The Potential of CRISPR/Cas9 to Circumvent the Risk Factor Neurotoxin β-N-oxalyl-L-α, β-diaminopropionic acid Limiting Wide Acceptance of the Underutilized Grass Pea (Lathyrus sativus L.)
Abstract
:1. Background
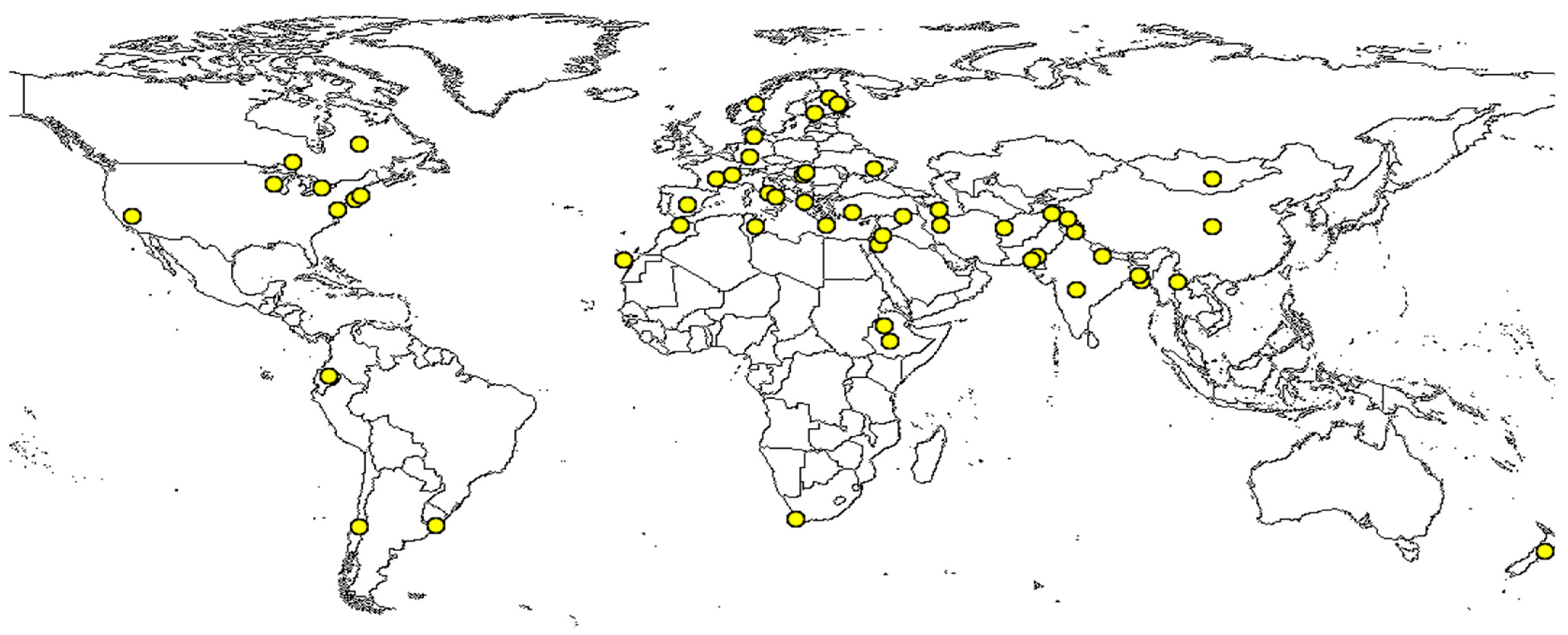
2. Nutritional Benefits and Side Effects of Grass Pea
3. Conventional Improvement for β-ODAP Reduction and Influence of Environmental Factors
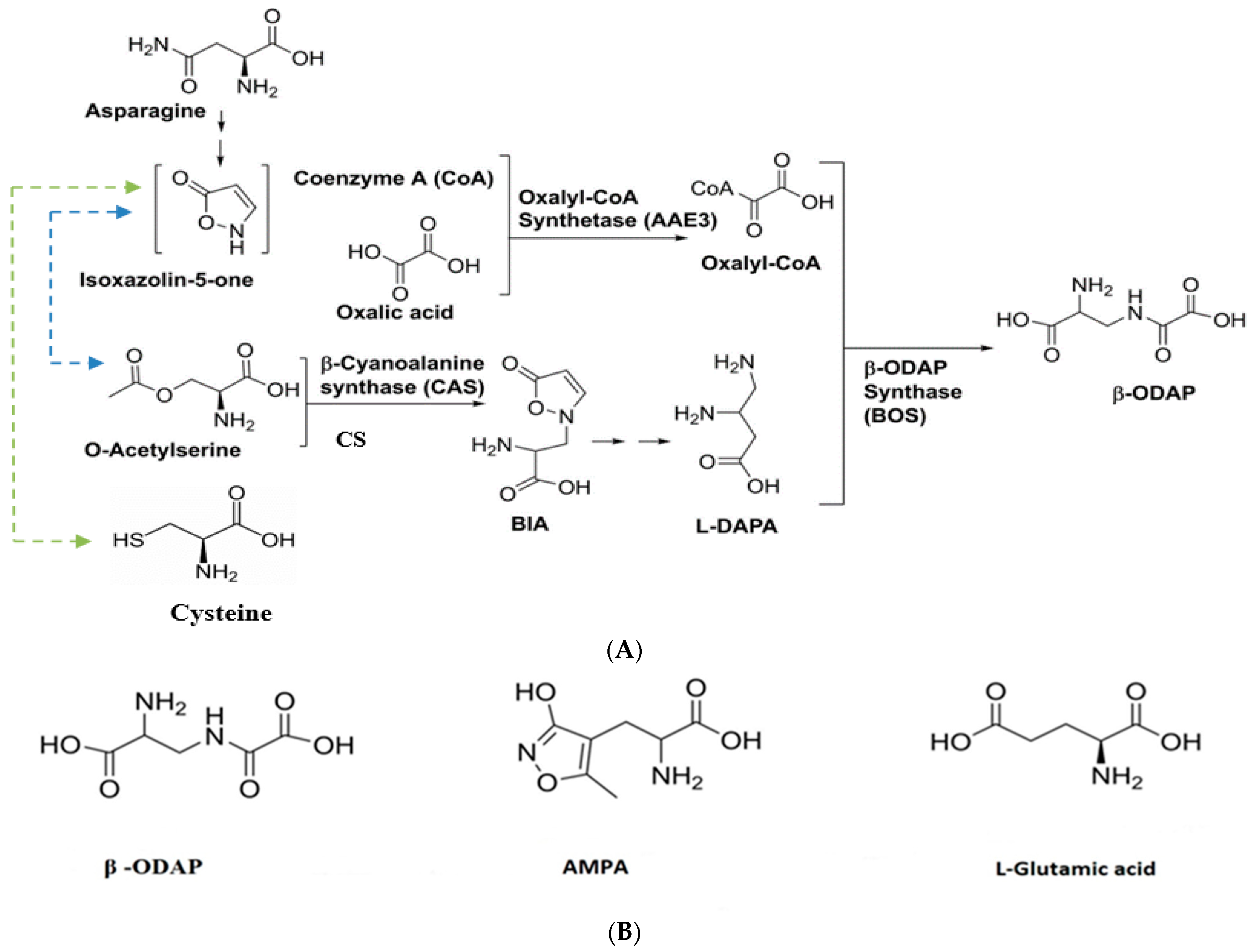
4. Biosynthesis of β-ODAP in Grass Pea
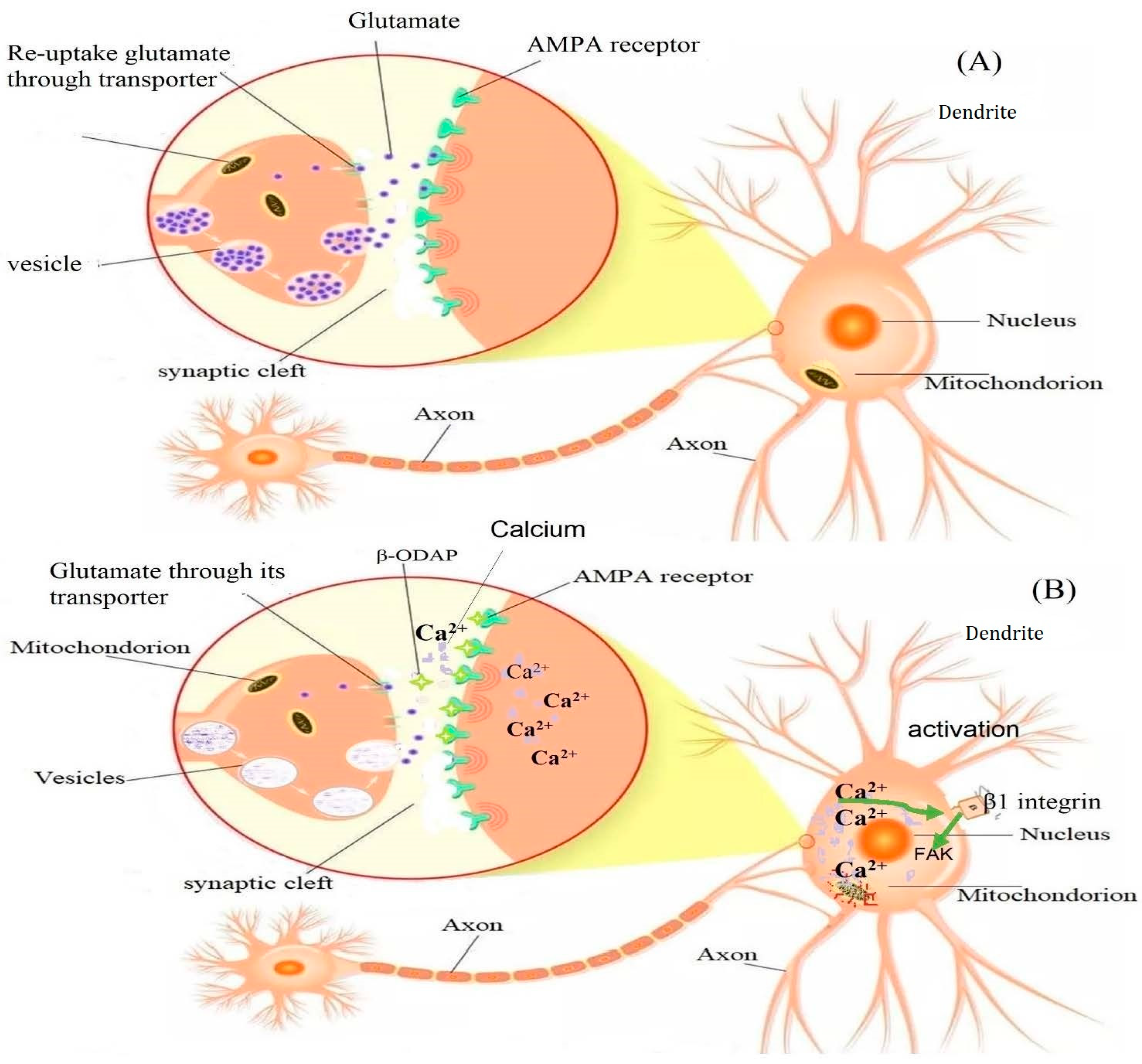
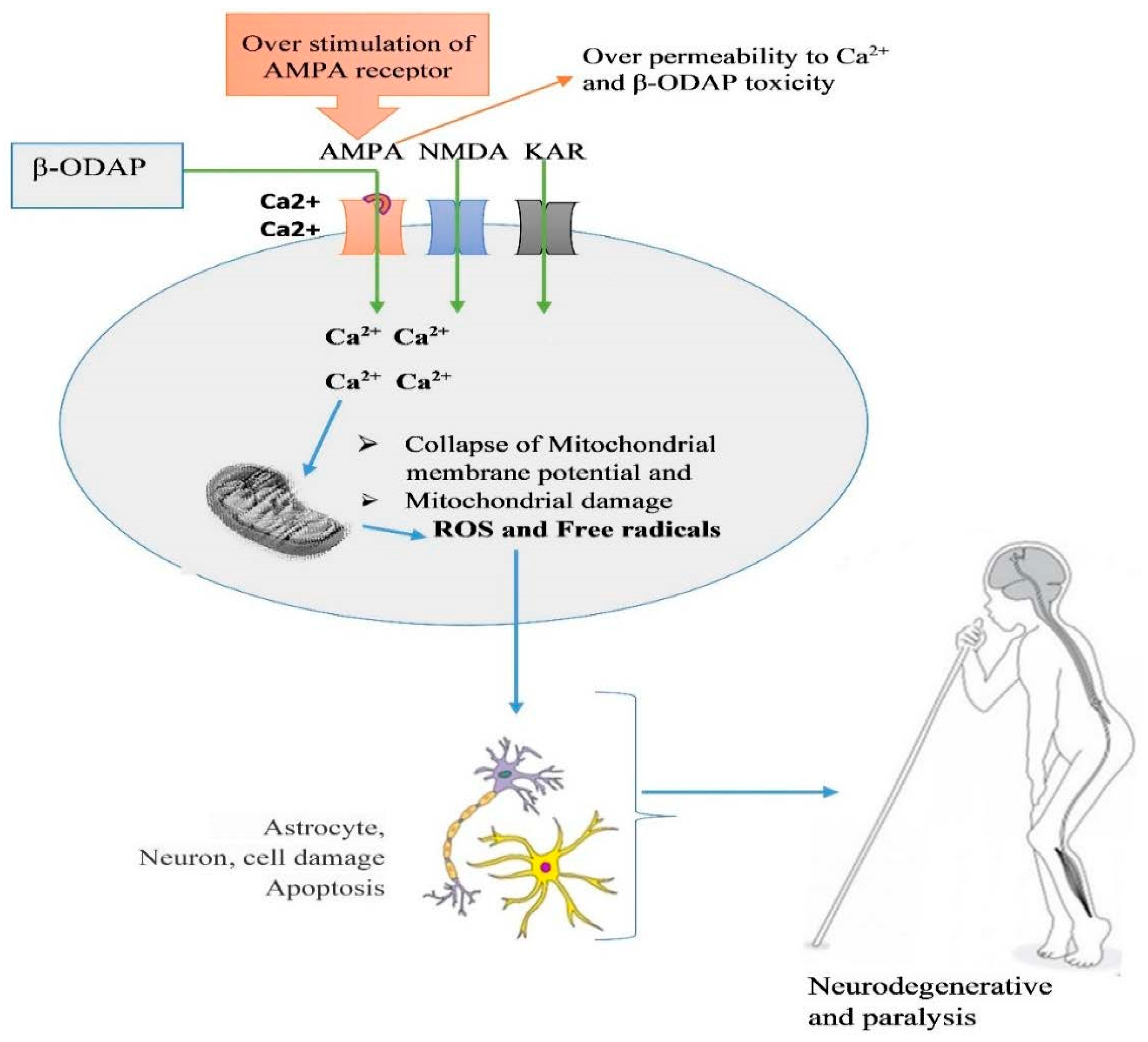
5. Mechanism of β-ODAP Action and Toxicity
6. Reducing β-ODAP Using Traditional Food Processing Strategies
7. Enhancing Sulfur-Containing Amino Acids (SCA) through Genetic Modification of Grass Pea
8. The Potential of the Genome Editing Approach for Antinutritional Factors in Crops
9. The Potential of CRISPR/Cas9 to Target β-ODAP Biosynthesis in Grass Pea
10. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tan, R.-Y.; Xing, G.-Y.; Zhou, G.-M.; Li, F.-M.; Hu, W.-T.; Lambein, F.; Xiong, J.-L.; Zhang, S.-X.; Kong, H.-Y.; Zhu, H.; et al. Plant toxin β-ODAP activates integrin β1 and focal adhesion: A critical pathway to cause neurolathyrism. Sci. Rep. 2017, 7, 40677. [Google Scholar] [CrossRef] [PubMed]
- Lambein, F.; Travella, S.; Kuo, Y.-H.; Van Montagu, M.; Heijde, M. Grass pea (Lathyrus sativus L.): Orphan crop, nutraceutical or just plain food? Planta 2019, 250, 821–838. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, L.; Rubiales, D.; Bronze, M.R.; Vaz Patto, M.C. Grass Pea (Lathyrus sativus L.)—A Sustainable and Resilient Answer to Climate Challenges. Agronomy 2022, 12, 1324. [Google Scholar] [CrossRef]
- Smartt, J. Pulses of the classical world. In Grain Legumes: Evaluation and Genetic Resources; Cambridge University Press: Cambridge, UK, 1990; pp. 190–200. [Google Scholar]
- Abdipour, M.; Vaezi, B.; Khademi, K.; Ghasemi, S. An optimized artificial intelligence approach and sensitivity analysis for predicting the biological yield of grass pea (Lathyrus sativus L.). Arch. Agron. Soil Sci. 2019, 66, 1909–1924. [Google Scholar] [CrossRef]
- Sammour, R.H. Genetic diversity in Lathyrus sativus L. germplasm. Res. Rev. BioSci. 2014, 8, 325–336. [Google Scholar]
- Gupta, P.; Udupa, S.M.; Gupta, D.S.; Kumar, J.; Kumar, S. Population structure analysis and determination of neurotoxin content in a set of grass pea (Lathyrus sativus L.) accessions of Bangladesh origin. Crop J. 2018, 6, 435–442. [Google Scholar] [CrossRef]
- Verma, S.C.; Ohri, D. Chromosome and nuclear phenotype in the legume Lathyrus sativus L. Cytologia 1979, 44, 77–90. [Google Scholar] [CrossRef]
- Hanbury, C.; White, C.; Mullan, B.; Siddique, K. A review of the potential of Lathyrus sativus L. and L. cicera L. grain for use as animal feed. Anim. Feed. Sci. Technol. 2000, 87, 1–27. [Google Scholar] [CrossRef]
- Vaz Patto, M.C.; Skiba, B.; Pang, E.C.K.; Ochatt, S.J.; Lambein, F.; Rubiales, D. Lathyrus improvement for resistance against biotic and abiotic stresses: From classical breeding to marker assisted selection. Euphytica 2006, 147, 133–147. [Google Scholar] [CrossRef]
- Almeida, N.; Rubiales, D.; Vaz Patto, M. Grass Pea. In Grain Legumes. Handbook of Plant Breeding; De Ron, A., Ed.; Springer: New York, NY, USA, 2015; Volume 10. [Google Scholar] [CrossRef]
- Shiferaw, E.; Pè, M.E.; Porceddu, E.; Ponnaiah, M. Exploring the genetic diversity of Ethiopian grass pea (Lathyrus sativus L.) using EST-SSR markers. Mol. Breed. 2011, 30, 789–797. [Google Scholar] [CrossRef]
- Dadi, L.; Teklewold, H.; Aw-Hassan, A.; Moneim, A.A.; Bejiga, G. The Socioeconomic Factors Affecting Grass Pea Consumption and Influence of Lathyrism in Ethiopia; Integr. National Resources Management Res. Report Series, No. 4; ICARDA: Aleppo, Syria, 2003; pp. 7–11. [Google Scholar]
- Gabrekiristos, E.; Wondimu, M. Emerging and Reemerging Diseases of Common Bean (Phaseolus vulgaris L.) in major production areas: In the case of Ethiopia. J. Plant Pathol. Microbiol. 2022, 13, 619. [Google Scholar]
- Sarkar, A.; Emmrich, P.M.F.; Sarker, A.; Zong, X.; Martin, C.; Wang, T.L. Grass pea: Remodelling an ancient insurance crop for climate resilience. In Genomic Designing of Climate-Smart Pulse Crops; Kole, C., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 425–469. [Google Scholar]
- Fikre, A.; Van Moorhem, M.; Ahmed, S.; Lambein, F.; Gheysen, G. Studies on neurolathyrism in Ethiopia: Dietary habits, perception of risks and prevention. Food Chem. Toxicol. 2011, 49, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Arslan, M.; Basak, M.; Aksu, E.; Uzun, B.; Yol, E. Genotyping of Low β-ODAP Grass Pea (Lathyrus sativus L.) Germplasm with EST-SSR Markers. Braz. Arch. Biol. Technol. 2020, 63, e20190150. [Google Scholar] [CrossRef]
- Spencer, P.S.; Roy, D.N.; Ludolph, A.; Hugon, J.; Dwivedi, M.P.; Schaumburg, H.H. Lathyrism: Evidence for role of the neuroexcitatory amino acid BOAA. Lancet 1986, 2, 1066–1067. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Parihar, A.K.; Barpete, S.; Kumar, S.; Gupta, S. Current Perspectives on Reducing the β-ODAP Content and Improving Potential Agronomic Traits in Grass Pea (Lathyrus sativus L.). Front. Plant Sci. 2021, 12, 703275. [Google Scholar] [CrossRef]
- Banerjee, J.; Das, A.; Parihar, A.K.; Sharma, R.; Pramanik, K.; Barpete, S. Genomic Designing Towards Development of Abiotic Stress Tolerant Grass Pea for Food and Nutritional Security. In Genomic Designing for Abiotic Stress Resistant Pulse Crops; Kole, C., Ed.; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Rizvi, A.H.; Sarker, A.; Dogra, A. Enhancing grasspea (Lathyrus sativus L.) production in problematic soils of South Asia for nutritional security. Indian J. Genet. Plant Breed. 2016, 76, 583–592. [Google Scholar] [CrossRef]
- Rao, S.L.; Adiga, P.R.; Sarma, P.S. The Isolation and Characterization of β-N-Oxalyl-L-α,β-Diaminopropionic Acid: A Neurotoxin from the Seeds of Lathyrus sativus. Biochemistry 1964, 3, 432–436. [Google Scholar] [CrossRef]
- Goldsmith, M.; Barad, S.; Knafo, M.; Savidor, A.; Ben-Dor, S.; Brandis, A.; Mehlman, T.; Peleg, Y.; Albeck, S.; Dym, O.; et al. Identification and characterization of the key enzyme in the biosynthesis of the neurotoxin β-ODAP in grass pea. J. Biol. Chem. 2022, 298, 101806. [Google Scholar] [CrossRef]
- Edwards, A.; Njaci, I.; Sarkar, A.; Jiang, Z.; Kaithakottil, G.G.; Moore, C.; Cheema, J.; Stevenson, C.E.M.; Rejzek, M.; Novák, P.; et al. Genomics and biochemical analyses reveal a metabolon key to β-L-ODAP biosynthesis in Lathyrus sativus. Nat. Commun. 2023, 14, 876. [Google Scholar] [CrossRef]
- Rathi, D.; Chakraborty, S.; Chakraborty, N. Grasspea, a critical recruit among neglected and underutilized legumes, for tapping genomic resources. Curr. Plant Biol. 2021, 26, 100200. [Google Scholar] [CrossRef]
- Urga, K.; Fufa, H.; Biratu, E.; Husain, A. Evaluation of Lathyrus sativus cultivated in Ethiopia for proximate composition, mineral and b-ODAP and antinutritional components. Afr. J. Food Agric. Nutr. Dev. 2005, 5, 1–15. [Google Scholar]
- Chandna, M.; Matta, N.K. Studies on Changing Protein Levels in Developing and Germinating Seeds of Lathyrus sativus L. J. Plant Biochem. Biotechnol. 1994, 3, 59–61. [Google Scholar] [CrossRef]
- Grela, E.R.; Rybiński, W.; Klebaniuk, R.; Matras, J. Morphological characteristics of some accessions of grass pea (Lathyrus sativus L.) grown in Europe and nutritional traits of their seeds. Genet. Resour. Crop Evol. 2010, 57, 693–701. [Google Scholar] [CrossRef]
- Arslan, M. Diversity for vitamin and amino acid content in grass pea (Lathyrus sativus L.). Legume Res. 2017, 40, 803–810. [Google Scholar] [CrossRef]
- Rao, S.L.N.; Ramachandran, L.K.; Adiga, P.R. The isolation and characterization of L-homoarginine from seeds of Lathyrus sativus. Biochemistry 1963, 2, 298–300. [Google Scholar] [CrossRef]
- Rao, S. A look at the brighter facets of β-N-oxalyl-L-α,β-diaminopropionic acid, homoarginine and the grass pea. Food Chem. Toxicol. 2011, 49, 620–622. [Google Scholar] [CrossRef]
- Tsikas, D.; Wu, G. Homoarginine, arginine, and relatives: Analysis, metabolism, transport, physiology, and pathology. Amino Acids 2015, 47, 1697–1702. [Google Scholar] [CrossRef]
- Rodionov, R.N.; Begmatov, H.; Jarzebska, N.; Patel, K.; Mills, M.T.; Ghani, Z.; Khakshour, D.; Tamboli, P.; Patel, M.N.; Abdalla, M.; et al. Homoarginine Supplementation Prevents Left Ventricular Dilatation and Preserves Systolic Function in a Model of Coronary Artery Disease. J. Am. Heart Assoc. 2019, 8, e012486. [Google Scholar] [CrossRef]
- Jammulamadaka, N.; Burgula, S.; Medisetty, R.; Ilavazhagan, G.; Rao, S.L.N.; Singh, S.S. β-N-oxalyl-l-α,β-diaminopropionic acid regulates mitogen-activated protein kinase signaling by down-regulation of phosphatidylethanolamine-binding protein 1. J. Neurochem. 2011, 118, 176–186. [Google Scholar] [CrossRef]
- May, M.; Kayacelebi, A.A.; Batkai, S.; Jordan, J.; Tsikas, D.; Engeli, S. Plasma and tissue homoarginine concentrations in healthy and obese humans. Amino Acids 2015, 47, 1847–1852. [Google Scholar] [CrossRef]
- Tsikas, D.; Bollenbach, A.; Hanff, E.; Kayacelebi, A.A. Asymmetric dimethylarginine (ADMA), symmetric dimethylarginine (SDMA) and homoarginine (hArg): The ADMA, SDMA and hArg paradoxes. Cardiovasc. Diabetol. 2018, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Llorent-Martínez, E.J.; Zengin, G.; Fernández-de Córdova, M.L.; Bender, O.; Atalay, A.; Ceylan, R.; Mollica, A.; Mocan, A.; Uysal, S.; Guler, G.O.; et al. Traditionally used Lathyrus species: Phytochemical composition, antioxidant activity, enzyme inhibitory properties, cytotoxic effects, and in silico studies of L. czeczottianus and L. nissolia. Front. Pharmacol. 2017, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Striefler, M.; Cohn, D.; Hirano, A.; Schujman, E. The central nervous system in a case of neurolathyrism. Neurology 1977, 27, 1176–1178. [Google Scholar] [CrossRef] [PubMed]
- Haimanot, H.; Kidane, Y.; Wuhib, E.; Kalissa, A.; Alemu, T.; Zein, Z.; Spencer, P. Lathyrism in rural northwestern Ethiopia: A highly prevalent neurotoxic disorder. Int. J. Epidemiol. 1990, 19, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Van Moorhem, M.; Lambein, F.; Leybaert, L. Unraveling the mechanism of β-N-oxalyl-α,β-diaminopropionic acid (β-ODAP) induced excitotoxicity and oxidative stress, relevance for neurolathyrism prevention. Food Chem. Toxicol. 2011, 49, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Getahun, H.; Mekonnen, A.; TekleHaimanot, R.; Lambein, F. Epidemic of neurolathyrism in Ethiopia. Lancet 1999, 354, 306–307. [Google Scholar] [CrossRef]
- Woldeamanuel, Y.W.; Hassan, A.; Zenebe, G. Neurolathyrism: Two Ethiopian case reports and review of the literature. J. Neurol. 2011, 259, 1263–1268. [Google Scholar] [CrossRef]
- Giménez-Roldán, S.; Spencer, P.S. Azañón’s disease. A 19th century epidemic of neurolathyrism in Spain. Rev. Neurol. 2016, 172, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Giménez-Roldán, S.; Palmer, V.S.; Spencer, P.S. Lathyrism in Spain: Lessons from 68 publications following the 1936–39 Civil War. J. Hist. Neurosci. 2023, 32, 423–455. [Google Scholar] [CrossRef]
- Girma, A.; Tefera, B.; Dadi, L. Grass Pea and Neurolathyrism: Farmers’ perception on its consumption and protective measure in North Shewa, Ethiopia. Food Chem. Toxicol. 2011, 49, 668–672. [Google Scholar] [CrossRef]
- Hoque, H.; Jamali, S.; Akther, J.; Prodhan, S.H. Computational analysis of milk sources from different domestic animals as supplementary food source to protect Lathyrism. Int. J. Biosci. 2012, 2, 74–82. [Google Scholar]
- Girma, D.; Korbu, L. Genetic improvement of grass pea (Lathyrus sativus) in Ethiopia: An unfulfilled promise. Plant Breed. 2012, 131, 231–236. [Google Scholar] [CrossRef]
- Bell, E.; O’Donovan, J.P. The isolation of α- and γ-oxalyl derivatives of α,γ-diaminobutyric acid from seeds of Lathyrus latifolius, and the detection of the α-oxalyl isomer of the neurotoxin α-amino-β-oxalylaminopropionic acid which occurs together with the neurotoxin in this and other species. Phytochemistry 1966, 5, 1211–1219. [Google Scholar] [CrossRef]
- Vaz Patto, M.C.; Rubiales, D. Lathyrus diversity: Available resources with relevance to crop improvement—L. sativus and L. cicera as case studies. Ann. Bot. 2014, 113, 895–908. [Google Scholar] [CrossRef] [PubMed]
- Dixit, G.P.; Parihar, A.K.; Bohra, A.; Singh, N.P. Achievements and prospects of grass pea (Lathyrus sativus L.) improvement for sustainable food production. Crop J. 2016, 4, 407–416. [Google Scholar] [CrossRef]
- Song, Y.; Wang, L.; Liu, F.; Jiao, C.; Nan, H.; Shen, X.; Chen, H.; Li, Y.; Lei, B.; Jiang, J.; et al. β-Cyanoalanine Synthase Regulates the Accumulation of β-ODAP via Interaction with Serine Acetyltransferase in Lathyrus sativus. J. Agric. Food Chem. 2021, 69, 1953–1962. [Google Scholar] [CrossRef]
- Kusama-Eguchi, K.; Yoshino, N.; Minoura, A.; Watanabe, K.; Kusama, T.; Lambein, F.; Ikegami, F. Sulfur amino acids deficiency caused by grass pea diet plays an important role in the toxicity of L-β-ODAP by increasing the oxidative stress: Studies on a motor neuron cell line. Food Chem. Toxicol. 2011, 49, 636–643. [Google Scholar] [CrossRef]
- Kumar, S.; Bejiga, G.; Ahmed, S.; Nakkoul, H.; Sarker, A. Genetic improvement of grass pea for low neurotoxin (β-ODAP) content. Food Chem. Toxicol. 2011, 49, 589–600. [Google Scholar] [CrossRef]
- McCutchan, J.S. A brief history of grasspea and its use in crop improvement. Lathyrus Lathyrism Newsl. 2003, 3, 18–23. [Google Scholar]
- Rahman, M.; Ali, M.; Alam, F.; Banu, M.; Faruk, M.; Bhuiyan, M. Biocontrol of foot and root rot disease of grasspea (Lathyrus sativus) by dual inoculation with rhizobium and arbuscular mycorrhiza. Bangladesh J. Microbiol. 2017, 34, 109–117. [Google Scholar] [CrossRef]
- Campbell, C.G.; Briggs, C.J. Registration of low neurotoxin content Lathyrus germplasm LS 8246. Crop. Sci. 1987, 27, 821. [Google Scholar] [CrossRef]
- Yang, H.M.; Zhang, X.Y. Considerations on the reintroduction of grass pea in China. Lathyrus Lathyrism Newsl. 2005, 4, 22–26. [Google Scholar]
- Tadesse, W.; Bekele, E. Variation and association of morphological and biochemical characters in grass pea (Lathyrus sativus L.). Euphytica 2003, 130, 315–324. [Google Scholar] [CrossRef]
- Santha, I.M.; Mehta, S.L. Development of low ODAP somaclones of Lathyrus sativus. Lathyrus Lathyrism Newsl. 2001, 2, 42. [Google Scholar]
- Yadav, C.R. Genetic evaluation and varietal improvement of grasspea in Nepal. In Lathyrus Genetic Resources in Asia: Proceedings of a Regional Workshop; IPGRI Office for South Asia: New Delhi, India, 1996. [Google Scholar]
- Granati, E.; Bisignano, V.; Chiaretti, D.; Crinò, P.; Polignano, G.B. Characterization of Italian and exotic Lathyrus germplasm for quality traits. Genet. Resour. Crop Evol. 2003, 50, 273–280. [Google Scholar] [CrossRef]
- Krause, D.; Krause, I. New green manuring Lathyrus sativus variety AC Greenfix available in USA. Lathyrus Lathyrism Newsl. 2003, 3, 13–14. [Google Scholar]
- Mera, M.; Tay, J.; France, A.; Montenegro, A.; Espinoza, N.; Gaete, N. Luanco-INIA, a large-seeded cultivar of Lathyrus sativus released in Chile. Lathyrus Lathyrism Newsl. 2003, 3, 26. [Google Scholar]
- Sen Gupta, D.; Barpete, S.; Kumar, J.; Kumar, S. Breeding for better grain quality in lathyrus. In Breeding for Enhanced Nutrition and Bio-Active Compounds in Food Legumes; Gupta, D.S., Gupa, S., Kumar, J., Eds.; Springer: Cham, Switzerland, 2021; pp. 131–156. [Google Scholar] [CrossRef]
- Malek, M.A.; Sarwar, C.D.M.; Sarker, A.; Hassan, M.S. Status of grass pea research and future strategy in Bangladesh. In Lathyrus Genetic Resources in Asia; Arora, R.K., Mathur, P.N., Riley, K.W., Adham, Y., Eds.; International Plant Genetic Resources Institute: Rome, Italy, 1996; pp. 7–12. [Google Scholar]
- Kumar, S.; Gupta, P.; Barpete, S.; Choukri, H.; Maalouf, F.; Sarkar, A. Grass pea. In The Beans and the Peas; Woodhead Publishing: Duxford, UK, 2021; pp. 273–287. [Google Scholar]
- ICARDA. ICARDA Annual Report; International Center for Agricultural Research in the Dry Areas: Aleppo, Syria, 2007. [Google Scholar]
- Mehta, S.L.; Santha, I.M. Somaclonal variation and genetic transformation in Lathyrus sativus. In Handbook of New Technologies for Genetic Improvement of Legumes; Kirti, P.B., Ed.; CRS Press: New York, NY, USA, 2008; pp. 177–184. [Google Scholar]
- Campbell, C.G.; Mehra, R.B.; Agrawal, S.K.; Chen, Y.Z.; El Moneim, A.M.A.; Khawaja, H.I.T.; Yadov, C.R.; Tay, J.U.; Araya, W.A. Current status and future strategy in breeding grasspea (Lathyrus sativus). Euphytica 1993, 73, 167–175. [Google Scholar] [CrossRef]
- MoARD. Animal and Plant Health Regulatory Directorate; Crop Variety Register: Addis Ababa, Ethiopia, 2008; p. 11. [Google Scholar]
- Hussain, M.; Chowdhury, B.; Hoque, R.; Lambein, F. Effect of water stress, salinity, interaction of cations, stage of maturity of seeds and storage devices on the ODAP content of Lathyrus sativus. In Lathyrus and Lathyrism a Decade of Progress; Tekle Haimanot, R., Lambein, F., Eds.; University of Ghent: Ghent, Belgium, 1997; pp. 107–110. [Google Scholar]
- Xing, G.; Cui, K.; Li, J.; Wang, Y.; Li, Z. Water stress and accumulation of β-N-oxalyl-L-α,β-diaminopropionic acid in grass pea (Lathyrus sativus). J. Agric. Food Chem. 2001, 49, 216–220. [Google Scholar] [CrossRef]
- Jiao, C.J.; Xu, Q.L.; Wang, C.Y.; Li, Z.X.; Wang, Y.F. Accumulation pattern of toxin β-ODAP during lifespan and effect of nutrient elements on β-ODAP content in Lathyrus sativus seedlings. J. Agric. Sci. 2006, 144, 369–375. [Google Scholar] [CrossRef]
- Haque, R.M.; Kuo, Y.-H.; Lambein, F.; Hussain, M. Effect of environmental factors on the biosynthesis of the neuro-excitatory amino acid β-ODAP (β-N-oxalyl-l-α,β-diaminopropionic acid) in callus tissue of Lathyrus sativus. Food Chem. Toxicol. 2011, 49, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Tokarz, B.; Wójtowicz, T.; Makowski, W.; Jędrzejczyk, R.J.; Tokarz, K.M. What is the Difference between the Response of Grass Pea (Lathyrus sativus L.) to Salinity and Drought Stress?—A Physiological Study. Agronomy 2020, 10, 833. [Google Scholar] [CrossRef]
- Malathi, K.; Padmanaban, G.; Rao, S.L.; Sarma, P.S. Studies on the biosynthesis of β-N-oxalyl-L-α, β-diaminopropionic acid, the Lathyrus sativus neurotoxin. Biochim. Biophys. Acta 1967, 141, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Malathi, K.; Padmanaban, G.; Sarma, P.S. Biosynthesis of β-N-Oxalyl-L-α,β-Diaminopropionic acid, Lathyrus sativus neurotoxin. Phytochemistry 1970, 9, 1603–1610. [Google Scholar] [CrossRef]
- Ikegami, F.; Ongena, G.; Sakai, R.; Itagaki, S.; Kobori, M.; Ishikawa, T.; Kuo, Y.-H.; Lambein, F.; Murakoshi, I. Biosynthesis of β-(isoxazolin-5-on-2-yl)-l-alanine by cysteine synthase in Lathyrus sativus. Phytochemistry 1993, 33, 93–98. [Google Scholar] [CrossRef]
- Kuo, J.M.; Raushel, F.M. Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed mutagenesis. Biochemistry 1994, 33, 4265–4272. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, F.; Chen, P.; Jez, J.M.; Krishnan, H.B. β-N-Oxalyl-l-α,β-diaminopropionic acid (β-ODAP) content in Lathyrus sativus: The integration of nitrogen and sulfur metabolism through β-cyanoalanine synthase. Int. J. Mol. Sci. 2017, 18, 526. [Google Scholar] [CrossRef]
- Yan, Z.-Y.; Spencer, P.S.; Li, Z.-X.; Liang, Y.-M.; Wang, Y.-F.; Wang, C.-Y.; Li, F.-M. Lathyrus sativus (grass pea) and its neurotoxin ODAP. Phytochemistry 2006, 67, 107–121. [Google Scholar] [CrossRef]
- Feinberg, A.; Stenke, A.; Peter, T.; Hinckley, E.-L.S.; Driscoll, C.T.; Winkel, L.H.E. Reductions in the deposition of sulfur and selenium to agricultural soils pose risk of future nutrient deficiencies. Commun. Earth Environ. 2021, 2, 101. [Google Scholar] [CrossRef]
- Ross, S.M.; Roy, D.N.; Spencer, P.S. β-N-Oxalylamino-L-alanine: Action on high-affinity transport of neurotransmitters in rat brain and spinal cord synaptosomes. J. Neurochem. 1985, 44, 886–892. [Google Scholar] [CrossRef] [PubMed]
- Krogsgaard-Larsen, P.; Hansen, J.J. Naturally-occurring excitatory amino acids as neurotoxins and leads in drug design. Toxicol. Lett. 1992, 64–65, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Grosskreutz, J.; Van Den Bosch, L.; Keller, B. Calcium dysregulation in amyotrophic lateral sclerosis. Cell Calcium 2010, 47, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Kusama-Eguchi, K.; Miyano, T.; Yamamoto, M.; Suda, A.; Ito, Y.; Ishige, K.; Ishii, M.; Ogawa, Y.; Watanabe, K.; Ikegami, F.; et al. New insights into the mechanism of neurolathyrism: L-β-ODAP triggers [Ca2+]i accumulation and cell death in primary motor neurons through transient receptor potential channels and metabotropic glutamate receptors. Food Chem. Toxicol. 2014, 67, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Khandare, A.L.; Kalakumar, B.; Validandi, V.; Ssyh, Q.; Harishankar, N.; Singh, S.S.; Kodali, V. Neurolathyrism in goat (Capra hircus) kid: Model development. Res. Vet. Sci. 2020, 132, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Doble, A. The role of excitotoxicity in neurodegenerative disease: Implications for therapy. Pharmacol. Ther. 1999, 81, 163–221. [Google Scholar] [CrossRef] [PubMed]
- Hertz, L. Glutamate, a neurotransmitter–and so much more A synopsis of Wierzba III. Neurochem. Int. 2006, 48, 416–425. [Google Scholar] [CrossRef]
- Saeed, U.; Durgadoss, L.; Valli, R.K.; Joshi, D.C.; Joshi, P.G.; Ravindranath, V. Knockdown of Cytosolic Glutaredoxin 1 Leads to Loss of Mitochondrial Membrane Potential: Implication in Neurodegenerative Diseases. PLoS ONE 2008, 3, e2459. [Google Scholar] [CrossRef]
- Ravindranath, V. Neurolathyrism: Mitochondrial dysfunction in excitotoxicity mediated by L-β-oxalyl aminoalanine. Neurochem. Int. 2002, 40, 505–509. [Google Scholar] [CrossRef]
- López-Martín, M.C.; Becana, M.; Romero, L.C.; Gotor, C. Knocking out cytosolic cysteine synthesis compromises the antioxidant capacity of the cytosol to maintain discrete concentrations of hydrogen peroxide in arabidopsis. Plant Physiol. 2008, 147, 562–572. [Google Scholar] [CrossRef]
- Hailu, D.; Abera, S.; Abera, T. Effects of Processing on Nutritional Composition and Anti-Nutritional Factors of Grass pea (Lathyrus sativus L.): A review. Food Sci. Qual. Manag. 2015, 36, 61–70. [Google Scholar]
- Getahun, H.; Lambein, F.; Vanhoorne, M.; Stuyft, P.V.D. Neurolathyrism risk depends on type of grass pea preparation and on mixing with cereals and antioxidants. Trop. Med. Int. Health 2005, 10, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Tadele, D.; Alemu, Y.; Nigusie, D.; Peters, K.J. Evaluation of Processing Methods on the Feeding Value of Grass Pea to Broilers. Int. J. Poult. Sci. 2003, 2, 120–127. [Google Scholar]
- Akalu, G.; Johansson, G.; Nair, B.M. Effect of processing on the content of β-N-oxalyl-α, β-diaminopropionic acid (gb-ODAP) in grass pea (Lathyrus sativus) seeds and flour as determined by flow injection analysis. Food Chem. 1998, 62, 233–237. [Google Scholar] [CrossRef]
- Buta, M.B.; Emire, S.A.; Posten, C.; Andrée, S.; Greiner, R. Reduction of β-ODAP and IP6 contents in Lathyrus sativus L. seed by high hydrostatic pressure. Food Research International 2019, 120, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Barik, D.P.; Mohapatra, U.; Chand, P.K. Transgenic grasspea (Lathyrus sativus L.): Factors influencing Agrobacterium-mediated transformation and regeneration. Plant Cell Rep. 2005, 24, 523–531. [Google Scholar] [CrossRef]
- Barna, K.S.; Mehta, S.L. Genetic Transformation and Somatic Embryogenesis in Lathyrus sativus. J. Plant Biochem. Biotechnol. 1995, 4, 67–71. [Google Scholar] [CrossRef]
- Girma, D. Ethiopian Grass Pea (Lathyrus sativus L.) Started the Genomics Era: Transient Genetic Transformation of Grass Pea; LAP Lambert Academic Publishing: Köln, Germany, 2010. [Google Scholar]
- Krishnan, H.B. Engineering soybean for enhanced sulfur amino acid content. Crop Sci. 2005, 45, 454–461. [Google Scholar] [CrossRef]
- Kortt, A.A.; Caldwell, J.B.; Lilley, G.G.; Higgins, T.J.V. Amino acid and cDNA sequences of a methionine-rich 2S protein from sunflower seed (Helianthus annuus L.). Eur. J. Biochem. 1991, 195, 329–334. [Google Scholar] [CrossRef]
- Kirihara, J.A.; Petri, J.B.; Messing, J. Isolation and sequence of a gene encoding a methionine-rich 10-kDa zein protein from maize. Gene 1988, 71, 359–370. [Google Scholar] [CrossRef]
- Chui, C.F.; Falco, S.C. A new methionine-rich seed storage protein from maize. Plant Physiol. 1995, 107, 291. [Google Scholar] [CrossRef]
- Planta, J.; Xiang, X.; Leustek, T.; Messing, J. Engineering sulfur storage in maize seed proteins without apparent yield loss. Proc. Natl. Acad. Sci. USA 2017, 114, 11386–11391. [Google Scholar] [CrossRef] [PubMed]
- Dinkins, R.D.; Reddy, M.S.S.; Meurer, C.A.; Yan, B.; Trick, H.; Thibaud-Nissen, F.; Finer, J.J.; Parrott, W.A.; Collins, G.B. Increased sulfur amino acids in soybean plants overexpressing the maize 15 kDa zein protein. In Vitr Cell. Dev. Biol.-Plant 2001, 37, 742–747. [Google Scholar] [CrossRef]
- Kim, W.S.; Krishnan, H.B. Allelic variation and differential expression of methionine-rich δ-zeins in maize inbred lines B73 and W23a1. Planta 2003, 217, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Liu, X.; Chen, L.; Cai, Y.; Yao, W.; Yuan, S.; Wu, C.; Han, T.; Sun, S.; Hou, W. Elevated methionine content in soybean seed by overexpressing maize β-zein protein. Oil Crop Sci. 2020, 5, 11–16. [Google Scholar] [CrossRef]
- Kim, W.-S.; Sun-Hyung, J.; Oehrle, N.W.; Jez, J.M.; Krishnan, H.B. Overexpression of ATP sulfurylase improves the sulfur amino acid content, enhances the accumulation of Bowman–Birk protease inhibitor and suppresses the accumulation of the β-subunit of β-conglycinin in soybean seeds. Sci. Rep. 2020, 10, 14989. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-S.; Chronis, D.; Juergens, M.; Schroeder, A.C.; Hyun, S.W.; Jez, J.M.; Krishnan, H.B. Transgenic soybean plants overexpressing O-acetylserine sulfhydrylase accumulate enhanced levels of cysteine and Bowman–Birk protease inhibitor in seeds. Planta 2011, 235, 13–23. [Google Scholar] [CrossRef]
- Avraham, T.; Badani, H.; Galili, S.; Amir, R. Enhanced levels of methionine and cysteine in transgenic alfalfa (Medicago sativa L.) plants over-expressing the Arabidopsis cystathionine γ-synthase gene. Plant Biotechnol. J. 2005, 3, 71–79. [Google Scholar] [CrossRef]
- Endo, M.; Mikami, M.; Toki, S. Biallelic gene targeting in rice. Plant Physiol. 2016, 170, 667–677. [Google Scholar] [CrossRef]
- Hilscher, J.; Bürstmayr, H.; Stoger, E. Targeted modification of plant genomes for precision crop breeding. Biotechnol. J. 2017, 12, 1600173. [Google Scholar] [CrossRef] [PubMed]
- Friedrichs, S.; Takasu, Y.; Kearns, P.; Dagallier, B.; Oshima, R.; Schofield, J.; Moreddu, C. An overview of regulatory approaches to genome editing in agriculture. Biotechnol. Res. Innov. 2019, 3, 208–220. [Google Scholar] [CrossRef]
- Zannoni, L. Evolving Regulatory Landscape for Genome-Edited Plants. CRISPR J. 2019, 2, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Hussain, A.; Manghwar, H.; Xie, K.; Xie, S.; Zhao, S.; Larkin, R.M.; Qing, P.; Jin, S.; Ding, F. Genome editing with the CRISPR-Cas system: An art, ethics and global regulatory perspective. Plant Biotechnol. J. 2020, 18, 1651–1669. [Google Scholar] [CrossRef] [PubMed]
- Emmrich, P.M.; Sarkar, A.; Njaci, I.; Kaithakottil, G.G.; Ellis, N.; Moore, C.; Edwards, A.; Heavens, D.; Waite, D.; Cheema, J.; et al. A draft genome of grass pea (Lathyrus sativus), a resilient diploid legume. BioRxiv 2020. [Google Scholar] [CrossRef]
- Friedrichs, S.; Takasu, Y.; Kearns, P.; Dagallier, B.; Oshima, R.; Schofield, J.; Moreddu, C. Meeting report of the OECD conference on “genome editing: Applications in agriculture—Implications for health, environment and regulation”. Transgenic Res. 2019, 28, 419–463. [Google Scholar] [CrossRef] [PubMed]
- Chilcoat, D.; Liu, Z.B.; Sander, J. Use of CRISPR/Cas9 for crop improvement in maize and soybean. Prog. Mol. Biol. Transl. Sci. 2017, 149, 27–46. [Google Scholar] [PubMed]
- Fiaz, S.; Ahmad, S.; Noor, M.A.; Wang, X.; Younas, A.; Riaz, A.; Riaz, A.; Ali, F. Applications of the CRISPR/Cas9 system for rice grain quality improvement: Perspectives and opportunities. Int. J. Mol. Sci. 2019, 20, 888. [Google Scholar] [CrossRef]
- Wang, H.; Wu, Y.; Zhang, Y.; Yang, J.; Fan, W.; Zhang, H.; Zhao, S.; Yuan, L.; Zhang, P. CRISPR/Cas9-Based Mutagenesis of Starch Biosynthetic Genes in Sweet Potato (Ipomoea Batatas) for the Improvement of Starch Quality. Int. J. Mol. Sci. 2019, 20, 4702. [Google Scholar] [CrossRef]
- Tuncel, A.; Corbin, K.R.; Ahn-Jarvis, J.; Harris, S.; Hawkins, E.; Smedley, M.A.; Harwood, W.; Warren, F.J.; Patron, N.J.; Smith, A.M. Cas9-mediated mutagenesis of potato starch-branching enzymes generates a range of tuber starch phenotypes. Plant Biotechnol. J. 2019, 17, 2259–2271. [Google Scholar] [CrossRef]
- Li, J.; Jiao, G.; Sun, Y.; Chen, J.; Zhong, Y.; Yan, L.; Jiang, D.; Ma, Y.; Xia, L. Modification of starch composition, structure and properties through editing of TaSBEIIa in both winter and spring wheat varieties by CRISPR/Cas9. Plant Biotechnol. J. 2021, 19, 937–951. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Mohamed, S.; Tanoi, K.; Kobayashi, N.I.; Takagi, K.; Vernet, A.; Guiderdoni, E.; Périn, C.; Sentenac, H.; Véry, A. Production of low-Cs+ rice plants by inactivation of the K+ transporter OsHAK1 with the CRISPR-Cas system. Plant J. 2017, 92, 43–56. [Google Scholar] [CrossRef]
- Gomez, M.A.; Berkoff, K.C.; Gill, B.K.; Iavarone, A.T.; Lieberman, S.E.; Ma, J.M.; Schultink, A.; Karavolias, N.G.; Wyman, S.K.; Chauhan, R.D.; et al. CRISPR-Cas9-mediated knockout of CYP79D1 and CYP79D2 in cassava attenuates toxic cyanogen production. Front. Plant Sci. 2023, 13, 1079254. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Shin, G.; Kim, H.J.; Lee, S.B.; Moon, J.Y.; Jeong, J.C.; Choi, H.-K.; Kim, I.A.; Song, H.J.; Kim, C.Y.; et al. Mutation of GmIPK1 gene using CRISPR/Cas9 reduced phytic acid content in soybean seeds. Int. J. Mol. Sci. 2022, 23, 10583. [Google Scholar] [CrossRef] [PubMed]
- Sashidhar, N.; Harloff, H.J.; Potgieter, L.; Jung, C. Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol. J. 2020, 18, 2241–2250. [Google Scholar] [CrossRef] [PubMed]
- Raffan, S.; Sparks, C.; Huttly, A.; Hyde, L.; Martignago, D.; Mead, A.; Hanley, S.J.; Wilkinson, P.A.; Barker, G.; Edwards, K.J.; et al. Wheat with greatly reduced accumulation of free asparagine in the grain, produced by CRISPR/Cas9 editing of asparagine synthetase gene TaASN2. Plant Biotechnol. J. 2021, 19, 1602–1613. [Google Scholar] [CrossRef]
- Kenar, J.A. Reaction chemistry of gossypol and its derivatives. J. Am. Oil Chem. Soc. 2006, 83, 269–302. [Google Scholar] [CrossRef]
- Lin, J.-L.; Fang, X.; Li, J.-X.; Chen, Z.-W.; Wu, W.-K.; Guo, X.-X.; Liu, N.-J.; Huang, J.-F.; Chen, F.-Y.; Wang, L.-J.; et al. Dirigent gene editing of gossypol enantiomers for toxicity-depleted cotton seeds. Nat. Plants 2023, 9, 605–615. [Google Scholar] [CrossRef]
- Schachtsiek, J.; Stehle, F. Nicotine-free, nontransgenic tobacco (Nicotiana tabacum L.) edited by CRISPR-Cas9. Plant Biotechnol. J. 2019, 17, 2228–2230. [Google Scholar] [CrossRef]
- Zakaria, M.; Schemmerling, B.; Ober, D. CRISPR/Cas9-mediated genome editing in comfrey (Symphytum officinale) hairy roots results in the complete eradication of pyrrolizidine alkaloids. Molecules 2021, 26, 1498. [Google Scholar] [CrossRef]
- Ma, J.; Sun, S.; Whelan, J.; Shou, H. CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds. Int. J. Mol. Sci. 2021, 22, 3877. [Google Scholar] [CrossRef]
- Wang, Z.; Shea, Z.; Rosso, L.; Shang, C.; Li, J.; Bewick, P.; Li, Q.; Zhao, B.; Zhang, B. Development of new mutant alleles and markers for KTI1 and KTI3 via CRISPR/Cas9-mediated mutagenesis to reduce trypsin inhibitor content and activity in soybean seeds. Front. Plant Sci. 2023, 14, 1111680. [Google Scholar] [CrossRef] [PubMed]
- Rajarammohan, S.; Kaur, L.; Verma, A.; Singh, D.; Mantri, S.; Roy, J.K.; Sharma, T.R.; Pareek, A.; Kandoth, P.K. Genome sequencing and assembly of Lathyrus sativus—A nutrient-rich hardy legume crop. Sci. Data 2023, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Riepe, M.; Spencer, P.S.; Lambein, F.; Ludolph, A.C.; Allen, C.N. In vitro toxicological investigations of isoxazolinone amino acids of Lathyrus sativus. Nat. Toxins 1995, 3, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Saha, T.; Shee, R.; Sahid, S.; Shee, D.; Roy, C.; Sharma, R.; Pandey, A.; Paul, S.; Datta, R. Designer grass pea for transgene-free minimal neurotoxin-containing seeds with CRISPR-Cas9. bioRxiv 2023. [Google Scholar] [CrossRef]
- Verma, A.; Kaur, L.; Kaur, N.; Bhardwaj, A.; Pandey, A.K.; Kandoth, P.K. An efficient hairy root system for genome editing of a β-ODAP pathway gene in Lathyrus sativus. bioRxiv 2023. [Google Scholar] [CrossRef]
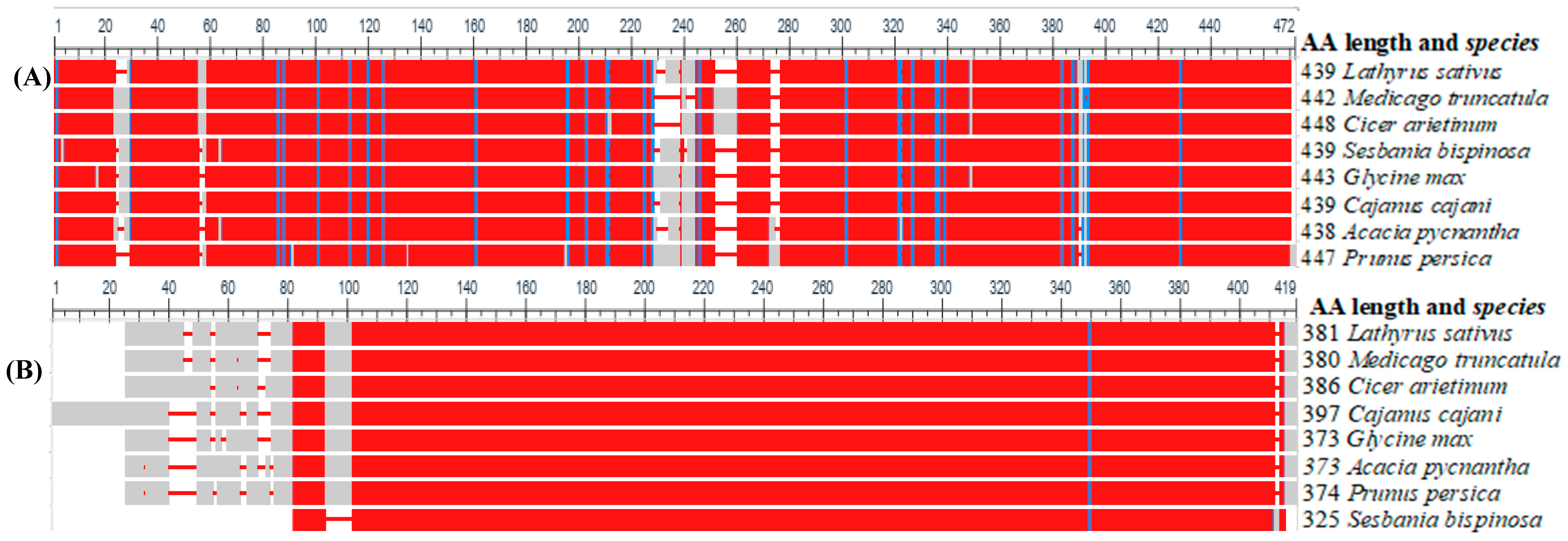
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef]
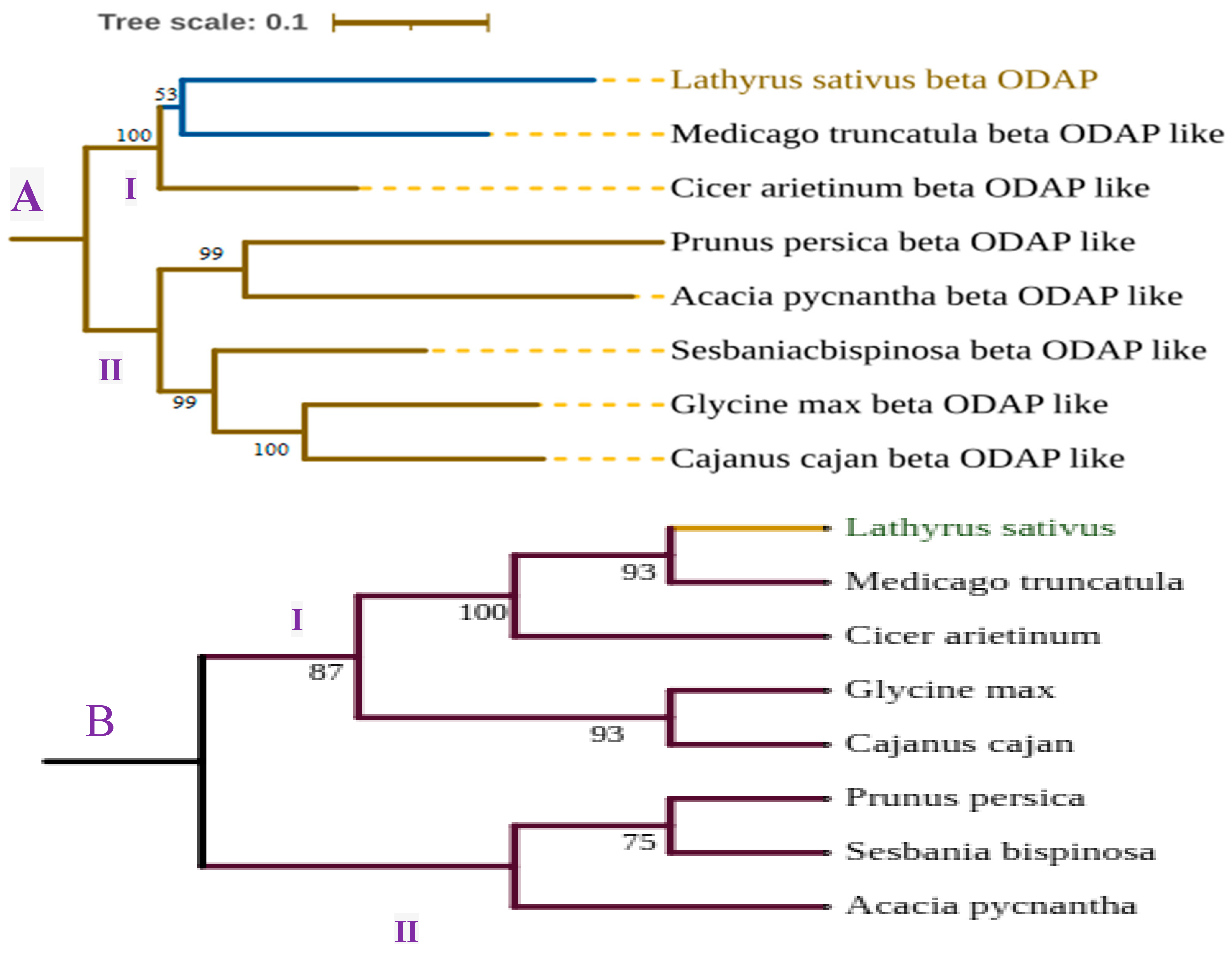
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bekele-Alemu, A.; Girma-Tola, D.; Ligaba-Osena, A. The Potential of CRISPR/Cas9 to Circumvent the Risk Factor Neurotoxin β-N-oxalyl-L-α, β-diaminopropionic acid Limiting Wide Acceptance of the Underutilized Grass Pea (Lathyrus sativus L.). Curr. Issues Mol. Biol. 2024, 46, 10570-10589. https://doi.org/10.3390/cimb46090626
Bekele-Alemu A, Girma-Tola D, Ligaba-Osena A. The Potential of CRISPR/Cas9 to Circumvent the Risk Factor Neurotoxin β-N-oxalyl-L-α, β-diaminopropionic acid Limiting Wide Acceptance of the Underutilized Grass Pea (Lathyrus sativus L.). Current Issues in Molecular Biology. 2024; 46(9):10570-10589. https://doi.org/10.3390/cimb46090626
Chicago/Turabian StyleBekele-Alemu, Abreham, Deribew Girma-Tola, and Ayalew Ligaba-Osena. 2024. "The Potential of CRISPR/Cas9 to Circumvent the Risk Factor Neurotoxin β-N-oxalyl-L-α, β-diaminopropionic acid Limiting Wide Acceptance of the Underutilized Grass Pea (Lathyrus sativus L.)" Current Issues in Molecular Biology 46, no. 9: 10570-10589. https://doi.org/10.3390/cimb46090626
APA StyleBekele-Alemu, A., Girma-Tola, D., & Ligaba-Osena, A. (2024). The Potential of CRISPR/Cas9 to Circumvent the Risk Factor Neurotoxin β-N-oxalyl-L-α, β-diaminopropionic acid Limiting Wide Acceptance of the Underutilized Grass Pea (Lathyrus sativus L.). Current Issues in Molecular Biology, 46(9), 10570-10589. https://doi.org/10.3390/cimb46090626





