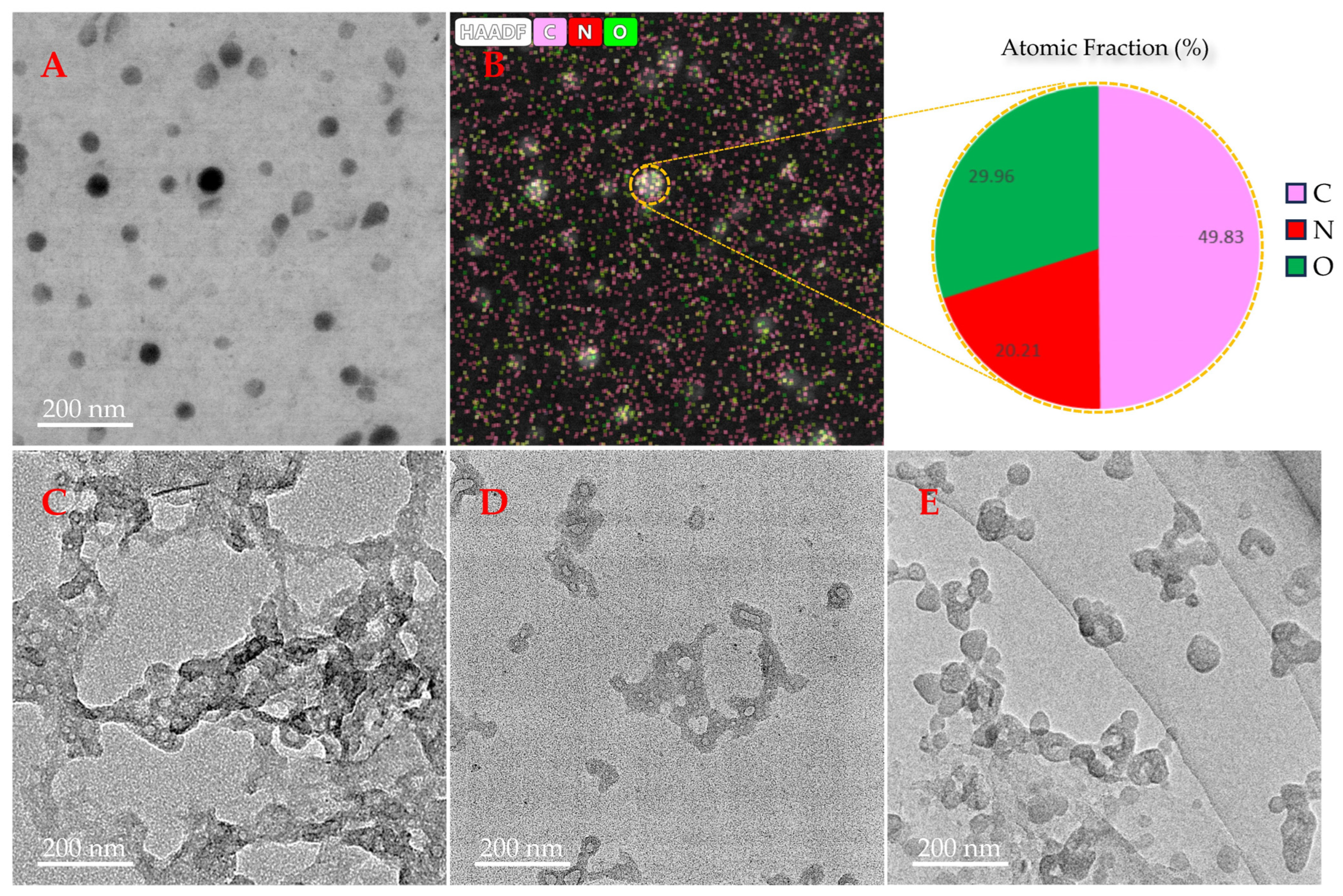
3.3. Observation of the Effect of TRRRR or ARRGR on Aβ42 Aggregation with a Transmission Electron Microscope (TEM)
3.4. ELISA Experiments to Investigate the Effects of TRRRR and ARRGR on Aβ42 Expression
3.5. Effect of Either TRRRR or ARRGR on Cell Death Induced by Secreted Aβ42
3.6. Effect of Either TRRRR or ARRGR on Reactive Oxygen Species Produced by SH-SY5Y Cells Secreting Aβ42
3.7. Protective Effect of Either TRRRR or ARRGR on Cell Apoptosis of SH-SY5Y Cells Secreting Aβ42
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hansen, R.A.; Gartlehner, G.; Webb, A.P.; Morgan, L.C.; Moore, C.G.; Jonas, D.E. Efficacy and safety of donepezil, galantamine, and rivastigmine for the treatment of Alzheimer’s disease: A systematic review and meta-analysis. Clin. Interv. Aging 2008, 3, 211–225. [Google Scholar] [PubMed]
- Alzheimer’s Association. 2023 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2023, 19, 1598–1695. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, A.; Norton, N.; Fast, T.; Frolich, L.; Georges, J.; Holzapfel, D.; Kirabali, T.; Krolak-Salmon, P.; Rossini, P.M.; Ferretti, M.T.; et al. Global estimates on the number of persons across the Alzheimer’s disease continuum. Alzheimer’s Dement. 2023, 19, 658–670. [Google Scholar] [CrossRef]
- Li, C.; Grajales, S.; Shuang, S.; Dong, C.; Nair, M. β-amyloid biomarker detection for Alzheimer’s disease. Zhonghua Yi Xue Za Zhi 2017, 1, 15–25. [Google Scholar] [CrossRef]
- Demuro, A.; Mina, E.; Kayed, R.; Milton, S.C.; Parker, I.; Glabe, C.G. Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. J. Biol. Chem. 2005, 280, 17294–17300. [Google Scholar] [CrossRef]
- Li, S.; Hong, S.; Shepardson, N.E.; Walsh, D.M.; Shankar, G.M.; Selkoe, D. Soluble oligomers of amyloid Beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron 2009, 62, 788–801. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Lauderback, C.M. Lipid peroxidation and protein oxidation in Alzheimer’s disease brain: Potential causes and consequences involving amyloid beta-peptide-associated free radical oxidative stress. Free Radic. Biol. Med. 2002, 32, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.A.; Lee, H.P.; Rolston, R.K.; Zhu, X.; Marlatt, M.W.; Castellani, R.J.; Nunomura, A.; Casadesus, G.; Smith, M.A.; Lee, H.G.; et al. Oxidative Stress and its Implications for Future Treatments and Management of Alzheimer Disease. Int. J. Biomed. Sci. IJBS 2010, 6, 225–227. [Google Scholar] [PubMed]
- Bhattacharjee, S.; Bhattacharyya, R. PRFF Peptide Mimic Interferes with Toxic Fibrin-Abeta(42) Interaction by Emulating the Abeta Binding Interface on Fibrinogen. ACS Chem. Neurosci. 2021, 12, 4144–4152. [Google Scholar] [CrossRef]
- Goyal, D.; Shuaib, S.; Mann, S.; Goyal, B. Rationally Designed Peptides and Peptidomimetics as Inhibitors of Amyloid-beta (Abeta) Aggregation: Potential Therapeutics of Alzheimer’s Disease. ACS Comb. Sci. 2017, 19, 55–80. [Google Scholar] [CrossRef]
- Lei, L.; Zou, Z.; Liu, J.; Xu, Z.; Fu, Y.; Tian, Y.; Zhang, W. Multifunctional peptide assembled micelles for simultaneously reducing amyloid–β and reactive oxygen–β species. Chem. Sci. 2021, 12, 6449–6459. [Google Scholar] [CrossRef] [PubMed]
- Mondal, P.; Khan, J.; Gupta, V.; Ghosh, S. In Silico Approach for Designing Potent Neuroprotective Hexapeptide. ACS Chem. Neurosci. 2019, 10, 3018–3030. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Tang, Y.; Zhang, D.; Liu, Y.; Zhang, Y.; Chen, H.; Hu, R.; Zhang, M.; Zheng, J. Conformational-specific self-assembled peptides as dual-mode, multi-target inhibitors and detectors for different amyloid proteins. J. Mater. Chem. B 2022, 10, 1754–1762. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Pradhan, K.; Khan, J.; Das, G.; Mukherjee, N.; Das, D.; Ghosh, S. Human Serum Albumin-Inspired Glycopeptide-Based Multifunctional Inhibitor of Amyloid-beta Toxicity. ACS Omega 2020, 5, 18628–18641. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.; Patel, B.; Makwana, V.; Jadhav, H.R.; Kiefel, M.; Davey, A.; Reekie, T.A.; Rudrawar, S.; Kassiou, M. Peptides, Peptidomimetics, and Carbohydrate-Peptide Conjugates as Amyloidogenic Aggregation Inhibitors for Alzheimer’s Disease. ACS Chem. Neurosci. 2018, 9, 1530–1551. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef]
- Delaunay, M.; Ha-Duong, T. Computational Tools and Strategies to Develop Peptide-Based Inhibitors of Protein-Protein Interactions, Computational Peptide Science: Methods and Protocols; Springer: New York, NY, USA, 2021. [Google Scholar]
- Rehman, A.U.; Khurshid, B.; Ali, Y.; Rasheed, S.; Wadood, A.; Ng, H.L.; Chen, H.F.; Wei, Z.; Luo, R.; Zhang, J. Computational approaches for the design of modulators targeting protein-protein interactions. Expert Opin. Drug Discov. 2023, 18, 315–333. [Google Scholar] [CrossRef]
- Chandrasekhar, G.; Srinivasan, E.; Nandhini, S.; Pravallika, G.; Sanjay, G.; Rajasekaran, R. Computer aided therapeutic tripeptide design, in alleviating the pathogenic proclivities of nocuous alpha-synuclein fibrils. J. Biomol. Struct. Dyn. 2024, 42, 483–494. [Google Scholar] [CrossRef]
- Han, Y.; Král, P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano 2020, 14, 5143–5147. [Google Scholar] [CrossRef]
- Jarmula, A.; Zubalska, M.; Stepkowski, D. Consecutive Aromatic Residues Are Required for Improved Efficacy of beta-Sheet Breakers. Int. J. Mol. Sci. 2022, 23, 5247. [Google Scholar] [CrossRef]
- Kanchi, P.K.; Dasmahapatra, A.K. Enhancing the binding of the beta-sheet breaker peptide LPFFD to the amyloid-beta fibrils by aromatic modifications: A molecular dynamics simulation study. Comput. Biol. Chem. 2021, 92, 107471. [Google Scholar] [CrossRef] [PubMed]
- Luan, B.; Huynh, T. Crystal-structures-guided design of fragment-based drugs for inhibiting the main protease of SARS-CoV-2. Proteins 2022, 90, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- VK, P.; Rath, S.P.; Abraham, P. Computational designing of a peptide that potentially blocks the entry of SARS-CoV, SARS-CoV-2 and MERS-CoV. PLoS ONE 2021, 16, e0251913. [Google Scholar] [CrossRef]
- Abel, R.; Wang, L.; Harder, E.D.; Berne, B.J.; Friesner, R.A. Advancing Drug Discovery through Enhanced Free Energy Calculations. Acc. Chem. Res. 2017, 50, 1625–1632. [Google Scholar] [CrossRef]
- Kuhn, B.; Tichý, M.; Wang, L.; Robinson, S.; Martin, R.E.; Kuglstatter, A.; Benz, J.; Giroud, M.; Schirmeister, T.; Abel, R.; et al. Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors. J. Med. Chem. 2017, 60, 2485–2497. [Google Scholar] [CrossRef]
- Meier, K.; Bluck, J.P.; Christ, C.D. Free Energy Methods in Drug Discovery: Current State and Future Directions. In ACS Symposium Series; Amarcost, K.A., Thompson, D.C., Eds.; ACS Publications: Washington, DC, USA, 2021; Chapter 2; pp. 39–66. [Google Scholar]
- Ross, G.A.; Lu, C.; Scarabelli, G.; Albanese, S.K.; Houang, E.; Abel, R.; Harder, E.D.; Berne, B.J.; Friesner, R.A. The maximal and current accuracy of rigorous protein-ligand binding free energy calculations. Acc. Chem. Res. 2017, 50, 1625–1632. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, B.; Pei, J.; Luo, Y.; Yuan, N.; Xiao, Z.; Wu, H.; Luo, C.; Wang, J.; Wei, S.; et al. Molecular dynamic and pharmacological studies on protein-engineered hirudin variants of Hirudinaria manillensis and Hirudo medicinalis. Br. J. Pharmacol. 2022, 179, 3740–3753. [Google Scholar] [CrossRef]
- Yuan, N.; Ye Lz Sun, Y.; Wu, H.; Xiao, Z.; Fu, W.; Chen, Z.; Pei, Y.; Min, Y.; Wang, D. Molecular Integrative Analysis of the Inhibitory Effects of Dipeptides on Amyloid Peptide 1–42 Polymerization. Int. J. Mol. Sci. 2023, 24, 7673. [Google Scholar] [CrossRef]
- Summa, V.; Petrocchi, A.; Bonelli, F.; Crescenzi, B.; Donghi, M.; Ferrara, M.; Fiore, F.; Gardelli, C.; Gonzalez Paz, O.; Hazuda, D.J.; et al. Discovery of Raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 2008, 51, 5843–5855. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Limon, A.; Aguilar-Toalá, J.E.; Liceaga, A.M. Integration of Molecular Docking Analysis and Molecular Dynamics Simulations for Studying Food Proteins and Bioactive Peptides. J. Agric. Food Chem. 2022, 70, 934–943. [Google Scholar] [CrossRef]
- Guallar, V.; Wallrapp, F.H. QM/MM methods: Looking inside heme proteins biochemisty. Biophys. Chem. 2010, 149, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Schames, J.R.; Henchman, R.H.; Siegel, J.S.; Sotriffer, C.A.; Ni, H.; McCammon, J.A. Discovery of a novel binding trench in HIV integrase. J. Med. Chem. 2004, 47, 1879–1881. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Tam, B.; Wang, S.M. Applications of Molecular Dynamics Simulation in Protein Study. Membranes 2022, 12, 844. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Berendsen, H.J. Computer Simulation of Molecular Dynamics: Methodology, Applications, and Perspectives in Chemistry. Angew. Chem. Int. Ed. Engl. 1990, 29, 992–1023. [Google Scholar] [CrossRef]
- Dror, R.O.; Dirks, R.M.; Grossman, J.P.; Xu, H.; Shaw, D.E. Biomolecular simulation: A computational microscope for molecular biology. Annu. Rev. Biophys. 2012, 41, 429–452. [Google Scholar] [CrossRef]
- Gera, J.; Szogi, T.; Bozso, Z.; Fulop, L.; Barrera, E.E.; Rodriguez, A.M.; Mendez, L.; Delpiccolo, C.M.L.; Mata, E.G.; Cioffi, F.; et al. Searching for improved mimetic peptides inhibitors preventing conformational transition of amyloid-beta(42) monomer. Bioorg. Chem. 2018, 81, 211–221. [Google Scholar] [CrossRef]
- Liu, F.F.; Dong, X.Y.; He, L.; Middelberg, A.P.; Sun, Y. Molecular insight into conformational transition of amyloid beta-peptide 42 inhibited by (-)-epigallocatechin-3-gallate probed by molecular simulations. J. Phys. Chem. B 2011, 115, 11879–11887. [Google Scholar] [CrossRef]
- Chang, S.Y.; Liu, F.F.; Dong, X.Y.; Sun, Y. Molecular insight into conformational transmission of human P-glycoprotein. J. Chem. Phys. 2013, 139, 225102–225112. [Google Scholar] [CrossRef] [PubMed]
- Reddy, G.; Straub, J.E.; Thirumalai, D. Dynamics of locking of peptides onto growing amyloid fibrils. Proc. Natl. Acad. Sci. USA 2009, 106, 11948–11953. [Google Scholar] [CrossRef]
- Tonali, N.; Dodero, V.I.; Kaffy, J.; Hericks, L.; Ongeri, S.S. Real-time BODIPY-Binding assay to screen inhibitors of the early oligomerization process of Aβ1-42 peptide. Chembiochem 2020, 21, 1129–1135. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, H.; Li, R.; Sterling, K.; Song, W. Amyloid beta-based therapy for Alzheimer’s disease: Challenges, successes and future. Signal Transduct. Target. Ther. 2023, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- Haass, C.; Selkoe, D.J. Cellular processing of β-amyloid precursor protein and the genesis of amyloid β-peptide. Cell 1993, 75, 1039–1042. [Google Scholar] [CrossRef] [PubMed]
- Conti Filho, C.E.; Loss, L.B.; Marcolongo-Pereira, C.; Rossoni Junior, J.V.; Barcelos, R.M.; Chiarelli-Neto, O.; da Silva, B.S.; Passamani Ambrosio, R.; Castro, F.; Teixeira, S.F.; et al. Advances in Alzheimer’s disease’s pharmacological treatment. Front. Pharmacol. 2023, 14, 1101452. [Google Scholar] [CrossRef]
- Jokar, S.; Khazaei, S.; Gameshgoli, X.E.; Khafaji, M.; Yarani, B.; Sharifzadeh, M.; Beiki, D.; Bavi, O. Amyloid beta-Targeted Inhibitory Peptides for Alzheimer’s Disease: Current State and Future Perspectives. In Alzheimer’s Disease: Drug Discovery; Huang, X., Ed.; Exon Publications: Brisbane, AU, Australia, 2020. [Google Scholar]
- Cebers, G.; Alexander, R.C.; Haeberlein, S.B.; Han, D.; Goldwater, R.; Ereshefsky, L.; Olsson, T.; Ye, N.; Rosen, L.; Russell, M.; et al. AZD3293: Pharmacokinetic and Pharmacodynamic Effects in Healthy Subjects and Patients with Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 55, 1039–1053. [Google Scholar] [CrossRef]
- McDade, E.; Voytyuk, I.; Aisen, P.; Bateman, R.J.; Carrillo, M.C.; De Strooper, B.; Haass, C.; Reiman, E.M.; Sperling, R.; Tariot, P.N.; et al. The case for low-level BACE1 inhibition for the prevention of Alzheimer disease. Nat. Rev. Neurol. 2021, 17, 703–714. [Google Scholar] [CrossRef]
- Yang, G.; Zhou, R.; Guo, X.; Yan, C.; Lei, J.; Shi, Y. Structural basis of gamma-secretase inhibition and modulation by small molecule drugs. Cell 2021, 184, 521–533 e514. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dong, X.; Liu, F.; Zheng, J.; Sun, Y. Iminodiacetic acid-conjugated nanoparticles as a bifunctional modulator against Zn2+-mediated amyloid protein aggregation and cytotoxicity. J. Colloid Interface Sci. 2017, 505, 973–982. [Google Scholar] [CrossRef]
- Wang, W.; Han, Y.; Fan, Y.; Wang, Y. Effects of Gold Nanospheres and Nanocubes on Amyloid-beta Peptide Fibrillation. Langmuir 2019, 35, 2334–2342. [Google Scholar] [CrossRef]
- Brahmachari, S.; Paul, A.; Segal, D.; Gazit, E. Inhibition of amyloid oligomerization into different supramolecular architectures by small molecules: Mechanistic insights and design rules. Future Med. Chem. 2017, 9, 797–810. [Google Scholar] [CrossRef]
- Jokar, S.; Khazaei, S.; Behnammanesh, H.; Shamloo, A.; Erfani, M.; Beiki, D.; Bavi, O. Recent advances in the design and applications of amyloid-beta peptide aggregation inhibitors for Alzheimer’s disease therapy. Biophys. Rev. 2019, 11, 901–925. [Google Scholar] [CrossRef]
- Rahimi, F.; Li, H.; Sinha, S.; Bitan, G. Modulators of amyloid β-protein self-assembly. In Developing Therapeutics for Alzheimer’s Disease; Elsevier: Boston, MA, USA, 2016; pp. 97–191. [Google Scholar] [CrossRef]
- Lee, A.C.; Harris, J.L.; Khanna, K.K.; Hong, J.H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef] [PubMed]
- Danho, W.; Swistok, J.; Khan, W.; Chu, X.J.; Cheung, A.; Fry, D.; Sun, H.; Kurylko, G.; Rumennik, L.; Cefalu, J.; et al. Opportunities and challenges of developing peptide drugs in the pharmaceutical industry. Adv. Exp. Med. Biol. 2009, 611, 467–469. [Google Scholar] [CrossRef] [PubMed]
- Jokar, S.; Erfani, M.; Bavi, O.; Khazaei, S.; Sharifzadeh, M.; Hajiramezanali, M.; Beiki, D.; Shamloo, A. Design of peptide-based inhibitor agent against amyloid-beta aggregation: Molecular docking, synthesis and in vitro evaluation. Bioorg. Chem. 2020, 102, 104050. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Ye, L.; Yuan, N.; Che Ajuyo, N.M.; Xiao, Z.; Liu, L.; Chen, Z.; Pei, Y.; Min, Y.; Wang, D. A Molecular Integrative Study on the Inhibitory Effects of WRR and ERW on Amyloid Peptide (1–42) Polymerization and Cell Toxicity. Polymers 2023, 15, 4356. [Google Scholar] [CrossRef]
- Funke, S.A.; Willbold, D. Peptides for therapy and diagnosis of Alzheimer’s disease. Curr. Pharm. Des. 2012, 18, 755–767. [Google Scholar] [CrossRef]
- Kumar, J.; Sim, V. D-amino acid-based peptide inhibitors as early or preventative therapy in Alzheimer disease. Prion 2014, 8, 119–124. [Google Scholar] [CrossRef]
- Allolio, C.; Magarkar, A.; Jurkiewicz, P.; Baxova, K.; Javanainen, M.; Mason, P.E.; Sachl, R.; Cebecauer, M.; Hof, M.; Horinek, D.; et al. Arginine-rich cell-penetrating peptides induce membrane multilamellarity and subsequently enter via formation of a fusion pore. Proc. Natl. Acad. Sci. USA 2018, 115, 11923–11928. [Google Scholar] [CrossRef]
- Mitchell, D.J.; Kim, D.T.; Steinman, L.; Fathman, C.G.; Rothbard, J.B. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Pept. Res. 2000, 56, 318–325. [Google Scholar] [CrossRef]
- Schmidt, N.; Mishra, A.; Lai, G.H.; Wong, G.C. Arginine-rich cell-penetrating peptides. FEBS Lett. 2010, 584, 1806–1813. [Google Scholar] [CrossRef]
- Habault, J.; Poyet, J.L. Recent Advances in Cell Penetrating Peptide-Based Anticancer Therapies. Molecules 2019, 24, 927. [Google Scholar] [CrossRef]
- Cheng, Y.S.; Chen, Z.T.; Liao, T.Y.; Lin, C.; Shen, H.C.H.; Wang, Y.H.; Chang, C.W.; Liu, R.S.; Chen, R.P.Y.; Tu, P.H. An intranasally delivered peptide drug ameliorates cognitive decline in Alzheimer transgenic mice. EMBO Mol. Med. 2017, 9, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Henning-Knechtel, A.; Kumar, S.; Wallin, C.; Król, S.; Wärmländer, S.K.; Jarvet, J.; Esposito, G.; Kirmizialtin, S.; Gräslund, A.; Hamilton, A.D.; et al. Designed Cell-Penetrating Peptide Inhibitors of Amyloid-beta Aggregation and Cytotoxicity. Cell Rep. Physic. Sci. 2020, 1, 100014–100035. [Google Scholar] [CrossRef]
- Kawasaki, T.; Onodera, K.; Kamijo, S. Identification of novel short peptide inhibitors of soluble 37/48 kDa oligomers of amyloid beta42. Biosci. Biotechnol. Biochem. 2011, 75, 1496–1501. [Google Scholar] [CrossRef]
- Parthsarathy, V.; McClean, P.L.; Holscher, C.; Taylor, M.; Tinker, C.; Jones, G.; Kolosov, O.; Salvati, E.; Gregori, M.; Masserini, M.; et al. A novel retro-inverso peptide inhibitor reduces amyloid deposition, oxidation and inflammation and stimulates neurogenesis in the APPswe/PS1DeltaE9 mouse model of Alzheimer’s disease. PLoS ONE 2013, 8, e54769. [Google Scholar] [CrossRef]
- Xu, B.; Jacobs, M.I.; Kostko, O.; Ahmed, M. Guanidinium Group Remains Protonated in a Strongly Basic Arginine Solution. Chemphyschem 2017, 18, 1503–1506. [Google Scholar] [CrossRef]
- Sharma, S.; Sarkar, S.; Paul, S.S.; Roy, S.; Chattopadhyay, K. A small molecule chemical chaperone optimizes its unfolded state contraction and denaturant like properties. Sci. Rep. 2013, 3, 3525. [Google Scholar] [CrossRef]
- Arakawa, T.; Kita, Y.; Ejima, D.; Tsumoto, K.; Fukada, H. Aggregation suppression of proteins by arginine during thermal unfolding. Protein Pept. Lett. 2006, 13, 921–927. [Google Scholar] [CrossRef]
- Ghosh, R.; Sharma, S.; Chattopadhyay, K. Effect of arginine on protein aggregation studied by fluorescence correlation spectroscopy and other biophysical methods. Biochemistry 2009, 48, 1135–1143. [Google Scholar] [CrossRef]
- Golovanov, A.P.; Hautbergue, G.M.; Wilson, S.A.; Lian, L.Y. A simple method for improving protein solubility and long-term stability. J. Am. Chem. Soc. 2004, 126, 8933–8939. [Google Scholar] [CrossRef]
- Kaur, A.; Goyal, B. Identification of new pentapeptides as potential inhibitors of amyloid-beta(42) aggregation using virtual screening and molecular dynamics simulations. J. Mol. Graph. Model. 2023, 124, 108558. [Google Scholar] [CrossRef]
- Antzutkin, O.N.; Leapman, R.D.; Balbach, J.J.; Tycko, R. Supramolecular structural constraints on Alzheimer’s beta-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry 2002, 41, 15436–15450. [Google Scholar] [CrossRef] [PubMed]
- Balbach, J.J.; Petkova, A.T.; Oyler, N.A.; Antzutkin, O.N.; Gordon, D.J.; Meredith, S.C.; Tycko, R. Supramolecular structure in full-length Alzheimer’s beta-amyloid fibrils: Evidence for a parallel beta-sheet organization from solid-state nuclear magnetic resonance. Biophys. J. 2002, 83, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, L.; Ajuyo, N.M.C.; Wu, Z.; Yuan, N.; Xiao, Z.; Gu, W.; Zhao, J.; Pei, Y.; Min, Y.; Wang, D. Molecular Integrative Study on Inhibitory Effects of Pentapeptides on Polymerization and Cell Toxicity of Amyloid-β Peptide (1–42). Curr. Issues Mol. Biol. 2024, 46, 10160-10179. https://doi.org/10.3390/cimb46090606
Ye L, Ajuyo NMC, Wu Z, Yuan N, Xiao Z, Gu W, Zhao J, Pei Y, Min Y, Wang D. Molecular Integrative Study on Inhibitory Effects of Pentapeptides on Polymerization and Cell Toxicity of Amyloid-β Peptide (1–42). Current Issues in Molecular Biology. 2024; 46(9):10160-10179. https://doi.org/10.3390/cimb46090606
Chicago/Turabian StyleYe, Lianmeng, Nuela Manka’a Che Ajuyo, Zhongyun Wu, Nan Yuan, Zhengpan Xiao, Wenyu Gu, Jiazheng Zhao, Yechun Pei, Yi Min, and Dayong Wang. 2024. "Molecular Integrative Study on Inhibitory Effects of Pentapeptides on Polymerization and Cell Toxicity of Amyloid-β Peptide (1–42)" Current Issues in Molecular Biology 46, no. 9: 10160-10179. https://doi.org/10.3390/cimb46090606
APA StyleYe, L., Ajuyo, N. M. C., Wu, Z., Yuan, N., Xiao, Z., Gu, W., Zhao, J., Pei, Y., Min, Y., & Wang, D. (2024). Molecular Integrative Study on Inhibitory Effects of Pentapeptides on Polymerization and Cell Toxicity of Amyloid-β Peptide (1–42). Current Issues in Molecular Biology, 46(9), 10160-10179. https://doi.org/10.3390/cimb46090606






