How “Omics” Studies Contribute to a Better Understanding of Fuchs’ Endothelial Corneal Dystrophy
Abstract
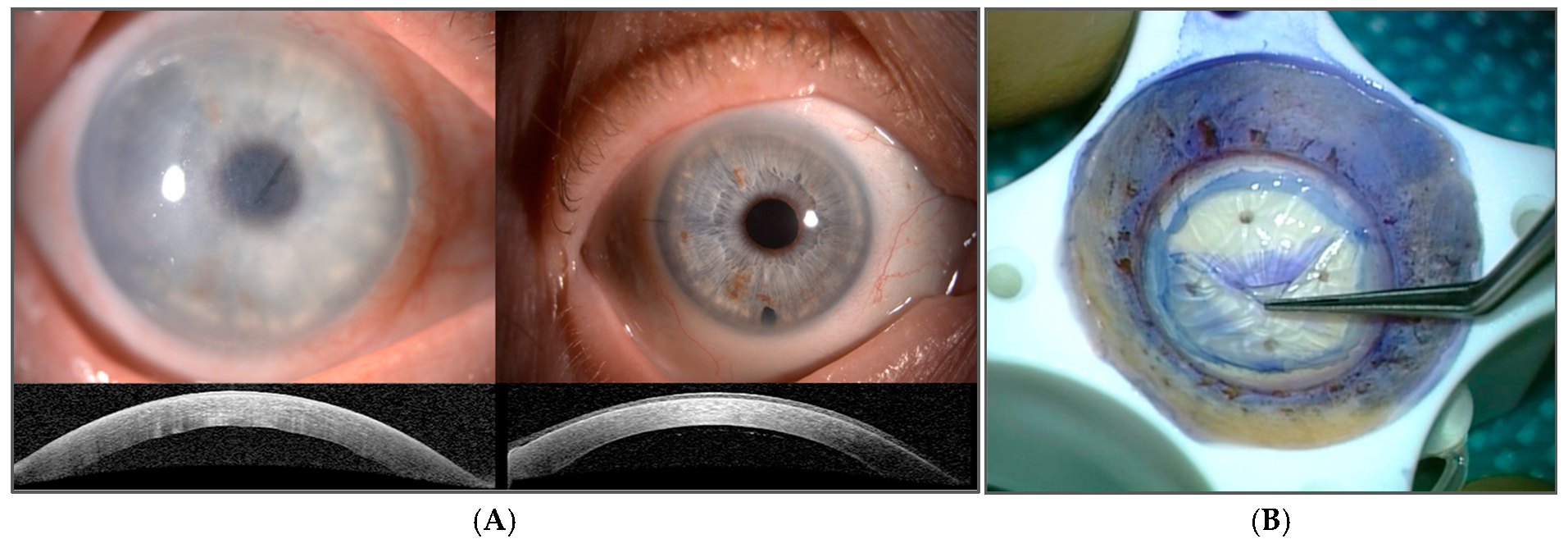
1. Introduction
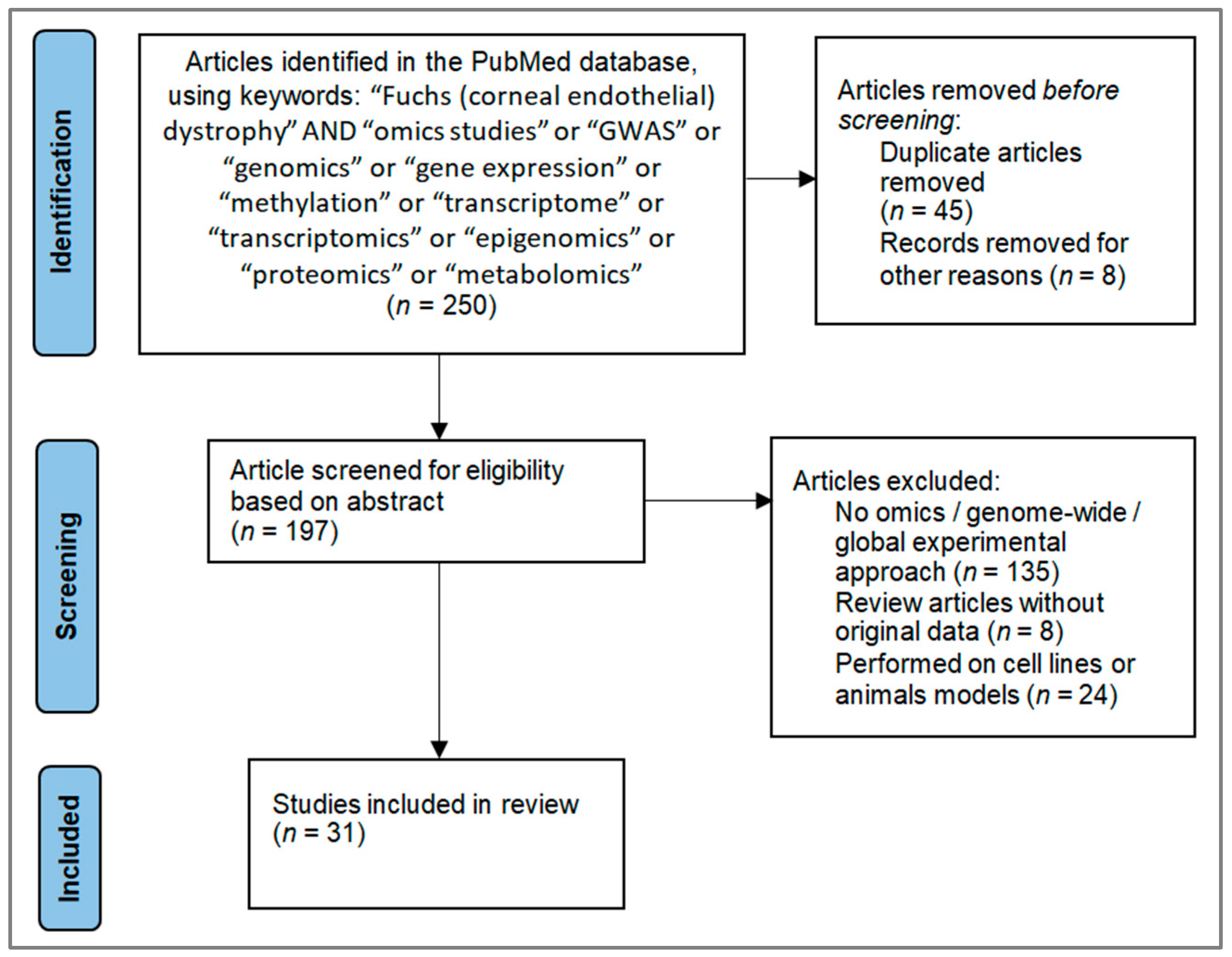
2. Literature Selection and Study Overview
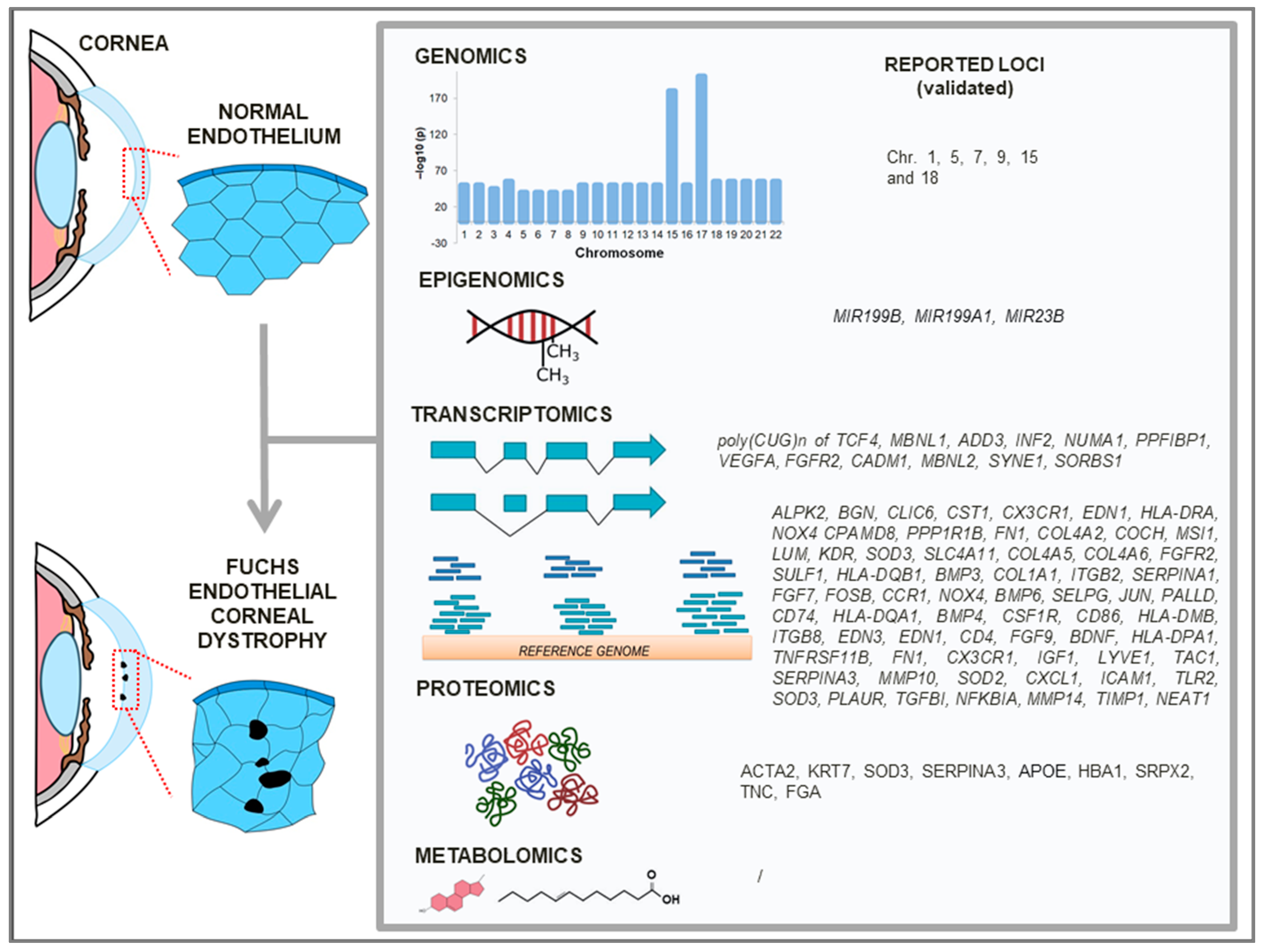
3. Genomics of FECD
3.1. Family-Based Studies
3.2. Population-Based Studies
3.3. Mitochondrial DNA (mtDNA)
| Ref. | Relation of Participants (Raw Data Accession) | Discovery Group | Replication Group | Genomic Approach | Genome-Wide Platform | Statistical Approach | Reported Loci (SNPs) Associated with FECD; Statistical Significance | Validation of Identified Loci | ||
|---|---|---|---|---|---|---|---|---|---|---|
| FECD Cases | Controls | FECD Cases | Controls | |||||||
| [8] | European population (datasets from [39] and dbGaP database: phs000421.v1.p1and phs000429.v1.p1) | 530 + 457 | 498 + 498 | 857 + 857 | 2342 + 2186 | Whole-genome | / | Logistic regression model | rs2853826 (the variant A10398G) of ND3 (p = 0.034) and Haplogroup I (p = 0.041) | / |
| [33] | Three-generation family | 11 | 6 | / | / | Whole-genome | Affymetrx SNP microarry ver. 5.0 | Parametric two-point linkage and haplotype analyses; autosomal dominant model | Chr5 (rs13173656, rs9313417, rs2116736, rs17451810, rs4958561, rs778816); LOD > 2 | Confirmed by short-tandem-repeat microsatellite markers |
| [34] | Large multigenerational family and 21 small multiplex families | 56 | 36 | / | / | Whole-genome | Illumina SNP linkage panel IV | Parametric two-point and nonparametric multipoint linkage analyses; autosomal dominant and recessive models | Two-point analysis: Chr1 (rs760594), Chr7 (rs257376, rs1047035), Chr15 (rs352476, rs1075991), Chr17 (rs938350), ChrX (rs548996); HLOD > 1.5. Multipoint analysis: Chr7 (rs740295, rs918980), Chr17 (rs1530348); LOD > 1.5. | / |
| [35] | Three-generation family | 12 | 3 | / | / | Whole-exome | Illumina HiSeq2000 Genome Analyzer | In-house pipeline | Chr15 (nonsense mutation c.3082C > T in AGBL1) | Confirmed by dideoxy sequencing and low gene expression in CE by qPCR |
| [36] | Five-generation Chinese family | 9 + 5 (coexisted APC) | 19 | / | / | Whole-exome | Illumina HiSeq X-Ten | Multipoint parametric and nonparametric linkage analyses; autosomal dominant models | Linkage analyses: no candidate region with LOD > 2. After additional data filtering: Chr7 (INTS1) and Chr9 (SH3GL2) | Confirmed by PCR and Sanger sequencing |
| [39] | Caucasian 64 multiplex families | 165 | 50 | / | / | Whole-genome | Illumina Golden- Gate linkage panel IVB and Infinium Human- Linkage12 | Parametric and nonparametric two-point and multipoint analyses; dominant and recessive models | Multipoint dominant model: Chr18 (rs1145315); HLOD = 2.5. Two-point analysis: Dominant model: Chr10 (rs1889974), Chr15 (rs235512). Recessive model Chr19 (rs893186); HLOD > 3. | / |
| Caucasian population | 450 | 340 | / | / | Whole-genome | / | Association analysis; dominant, additive and recessive models | Chr18 (rs613872 TCF4) for all three genetic models (p < 0.05; with p = 9.33 × 10−35 in dominant model) | Confirmed by PCR | |
| [38] | European population (available at dbGaP: phs000246.v2.p1) | 130 | 260 | 150 | 150 | Whole-genome | Illumina 370K Beadchip | Log-additive model (SAS software, v.9.1.) | Chr 18 (rs613872 TCF4); p = 2.34 × 10−26 | / |
| [40] | European population (available at dbGaP: phs000001.v3.p1) | 1404 | 2564 | 671 | 778 | Whole-genome | Illumina Omni2.5- 4v1_H array | Logistic regression with additive model | Chr1 (rs79742895 KANK4, rs3768617 LAMC1, rs1200114 LINC00970/ATP1B1), Chr18 (rs784257 TCF4); (p < 5 × 10−8) | Gene and protein expression in CE confirmed by RNA-seq and IHC, except KANK4 only by IHC. |
| [63] | European, African, Hispanic/Latino populations (datasets also obtained from [40]) | 2251 | 252,345 | 1404 | 2564 | Whole-genome | Thermo Fisher MVP 1.0 Axiom array | SAIGE v1.1.6.2, +mungle plug-in (bcftools v1.16) | Chr1 (rs79742895 KANK4, rs1200114 ATP1B1, rs2093985 LAMC1, rs11590557 SSBP3), Chr7 (rs74882680 THSD7A, rs150990106 LAMB1), Chr11 (rs1138714 PIDD1), Chr15 (rs12439253 RORA), Chr17 (rs9303111 HS3ST3B1), Chr18 (rs11659764 TCF4), Chr20 (rs141208202 LAMA5), Chr21 (rs114065856 COL18A1); p < 10−8 | / |
4. Transcriptomics of FECD
4.1. Alternative Splicing Events
| Ref. | Available Raw Data: Accession Number | Study Design | FECD Cases (Ancestry, Average Age) | Controls (Ancestry, Average Age) | Genome-Wide Platform | Bioinfor-Matics Tool/Software | Significant Loci Associated with FECD Cohort Relative to CO Comparison | Validation of Identified Locus/Loci | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F_RE+ | F_RE− | n.s. | CO_RE+ | CO | Differentially Expressed Genes (DEGs) | Differentially Spliced Genes (DSGs) or/and Alternative Splicing Events (ASEs) | ||||||
| [71] | Extracted from GEO: GSE142538, GSE112201; SRA: PRJNA524323 | Meta analysis | 25 | 8 | / | / | 19 | / | DEGs: AWFisher DSGs: rMATS turbo | 1184 ↑, 1018 ↓; FDR < 0.05 | DSGs: No data provided. Prevalence of exon skipping events among datasets; FDR < 0.05 | / |
| [72] | / | Differ. splicing analysis | 4 | / | / | / | 3 | Illumina HiSeq 2000 | Limma CASPER v1.9.0 | / | 342 DSGs; p < 0. 05. Identified poly(CUG)n TCF4 transcripts | Confirmed poly(CUG)n TCF4 by FISH. Confirmed ASE of MBNL1 (inclusion of exon 6), ADD3 (inclusion of exon 14) and INF2 (exclusion of exon 22) by RT-PCR |
| [73] | / | Differ. splicing analysis | 11 (72 years) | / | / | 4 (63 years) | Illumina HiSeq4000 | Mayo Analysis Pipeline for RNA-seq | / | 58 DSGs/61 ASE | Confirmed ASE for NUMA1 (exon exclusion), PPFIBP1 (exon exclusion), VEGFA (exon inclusion), FGFR2 (exon exclusion) by RT-PCR and MBNL2 (exon inclusion) by RT-PCR and Sanger sequencing | |
| [74] | Deposited to GEO: GSE112201 | Differ. splicing and gene expression analyses | 18 (71 years) | 6 (68 years) | / | / | / | Illumina HiSeq2000 or HiSeq4000 | DSGs: MISO DEGs: edgeR and z-test | 28 ↑, 11 ↓; log2 FC > 1 | 20 ASE | Confirmed ASE of ADD3 (exon inclusion), CADM1 (exon inclusion), INF2 (exon exclusion) by RT-PCR in F_RE+ group |
| [76] | Deposited to GEO: GSE171830 | Differ. gene expression analysis | / | / | 9 (pooled) | / | 3 | Illumina HumanHT-12 v4.0 | Limma | 126 ↑, 16 ↓; Log2 FC ≥ 1.5, FDR ≤ 0.05 | / | Confirmed ↑ of ALPK2, BGN, CLIC6, CST1, CX3CR1, EDN1, HLA-DRA, NOX4 and ↓ of CPAMD8 and PPP1R1B by qPCR |
| [77] | Extracted from GEO: GSE74123, GSE171830, GSE101872, GSE142538; SRA: PRJNA524323 | Meta analysis | 19 (Cauc-asians) | 9 (Cauc-asians) | 13 (Caucasia-ns) | / | 26 (Cauc-asians) | / | Combined effect size method. Random effect model. | 1103 ↑, 434 ↓; FDR < 0.05 | / | / |
| [78] | Deposited to DNA Data Bank of Japan: DRA015078 and Genomic Expression Archive: E-GEAD-564 | Differ. gene expression analysis | / | / | 10 (late-onsetCaucasia-ns, 67 years) | / | 7 (Cauc-asians; 61 years) | Illumina HiScanSQ | DESeq2 | 1092 ↑, 1274 ↓; Log2 FC > 2, FDR < 0.05 | / | / |
| [79] | Deposited to SRA: PRJNA524323 | Transcript. data generation | 8 | 4 | / | / | 6 | Illumina HiSeq 2500 | / | / | / | / |
| [80] | Retrieved from GEO: GSE171830 | Bioinfor. data reanalysis | / | / | 6 | / | 6 | / | Limma | 257 ↑, 46 ↓; log2 FC > 2; p < 0.01 | / | / |
| [75] | Deposited to GEO: GSE142538 | Differ. splicing and gene expression analyses | 6 | 4 | / | 6 (declared as pre-sympto-matic FECD) | 9 | Illumina Nextseq 500/550 | DSGs: rMATS (v.4.0) DEGs: Cuttdiff within Cufflinks | DEGs: 215 (CO_RE+), 1330 (F_RE+), 696 (F_RE−); FDR < 0.05 | ASE: Skipped exon: 313 (CO_RE+), 1030 (F_RE+), 737 (F_RE−). Alternative 5′ splice site: 30 (CO_RE+), 63 (F_RE+), 52 (F_RE−). Alternative 3′ splice site: 7 (CO_RE+), 51 (F_RE+), 37 (F_RE−). Mutually exclusive exon: 62 (CO_RE+), 237 (F_RE+), 140 (F_RE-). Retained intron: 37 (CO_RE+), 194 (F_RE+), 184 (F_RE−); FDR < 0.001. | ASE: Confirmed in CO_RE+/F_RE+ vs. CO for MBNL2 (exon 7 inclusion), SYNE1 (exon9 exclusion), INF2 (exon 22 exclusion), MBNL1 (exon 5 inclusion), NUMA1 (exon 16 exclusion), SORBS1 (exon 23 exclusion) by RT-PCR. DEGs: Confirmed FN1 ↑ in CO_RE+, COL4A2 ↑ in CO_RE+, COCH ↑ in CO_RE+, MSI1 ↑ in CO_RE+, LUM ↓ in F_RE+, KDR ↓ in F_RE+ and CO_RE+, SOD3 ↓ in F_RE+ by qPCR |
| [81] | Deposited to GEO: GSE75676 | Differ. gene expression analysis | / | / | 4 (late- onset, 75.5 yeras) | / | 4 (52.8 years) | Affymetrix GeneChip Human Gene 1.0 STA | Moderate t statistic | 487 ↑, 467 ↓; Log2 FC > 2, FDR < 0.05 | / | Confirmed ↑ SLC4A11, COL4A5, COL4A6, FGFR2, SULF1, HLA-DQB1, BMP3, COL1A1, ITGB2, SERPINA1, FGF7, FOSB, CCR1, NOX4, BMP6, SELPG, JUN, PALLD, CD74, HLA-DQA1, BMP4, CSF1R, CD86, HLA-DMB, ITGB8, EDN3, EDN1, CD4, FGF9, BDNF, HLA-DPA1, TNFRSF11B, FN1, CX3CR1, IGF1, LYVE1, and ↓ TAC1, SERPINA3, MMP10, SOD2, CXCL1, ICAM1, TLR2, SOD3, PLAUR, TGFBI, NFKBIA, MMP14, TIMP1 by qPCR. Confirmed ↑ HLA-DRA by qPCR/IHC. Confirmed ↑ ACTA2, KRT7, SOD3 and ↓ SERPINA3 by IHC [82] |
| [83] | Extracted from GEO: GSE74123 | Bioinfor. data reanalysis | / | / | 4 (late-onset, 75.5 years) | / | 4 (52.8 years) | / | Linear correlation | 592 ↑, 527 ↓; Log2 FC > 1; p < 0.05 | / | / |
| [84] | Retrieved from GEO: GSE10187 | Differ. gene expression | / | / | 5 | / | 2 | / | ARCHS4 gene expression matrix v5 | 50 DEGs | / | / |
| [85] | Retrieved from own study [78] and from SRA: DRP006678, DRA015078 | Bioinfor. data reanalysis | No data | No data | No data | No data | No data | / | DESeq2 package (v1.34.0) | / | 1 (TCF4-277 isoform of TCF4); Log2 FC ≥ 1.5; p < 0.05 | / |
| [86] | Extracted from GEO: GSE112201 | Differ. gene expression | / | / | 10 | / | 3 | Illumina NovaSeq 6000 | DESeq2 | lncRNA NEAT1 ↓; p < 0.01, log2 FC > 1.5 | / | Confirmed by qPCR |
4.2. Altered Gene-Expression Patterns
4.3. Reanalysis of Transcriptomics Data
4.4. Non-Coding RNA
4.5. Single-Cell RNA-Seq
5. Epigenomics of FECD
| Ref. | Raw Data: Accession Number | Study Design | Fuchs Group (Average Age) | Control Group (Average Age) | Genome-Wide Platform | Bioinformatics Tools/Program | Loci Associated with FECD (Hypo (↓) or Hyper (↑) Methylated) | Validation | |
|---|---|---|---|---|---|---|---|---|---|
| F_RE+ | n.s. | ||||||||
| [91] | Deposited to GEO: GSE94462 | Diff. methyated genes | / | 9 (64 years) | 4 (71 years) | Illumina Infinium HumanMethylation450 | Probe-wise linear model | 6439 probes ↑, 4531 probes ↓; FDR < 0.05 | Confirmed MIR199B ↑ |
| [92] | Extracted from GEO: GSE94462 | Diffe. methylated miRNAs | / | 9 | 4 | / | Sample pairwise correlation in R program | 154 probes ↑, 62 probes ↓; FDR < 0.01 | Confirmed miR-199A1 and miR-23B ↑ by qPCR |
| [93] | Deposited to GEO: GSE198917 | Differ. methylated CpG islands | 16 | / | 9 | Illumina Human Infinium MethylationEPIC array | Minfi package (v. 1.32) | 1505 CpGs ↑, 1983 CpGs ↓; FDR < 0.05 | / |
6. Proteomics of FECD
| PROTEOMICS of FECD | ||||||||
|---|---|---|---|---|---|---|---|---|
| Ref. | Tissue Source | Raw Data: Accession Number | FECD Group (Ancestry, Average Age) | Control Group (Average Age) | Genome-Wide Platform | Bioinformatics Tools/Program | Differentially (↑ = Up- and ↓ = Downregulated) Expressed Proteins/Metabolites Associated with FECD Group | Validation of Identified Loci |
| [95] | Aqueous humor | / | 12 (coexisted cataracts, 62.8 years) | 11 (with cataract, 64.0 years) | Liquid chromatography–tandem mass spectrometry | Student’s t-test | ↑: SBSN, hemoglobin fragment (n.s.), immunoglobulin kappa (n.s.), immunoglobulin lambda (n.s.), uncharacterized protein albumin (n.s.), ↓: AFM, C3, HRG, IGH (n.s.), FAM3C; p ≤ 0.01 | / |
| [96] | Descemet’s membrane | / | 3 (Caucasians, 72.3 years) | 3 (Caucasians, 85.3 years) | Nanoscale ultra-performance liquid chromatography–mass spectrometry | PLGS Expression Analysis Software (v.2.4.) Two-tailed Student t-test | APOE and IGHG1 ↓; p < 0.05 | Confirmed APOE ↓ gene (in CE) and protein expression by qPCR and IHC |
| [97] | Descemet’s membrane | Deposited to MassIVE: MSV000091078 | 1 (63 years old) | 1 (80 years old) | Liquid chromatography–tandem mass spectrometry | n.s. | 32 exclusive proteins | Confirmed exclusive expression of HBA1, SRPX2, TNC, hemoglobin γδεβ) (n.s.) and ↑ of FGA by immunofluorescence staining |
| METABOLOMICS of FECD | ||||||||
| [98] | Aqueous humor | / | 8 (56.8 years) | 10 (50.8 years) | Lipidomic ultra-performance liquid chromatography–mass spectrometry | Univariate analysis; Student’s t-test. | 23 ↑, 4 ↓ lipids; p < 0.05 | / |
7. Metabolomics of FECD
8. Data Synthesis
| Synthesized Highlighted Loci That Were Identified by Two-Group Comparison in Original Studies | Gene Ontology Database | GO_TERM/KEGG/Reactome Pathway | Annotated Genes | FDR Value | Omics Level |
|---|---|---|---|---|---|
| Comparison of GSEA results between F_n.s. vs. CO groups | |||||
| TCF4, KANK4, LAMC1, RORA, LAMA5, LINC00970, ATP1B1, LAMB1, PIDD1, COL18A1, SSBP3, THSD7A, HS3ST3B1, ND3 | GO_TERM_CC | GO:0043259~laminin-10 complex | LAMA5, LAMB1, LAMC1 | 3.17 × 10−5 | Genomics |
| GO_TERM_BP | GO:0007155~cell adhesion | LAMA5, COL18A1, LAMB1, LAMC1, ATP1B1 | 0.0232 | ||
| GO_TERM_MF | GO:0005201~extracellular-matrix structural constituent | LAMA5, LAMB1, LAMC1 | 0.0903 | ||
| KEGG_PATHWAY | hsa04512: ECM-receptor interaction | LAMA5, LAMB1, LAMC1 | 0.0287 | ||
| REACTOME_PATHWAY | R-HSA-3000157~Laminin interactions | LAMA5, COL18A1, LAMB1, LAMC1 | 1.04 × 10−4 | ||
| PDE11A, CCDC57, GNAS, MTUS2, COBL, SPG21, NME6, CDH4, MYADML, GUCY2C, BSN, CCDC124 | GO_TERM_CC | GO:0005813~centrosome | CCDC57, CCDC124, MTUS2 | 1 | Epigenomics |
| GO_TERM_BP | / | / | / | ||
| GO_TERM_MF | / | / | / | ||
| KEGG_PATHWAY | hsa00230: Purine metabolism | GUCY2C, PDE11A, NME6 | 0.1186 | ||
| REACTOME_PATHWAY | R-HSA-418346~Platelet homeostasis | PDE11A, GNAS | 1 | ||
| ALPK2, BGN, CLIC6, CST1, GPC3, CX3CR1M, EDN1, HLA-DRA, NOX4, CPAMD8, PPP1R1B, ANXA1, VCAN, TNC, IGFBP7, MATN3, SPARCL1 | GO_TERM_CC | GO:0062023~collagen-containing extracellular matrix | VCAN, ANXA1, BGN, TNC, GPC3, SPARCL1, IGFBP7, MATN3 | 3.11 × 10−7 | Transcriptomics |
| GO_TERM_BP | GO:0071385~cellular response to glucocorticoid stimulus | EDN1, ANXA1 | 1 | ||
| GO_TERM_MF | GO:0005201~extracellular-matrix structural constituent | BGN, TNC, IGFBP7, MATN3 | 0.0082 | ||
| KEGG_PATHWAY | hsa04933: AGE-RAGE signaling pathway in diabetic complications | EDN1, NOX4 | 1 | ||
| REACTOME_PATHWAY | R-HSA-8957275~Post-translational protein phosphorylation | VCAN, TNC, GPC3, SPARCL1, IGFBP7, MATN3 | 2.57 × 10−6 | ||
| ACTA2, KRT7, SOD3, SERPINA3, AFM, C3, HRG, FAM3C, SBSN, APOE, HBA1, SRPX2, TNC, COL6A2, COL8A1, COL18A1, LTBP2, LUM, MATN2, MATN, MUC6, PRELP, TNC, FGA | GO_TERM_CC | GO:0062023~collagen-containing extracellular matrix | FGA, COL18A1, SERPINA3, LUM, TNC, PRELP, LTBP2, SOD3, SRPX2, COL6A2, COL8A1, APOE, HRG, MATN2 | 3.67 × 10−16 | Proteomics |
| GO_TERM_BP | GO:0001525~angiogenesis | COL18A1, SRPX2, COL8A1, HRG | 0.6518 | ||
| GO_TERM_MF | GO:0005201~extracellular matrix structural constituent | FGA, SRPX2, LUM, TNC, LTBP2, PRELP, MATN2, MUC6 | 1.95 × 10−9 | ||
| KEGG_PATHWAY | hsa04974: Protein digestion and absorption | COL18A1, COL6A2, COL8A1 | 0.1611 | ||
| REACTOME_PATHWAY | R-HSA-216083~Integrin cell surface interactions | FGA, COL18A1, LUM, COL6A2, TNC, COL8A1 | 2.68 × 10−5 | ||
| Comparison of GSEA results between F_RE+ vs. CO groups | |||||
| TCF4, MBNL1, INF2, ITGA6, ADD3, SORBS1, NUMA1, KDR, PPFIBP1, MBNL2, INF2, SOD3, SORBS1, SYNE1, MBNLL1, MBN2, COCH, LUM | GO_TERM_CC | GO:0005938~cell cortex | PPFIBP1, NUMA1, ADD3 | 0.4548 | Transcriptomics |
| GO_TERM_BP | GO:0031589~cell–substrate adhesion | ITGA6, SORBS1 | 1 | ||
| GO_TERM_MF | GO:0045296~cadherin binding | PPFIBP1, KDR, ITGA6 | 0.8334 | ||
| KEGG_PATHWAY | hsa04820: Cytoskeleton in muscle cells | INF2, ITGA6, SYNE1 | 0.1618 | ||
| REACTOME_PATHWAY | R-HSA-216083~Integrin cell surface interactions | LUM, KDR, ITGA6 | 0.1557 | ||
| Comparison of GSEA results between F_RE+ vs. F_RE− groups | |||||
| MBNL1, NUMA1, APBB2, PPFIBP1, INF2, SCARB1, SYNE1, ADD3, MBNL2, TTC7A, ARVCF, TSPOAP1, NDUFV3, IFI44, EXOC1, ITGA6, CLASP1, COPZ2, CD46, CADM1 | GO_TERM_CC | GO:0005938~cell cortex | PPFIBP1, NUMA1, ADD3, CLASP1 | 0.0516 | Transcriptomics |
| GO_TERM_BP | GO:0007010~cytoskeleton organization | ADD3, CLASP1, SYNE1 | 0.9175 | ||
| GO_TERM_MF | GO:0045296~cadherin binding | PPFIBP1, ARVCF, ITGA6, CD46 | 0.1911 | ||
| KEGG_PATHWAY | hsa04820: Cytoskeleton in muscle cells | INF2, ITGA6, SYNE1 | 0.4008 | ||
| REACTOME_PATHWAY | R-HSA-380320~Recruitment of NuMA to mitotic centrosomes | NUMA1, CLASP1 | 1 | ||
| Comparison of GSEA results between CO_RE+ vs. CO groups | |||||
| INF2, NUMA1, SORBS1, SYNE1, MBNL1, MBN2, KDR, FN1, COL4A2, COCH, MSI1 | GOTERM_CC_DIRECT | GO:0062023~collagen-containing extracellular matrix | COL4A2, FN1, COCH | 0.9182 | Transcriptomics |
| GOTERM_BP_DIRECT | GO:0008360~regulation of cell shape | KDR, FN1, COCH | 0.2912 | ||
| GOTERM_MF_DIRECT | GO:0003779~actin binding | INF2, SORBS1, SYNE1 | 0.6847 | ||
| KEGG_PATHWAY | hsa04820: Cytoskeleton in muscle cells | INF2, COL4A2, FN1, SYNE1 | 0.0073 | ||
| REACTOME_PATHWAY | R-HSA-216083~Integrin cell surface interactions | COL4A2, KDR, FN1 | 0.0802 | ||
9. Discussion
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DEG | Differentially expressed gene |
| EMT | Epithelial to mesenchymal transition |
| ECM | Extracellular matrix |
| FECD | Fuchs’ endothelial corneal dystrophy |
| FDR | False discovery rate |
| GSEA | Gene-set enrichment analysis |
| GO | Gene Ontology |
| GEO | Gene-Expression Omnibus |
| GWAS | Genome-wide association study |
| ICH | Immunohistochemistry |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| mRNA | messenger RNA |
| miRNA | micro RNA |
| mtDNA | mitochondrial DNA |
| NGS | Next-generation sequencing |
| PCA | Principal component analysis |
| PCR | Polymerase chain reaction |
| RNA-seq | RNA-sequencing |
| ROS | Reactive oxygen species |
| sncRNA | Small non-coding RNA |
| lncRNA | Long non-coding RNA |
| SNP | Single nucleotide polymorphism |
| TNR | Trinucleotide repeats |
| UVA | Ultraviolet A |
| WES | Whole-exome sequencing |
| WGS | Whole-genome sequencing |
References
- Bonanno, J.A. Molecular mechanisms underlying the corneal endothelial pump. Exp. Eye Res. 2012, 95, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Delmonte, D.W.; Kim, T. Anatomy and physiology of the cornea. J. Cataract Refract. Surg. 2011, 37, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Weisenthal, R.W.; Daly, M.K.; De Freitas, D.; Feder, R.S.; Orlin, S.E.; Tu, E.Y.; Van Meter, W.S.; Verdier, D.D. Structure and function of the external eye and cornea. In 2020–2021 BCSC Section 8: External Disease and Cornea; American Academy of Ophthalmology: San Francisco, CA, USA, 2019; pp. 3–14. [Google Scholar]
- Bourne, W.M.; Nelson, L.R.; Hodge, D.O. Central corneal endothelial cell changes over a ten-year period. Investig. Ophthalmol. Vis. Sci. 1997, 38, 779–782. [Google Scholar] [CrossRef]
- Yee, R.W.; Matsuda, M.; Schultz, R.O.; Edelhauser, H.F.; Yee, R.W.; Matsuda, M.; Schultz, R.O.; Edelhauser, H.F. Changes in the normal corneal endothelial cellular pattern as a function of age. Curr. Eye Res. 1985, 4, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Price, M.O.; Mehta, J.S.; Jurkunas, U.V.; Price, F.W. Corneal endothelial dysfunction: Evolving understanding and treatment options. Prog. Retin. Eye Res. 2021, 82, 100904. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Cho, K.; Srikumaran, D. Fuchs dystrophy and cataract: Diagnosis, evaluation and treatment. Ophthalmol. Ther. 2023, 12, 691–704. [Google Scholar] [CrossRef]
- Li, Y.; Minear, M.A.; Qin, X.; Rimmler, J.; Hauser, M.A.; Allingham, R.R.; Igo, R.P.; Lass, J.H.; Iyengar, S.K.; Klintworth, G.K.; et al. Mitochondrial polymorphism A10398G and haplogroup I are associated with Fuchs’ endothelial corneal dystrophy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 4577–4584. [Google Scholar] [CrossRef][Green Version]
- Liu, C.; Miyajima, T.; Melangath, G.; Miyai, T.; Vasanth, S.; Deshpande, N.; Kumara, V.; Tonea, S.O.; Guptaa, R.; Zhua, S.; et al. Ultraviolet A light induces DNA damage and estrogen-DNA adducts in Fuchs endothelial corneal dystrophy causing females to be more affected. Proc. Natl. Acad. Sci. USA 2020, 117, 573–583. [Google Scholar] [CrossRef]
- Kumar, V.; Deshpande, N.; Parekh, M.; Wong, R.; Ashraf, S.; Zahid, M.; Hui, H.; Miall, A.; Kimpton, S.; Price, M.O.; et al. Estrogen genotoxicity causes preferential development of Fuchs endothelial corneal dystrophy in females. Redox Biol. 2023, 10, 102986. [Google Scholar] [CrossRef]
- Zwingelberg, S.B.; Lautwein, B.; Baar, T.; Gutenbrunner, M.H.; Von Brandenstein, M.; Nobacht, S.; Matthaei, M.; Bachmann, B.O. The influence of obesity, diabetes mellitus and smoking on fuchs endothelial corneal dystrophy (FECD). Sci. Rep. 2024, 14, 11596. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Kelliher, C.; Spitze, A.R.; Speck, C.L.; Eberhart, C.G.; Jun, A.S. Unfolded protein response in Fuchs endothelial corneal dystrophy: A unifying pathogenic pathway? Am. J. Ophthalmol. 2010, 149, 194. [Google Scholar] [CrossRef]
- Iliff, B.W.; Riazuddin, S.A.; Gottsch, J.D. The genetics of Fuchs’ corneal dystrophy. Expert Rev. Ophthalmol. 2012, 7, 363–375. [Google Scholar] [CrossRef]
- Jurkunas, U.V.; Bitar, M.S. Evidence of oxidative stress in the pathogenesis of Fuchs endothelial corneal dystrophy. Am. J. Pathol. 2010, 177, 2278–2289. [Google Scholar] [CrossRef] [PubMed]
- Matthaeia, M.; Zhua, A.Y.; Kallaya, L.; Eberharta, C.G.; Cursiefenb, C.; Jun, A.S. Transcript profile of cellular senescence-related genes in Fuchs endothelial corneal dystrophy. Exp. Eye Res. 2014, 129, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Méthot, S.J.; Proulx, S.; Brunette, I.; Rochette, P.J. Chronology of cellular events related to mitochondrial burnout leading to cell death in Fuchs endothelial corneal dystrophy. Sci. Rep. 2020, 10, 5811. [Google Scholar] [CrossRef] [PubMed]
- Tone, S.O.; Kocaba, V.; Böhm, M.; Wylegala, A.; White, T.L.; Jurkunas, U.V. Fuchs endothelial corneal dystrophy: The vicious cycle of Fuchs pathogenesis. Prog. Retin. Eye Res. 2021, 80, 100863. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, K.; Sasaki, H.; Shui, Y.; Chewc, S.J.; Chenga, H.-M.; Onof, M.; Morikawab, Y.; Sasaki, K. Prevalence of primary cornea guttata and morphology of corneal endothelium in aging Japanese and Singaporean subjects. Ophthalmic Res. 2002, 34, 135–138. [Google Scholar] [CrossRef]
- Zoega, G.M.; Fujisawa, A.; Sasaki, H.; Kubota, A.; Sasaki, K.; Kitagawa, K.; Jonasson, F. Prevalence and risk factors for cornea guttata in the Reykjavik eye study. Ophthalmology 2006, 113, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Eghrari, A.O.; McGlumphy, E.J.; Iliff, B.W.; Wang, J.; Emmert, D.; Riazuddin, S.A.; Katsanis, N.; Gottsch, J.D. Prevalence and severity of Fuchs corneal dystrophy in Tangier island. Am. J. Ophthalmol. 2012, 153, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Shen, L. Advances and trends in omics technology development. Front. Med. 2022, 1, 911861. [Google Scholar] [CrossRef]
- Moore, J.H.; Asselbergs, F.W.; Williams, S.M. Bioinformatics challenges for genome-wide association studies. Bioinformatics 2010, 26, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-cabrero, D.; Cervera, A.; Mcpherson, A.; Szcze, W.; Gaffney, D.J.; Elo, L.L.; Zhang, X. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Lauwen, S.; De Jong, E.K.; Lefeber, D.J.; Hollander, A.I. Den Omics biomarkers in ophthalmology. Investig. Ophthalmol. Vis. Sci. 2017, 58, BIO88–BIO98. [Google Scholar] [CrossRef]
- Altamirano, F.; Ortiz, G.; Connor, M.A.O.; Juan, C.; Sancén, P.; Zavala, J.; Valdez, J.E. Fuchs endothelial corneal dystrophy: An updated review. Int. Ophthalmol. 2024, 44, 61. [Google Scholar] [CrossRef]
- Kannabiran, C.; Chaurasia, S.; Ramappa, M.; Mootha, V.V. Update on the genetics of corneal endothelial dystrophies. Indian J. Ophthalmol. 2022, 70, 2239–2248. [Google Scholar] [CrossRef]
- Zhang, J.; Patel, D.V. The pathophysiology of Fuchs’ endothelial dystrophy—A review of molecular and cellular insights. Exp. Eye Res. 2015, 130, 97–105. [Google Scholar] [CrossRef]
- Gottsch, J.D.; Sundin, O.H.; Liu, S.H.; Jun, A.S.; Broman, K.W.; Stark, W.J.; Vito, E.C.L.; Narang, A.K.; Thompson, J.M.; Magovern, M. Inheritance of a novel COL8A2 mutation defines a distinct early-onset subtype of Fuchs corneal dystrophy. Investig. Ophthalmol. Vis. Sci. 2005, 46, 1934–1939. [Google Scholar] [CrossRef] [PubMed]
- Sundin, O.H.; Jun, A.S.; Broman, K.W.; Liu, S.H.; Sheehan, S.E.; Vito, E.C.L.; Stark, W.J.; Gottsch, J.D. Linkage of late-onset Fuchs corneal dystrophy to a novel locus at 13pTel-13q12.13. Investig. Ophthalmol. Vis. Sci. 2006, 47, 140–145. [Google Scholar] [CrossRef]
- Sundin, O.H.; Broman, K.W.; Chang, H.H.; Vito, E.C.L.; Stark, W.J.; Gottsch, J.D. A common locus for late-onset Fuchs corneal dystrophy maps to 18q21.2-q21.32. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3919–3926. [Google Scholar] [CrossRef] [PubMed]
- Riazuddin, S.A.; Eghrari, A.O.; Al-saif, A.; Davey, L.; Meadows, D.N.; Katsanis, N.; Gottsch, J.D. Linkage of a mild late-onset phenotype of Fuchs corneal dystrophy to a novel locus at 5q33.1-q35.2. Investig. Ophthalmol. Vis. Sci. 2009, 50, 5667–5671. [Google Scholar] [CrossRef] [PubMed]
- Afshari, N.A.; Li, Y.; Pericak-Vance, M.A.; Gregory, S.; Klintworth, G.K. Genome-wide linkage scan in Fuchs endothelial corneal dystrophy. Investig. Ophthalmol. Vis. Sci. 2009, 50, 1093–1097. [Google Scholar] [CrossRef]
- Riazuddin, S.A.; Vasanth, S.; Katsanis, N.; Gottsch, J.D. Mutations in AGBL1 cause dominant late-onset Fuchs corneal dystrophy and alter protein-protein interaction with TCF4. Am. J. Hum. Genet. 2013, 93, 758–764. [Google Scholar] [CrossRef]
- Jiang, X.; Jin, X.; Zhang, N.; Zhang, H. A family of fuchs endothelial corneal dystrophy and anterior polar cataract with an analysis of whole exome sequencing. Ophthalmic Genet. 2020, 41, 263–270. [Google Scholar] [CrossRef]
- Petersen, B.; Fredrich, B.; Hoeppner, M.P.; Ellinghaus, D.; Franke, A. Opportunities and challenges of whole-genome and -exome sequencing. BMC Genet. 2017, 18, 14. [Google Scholar] [CrossRef] [PubMed]
- Baratz, K.H.; Tosakulwong, N.; Ryu, E.; Brown, W.L.; Branham, K.; Chen, W.; Tran, K.D.; Schmid-Kubista, K.E.; Heckenlively, J.R.; Swaroop, A.; et al. E2-2 protein and Fuchs’s corneal dystrophy. N. Engl. J. Med. 2010, 363, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Minear, M.A.; Rimmler, J.; Zhao, B.; Balajonda, E.; Hauser, M.A.; Allingham, R.R.; Eghrari, A.O.; Riazuddin, S.A.; Katsanis, N.; et al. Replication of TCF4 through association and linkage studies in late-onset Fuchs endothelial corneal dystrophy. PLoS ONE 2011, 6, e18044. [Google Scholar] [CrossRef] [PubMed]
- Afshari, N.A.; Igo, R.P., Jr.; Morris, N.J.; Stambolian, D.; Sharma, S.; Pulagam, V.L.; Dunn, S.; Stamler, J.F.; Truitt, B.J.; Rimmler, J.; et al. Genome-wide association study identifies three novel loci in Fuchs endothelial corneal dystrophy. Nat. Commun. 2017, 30, 14898. [Google Scholar] [CrossRef]
- Biswas, S.; Munier, F.L.; Yardley, J.; Hart-holden, N.; Perveen, R.; Cousin, P.; Sutphin, J.E.; Noble, B.; Batterbury, M.; Kielty, C.; et al. Missense mutations in COL8A2, the gene encoding the α2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum. Mol. Genet. 2001, 10, 2415–2423. [Google Scholar] [CrossRef]
- Igo, R.P., Jr.; Kopplin, L.J.; Joseph, P.; Truitt, B.; Fondran, J.; Bardenstein, D.; Aldave, A.J.; Croasdale, C.R.; Price, M.O.; Rosenwasser, M.; et al. Differing roles for TCF4 and COL8A2 in central corneal thickness and Fuchs endothelial corneal dystrophy. PLoS ONE 2012, 7, e46742. [Google Scholar] [CrossRef] [PubMed]
- Synowiec, E.; Wojcik, K.A.; Izdebska, J.; Blasiak, J.; Szaflik, J.; Szaflik, J.P. Polymorphisms of the apoptosis-related FAS and FAS ligand genes in keratoconus and Fuchs endothelial corneal dystrophy. Tohoku J. Exp. Med. 2014, 234, 17–27. [Google Scholar] [CrossRef]
- Mootha, V.V.; Gong, X.; Ku, H.; Xing, C. Association and familial segregation of CTG18.1 trinucleotide repeat Expansion of TCF4 gene in Fuchs’ endothelial corneal dystrophy. Investig. Ophthalmol. Vis. Sci. 2014, 2, 33–42. [Google Scholar] [CrossRef]
- Polakowski, P. Variation in DNA base excision repair genes in Fuchs endothelial corneal dystrophy. Med. Sci. Monit. 2015, 21, 2809–2827. [Google Scholar] [CrossRef]
- Chaurasia, S.; Ramappa, M.; Annapurna, M.; Kannabiran, C. Coexistence of congenital hereditary endothelial dystrophy and Fuchs endothelial corneal dystrophy associated with SLC4A11 mutations in affected families. Cornea 2020, 39, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Viberg, A.; Westin, I.M.; Golovleva, I.; Bystrom, B. TCF4 trinucleotide repeat expansion in Swedish cases with Fuchs’ endothelial corneal dystrophy. Acta Ophthalmol. 2022, 100, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Tsedilina, T.R.; Sharova, E.; Iakovets, V.; Skorodumova, O.L. Systematic review of SLC4A11, variants in the development of Fuchs’ endothelial corneal dystrophy. Front. Med. 2023, 27, 1153122. [Google Scholar] [CrossRef]
- Kobayashi, A.; Fujiki, K.; Murakami, A.; Kato, T.; Chen, L.; Onoe, H.; Nakayasu, K.; Sakurai, M.; Takahashi, M.; Sugiyama, K.; et al. Analysis of COL8A2 gene mutation in Japanese patients with Fuchs’ endothelial dystrophy and posterior polymorphous dystrophy. Jpn. J. Ophthalmol. 2004, 48, 195–198. [Google Scholar] [CrossRef]
- Hemadevi, B.; Srinivasan, M.; Arunkumar, J.; Prajna, N.V. Genetic analysis of patients with Fuchs endothelial corneal dystrophy in India. BMC Ophthalmol. 2010, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Okumura, N.; Nakagawa, H.; Koizumi, N.; Ikeda, Y.; Ueno, M.; Yoshii, K.; Adachi, H.; Aleff, R.A.; Butz, M.L.; et al. Trinucleotide repeat expansion in the TCF4 gene in Fuchs’ endothelial corneal dystrophy in Japanese. Investig. Ophthalmol. Vis. Sci. 2015, 56, 4865–4869. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Zhang, W.; Yan, X.; Wang, L.; Dong, H.; Shou, T.; Lei, H.; Guo, Q. Analysis of SLC4A11, ZEB1, LOXHD1, COL8A2 and TCF4 gene sequences in a multi-generational family with late-onset Fuchs corneal dystrophy. Int. J. Mol. Med. 2016, 37, 1487–1500. [Google Scholar] [CrossRef][Green Version]
- Rao, B.S.; Ansar, S.; Arokiasamy, T.; Rachapalli, R.; Umashankar, V.; Rajagopal, R.; Soumittra, N. Analysis of candidate genes ZEB1 and LOXHD1 in late-onset Fuchs’ endothelial corneal dystrophy in an Indian cohort. Ophthalmic Genet. 2018, 39, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Wesdorp, M.; Schreur, V.; Beynon, A.J.; Oostrik, J.; van de Kamp, J.M.; Elting, M.W.; van den Boogaard, M.-J.H.; Feenstra, I.; Admiraal, R.J.C.; Kunst, H.P.M.; et al. Further audiovestibular characterization of DFNB77, caused by deleterious variants in LOXHD1, and investigation into the involvement of Fuchs corneal dystrophy. Clin. Genet. 2018, 94, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Okumura, N.; Hayashi, R.; Nakano, M.; Tashiro, K.; Yoshii, K.; Aleff, R.; Butz, M.; Highsmith, E.W.; Wieben, E.D.; Fautsch, M.P.; et al. Association of rs613872 and trinucleotide repeat expansion in the TCF4 gene of German patients with Fuchs endothelial corneal dystrophy. Cornea 2019, 38, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Moschos, M.M.; Diamantopoulou, A.; Gouliopoulos, N.; Droutsas, K.; Bagli, E.; Chatzistefanou, K.; Kitsos, G.; Kroupis, C. TCF4 and COL8A2 gene polymorphism screening in a Greek population of late-onset Fuchs endothelial corneal dystrophy. In Vivo 2019, 33, 963–971. [Google Scholar] [CrossRef]
- Stamler, J.F.; Roos, B.R.; Wagoner, M.D.; Goins, K.M.; Kitzmann, A.S.; Riley, J.B.; Stone, E.M.; Fingert, J.H. Confirmation of the association between the TCF4 risk allele and Fuchs endothelial corneal dystrophy in patients from the Midwestern United States. Ophthalmic Genet. 2013, 34, 32–34. [Google Scholar] [CrossRef] [PubMed]
- Kuot, A.; Hewitt, A.W.; Snibson, G.R.; Souzeau, E.; Mills, R.; Craig, J.E.; Burdon, K.P.; Sharma, S. TGC repeat expansion in the TCF4 gene increases the risk of Fuchs’ endothelial corneal dystrophy in Australian cases. PLoS ONE 2017, 23, e0183719. [Google Scholar] [CrossRef] [PubMed]
- Thalamuthu, A.; Khor, C.C.; Venkataraman, D.; Koh, L.W.; Tan, D.T.H.; Aung, T.; Mehta, J.S.; Vithana, E.N. Association of TCF4 gene polymorphisms with Fuchs’ corneal dystrophy in the Chinese. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5573–5578. [Google Scholar] [CrossRef] [PubMed]
- Okumura, N.; Puangsricharern, V.; Jindasak, R.; Koizumi, N.; Komori, Y.; Ryousuke, H.; Nakahara, M.; Masakazu, N.; Hiroko, A.; Tashiro, K.; et al. Trinucleotide repeat expansion in the transcription factor 4 (TCF4) gene in Thai patients with Fuchs endothelial corneal dystrophy. Eye 2020, 34, 880–885. [Google Scholar] [CrossRef]
- Wieben, E.D.; Aleff, R.A.; Tosakulwong, N.; Butz, M.L.; Highsmith, W.E.; Edwards, A.O.; Baratz, K.H. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS ONE 2012, 7, e49083. [Google Scholar] [CrossRef] [PubMed]
- Eghrari, A.O.; Vasanth, S.; Wang, J.; Riazuddin, S.A.; Gottsch, J.D. CTG18.1 expansion in TCF4 increases likelihood of transplantation in Fuchs corneal dystrophy. Cornea 2017, 36, 40–43. [Google Scholar] [CrossRef]
- Peachey, N.; Gorman, B.; Francis, M.; Nealon, C.; Halladay, C.; Duro, N.; Markianos, K.; Genovese, G.; Hysi, P.; Choquet, H.; et al. Multi-ancestry GWAS of Fuchs corneal dystrophy highlights roles of laminins, collagen, and endothelial cell regulation. Res. Sq. 2023, rs.3, rs-2762003. [Google Scholar]
- Czarny, P.; Seda, A.; Wielgorski, M.; Binczyk, E.; Markiewicz, B.; Kasprzak, E.; Jiménez-garcía, M.P.; Grabska-liberek, I.; Pawlowska, E.; Blasiak, J.; et al. Mutagenesis of mitochondrial DNA in Fuchs endothelial corneal dystrophy. Mutat. Res. 2014, 760, 42–47. [Google Scholar] [CrossRef]
- Pathak, D.; Nayak, B.; Singh, M.; Sharma, N.; Tandon, R.; Sinha, R.; Titiyal, J.S.; Dada, R. Mitochondrial complex 1 gene analysis in keratoconus. Mol. Vis. 2011, 17, 1514–1525. [Google Scholar] [PubMed]
- Kenney, M.C.; Hertzog, D.; Chak, G.; Atilano, S.R.; Khatibi, N.; Soe, K.; Nobe, A.; Yang, E.; Chwa, M.; Zhu, F.; et al. Mitochondrial DNA haplogroups confer differences in risk for age-related macular degeneration: A case control study. BMC Med. Genet. 2013, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Abu-Amero, K.K.; González, A.M.; Osman, E.A.; Larruga, J.M.; Cabrera, V.M. Mitochondrial DNA lineages of African origin confer susceptibility to primary open-angle glaucoma in Saudi patients. Mol. Vis. 2011, 3, 1468–1472. [Google Scholar]
- Chakraborty, M.; Jandhyam, H.; Kumar, S.; Das, S.; Alone, P.D. Intergenic variants, rs1200114 and rs1200108 are genetically associated along with a decreased ATP1B1 expression in Fuchs endothelial corneal dystrophy. Exp. Eye Res. 2023, 228, 109403. [Google Scholar] [CrossRef] [PubMed]
- Eghrari, A.O.; Vasanth, S.; Gapsis, B.C.; Bison, H.; Riazuddin, S.A.; Gottsch, J.D. Identification of a novel TCF4 isoform in human corneal endothelium. Cornea 2018, 37, 899–903. [Google Scholar] [CrossRef]
- Bhattacharyya, N.; Chai, N.; Hafford-tear, N.J.; Sadan, A.N.; Szabo, A.; Zarouchlioti, C.; Jedlickova, J.; Leung, S.K.; Liao, T.; Dudakova, L.; et al. Deciphering novel TCF4 -driven mechanisms underlying a common triplet repeat expansion-mediated disease. PLoS Genet. 2024, 20, e1011230. [Google Scholar] [CrossRef]
- Zhang, X.; Kumar, A.; Sathe, A.A.; Mootha, V.V.; Xing, C. Transcriptomic meta-analysis reveals ERRα-mediated oxidative phosphorylation is downregulated in Fuchs’ endothelial corneal dystrophy. PLoS ONE 2023, 18, e0295542. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Aleff, R.A.; Soragni, E.; Kalari, K.; Nie, J.; Tang, X.; Davila, J.; Kocher, J.; Patel, S.V.; Gottesfeld, J.M.; et al. RNA Toxicity and missplicing in the common eye disease Fuchs endothelial corneal dystrophy. J. Biol. Chem. 2015, 290, 5979–5990. [Google Scholar] [CrossRef] [PubMed]
- Wieben, E.D.; Aleff, R.A.; Tang, X.; Butz, M.L.; Kalari, K.R.; Edward, W.; Jen, J.; Vasmatzis, G.; Patel, S.V.; Maguire, L.J.; et al. Trinucleotide repeat expansion in the transcription factor 4 (TCF4) gene leads to widespread mRNA splicing changes in Fuchs’ endothelial corneal dystrophy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Wieben, E.D.; Aleff, R.A.; Tang, X.; Kalari, K.R.; Maguire, L.J.; Patel, V.; Baratz, K.H.; Fautsch, M.P. Gene expression in the corneal endothelium of Fuchs endothelial corneal dystrophy patients with and without expansion of a trinucleotide repeat in TCF4. PLoS ONE 2018, 13, e0200005. [Google Scholar] [CrossRef]
- Chu, Y.; Hu, J.; Liang, H.; Kanchwala, M.; Xing, C.; Beebe, W.; Bowman, C.B.; Gong, X.; Corey, D.R.; Mootha, V.V. Analyzing pre-symptomatic tissue to gain insights into the molecular and mechanistic origins of late-onset degenerative trinucleotide repeat disease. Nucleic Acids Res. 2020, 48, 6740–6758. [Google Scholar] [CrossRef] [PubMed]
- Kuot, A.; Corbett, M.A.; Mills, R.A.; Snibson, G.; Wiffen, S.; Loh, R.; Burdon, K.P.; Craig, J.E.; Sharma, S. Differential gene expression analysis of corneal endothelium indicates involvement of phagocytic activity in Fuchs’ endothelial corneal dystrophy. Exp. Eye Res. 2021, 210, 108692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dai, Y.; Li, Y.; Xu, J. Integrative analysis of gene expression datasets in corneal endothelium samples of Fuchs endothelial corneal dystrophy. Exp. Eye Res. 2023, 237, 109712. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Tokuda, Y.; Nakano, M.; Komori, Y.; Hanada, N.; Tourtas, T.; Schrehardt, U.S.; Kruse, F.; Tashiro, K.; Koizumi, N.; et al. RNA-Seq–based transcriptome analysis of corneal endothelial cells derived from patients with Fuchs endothelial corneal dystrophy. Sci. Rep. 2023, 13, 8647. [Google Scholar] [CrossRef]
- Nikitina, A.S.; Belodedova, A.V.; Malyugin, B.E.; Sharova, E.I.; Kostryukova, E.S.; Larin, A.K.; Veselovsky, V.A.; Antonova, O.P.; Skorodumova, L.O. Dataset on transcriptome profiling of corneal endothelium from patients with Fuchs endothelial corneal dystrophy. Data Brief 2019, 25, 104047. [Google Scholar] [CrossRef]
- Liu, C.; Gao, Z.; Li, J.; Zhou, Q. Identification of novel therapeutic targets for Fuchs’ endothelial corneal dystrophy based on gene bioinformatics analysis. PLoS ONE 2022, 17, e0264018. [Google Scholar] [CrossRef] [PubMed]
- De Roo, A.; Wouters, J.; Govaere, O.; Foets, B.; Van den Oord, J.J. Identification of circulating fibrocytes and dendritic derivatives in corneal endothelium of patients with Fuchs’ dystrophy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 670–681. [Google Scholar] [CrossRef][Green Version]
- De Roo, A.; Janssens, T.; Foets, B.; Van den Oord, J. Immunohistochemical profiling of corneas with Fuchs endothelial corneal dystrophy. Cornea 2017, 36, 866–874. [Google Scholar] [CrossRef]
- Cui, Z.; Zeng, Q.; Guo, Y.; Liu, S.; Wang, P.; Xie, M.; Chen, J. Pathological molecular mechanism of symptomatic late-onset Fuchs endothelial corneal dystrophy by bioinformatic analysis. PLoS ONE 2018, 13, e0197750. [Google Scholar] [CrossRef]
- Wen, H.; Gallo, R.A.; Huang, X.; Cai, J.; Mei, S.; Farooqi, A.A.; Zhao, J.; Tao, W. Incorporating differential gene expression analysis with predictive biomarkers to identify novel therapeutic drugs for Fuchs endothelial corneal dystrophy. J. Ophthalmol. 2021, 28, 5580595. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Nakagawa, T.; Yuasa, T.; Tokuda, Y.; Nakano, M.; Tashiro, K.; Tourtas, T.; Schlötzer-schrehardt, U.; Kruse, F.; Yamamoto, K.; et al. Dysregulation of the TCF4 isoform in corneal endothelial cells of patients with Fuchs endothelial corneal dystrophy. Investig. Ophthalmol. Vis. Sci. 2024, 65, 27. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Dou, S.; Zhang, B.; Jiang, H.; Qi, X.; Duan, H.; Wang, X.; Dong, C.; Zhang, B.N.; Xie, L.; et al. Heterogeneity of human corneal endothelium implicates lncRNA NEAT1 in Fuchs endothelial corneal dystrophy. Mol. Ther. Nucleic Acid 2022, 27, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Barrett, T. The Gene Expression Omnibus database. In Statistical Genomics; Methods in Molecular Biology; Humana: New York, NY, USA, 2016; pp. 93–110. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA modifications in gene expression regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Pukl Stunf, S. MicroRNA of epithelial to mesenchymal transition in Fuchs’ endothelial corneal dystrophy. Genes 2022, 13, 1711. [Google Scholar] [CrossRef] [PubMed]
- Matthaei, M.; Hu, J.; Kallay, L.; Eberhart, C.G.; Cursiefen, C.; Qian, J.; Lackner, E.; Jun, A.S. Endothelial cell microRNA expression in human late-onset Fuchs’ dystrophy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Khuc, E.; Bainer, R.; Wolf, M.; Clay, S.M.; Weisenberger, D.J.; Kemmer, J.; Weaver, V.M.; Hwang, D.G.; Chan, M.F. Comprehensive characterization of DNA methylation changes in Fuchs endothelial corneal dystrophy. PLoS ONE 2017, 16, e0175112. [Google Scholar] [CrossRef]
- Pan, P.; Weisenberger, D.J.; Zheng, S.; Wolf, M.; Hwang, D.G.; Rose-nussbaumer, J.R.; Jurkunas, U.V.; Chan, M.F. Aberrant DNA methylation of miRNAs in Fuchs endothelial corneal dystrophy. Sci. Rep. 2019, 9, 16385. [Google Scholar] [CrossRef] [PubMed]
- Westin, I.M.; Landfors, M.; Giannopoulos, A.; Viberg, A.; Osterman, P.; Bystrom, B.; Degerman, S.; Golovleva, I. DNA methylation changes and increased mRNA expression of coagulation proteins, factor V and thrombomodulin in Fuchs endothelial corneal dystrophy. Cell. Mol. Life Sci. 2023, 80, 62. [Google Scholar] [CrossRef]
- Soragni, E.; Petrosyan, L.; Rinkoski, T.A.; Wieben, E.D.; Baratz, K.H.; Fautsch, M.P.; Gottesfeld, J.M. Repeat-associated non-ATG (RAN) translation in Fuchs’ endothelial corneal dystrophy. Investig. Ophthalmol. Vis. Sci. 2018, 59, 1888–1896. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.R.; Segu, Z.M.; Price, M.O.; Lai, X.; Witzmann, F.A.; Mechref, Y.; Yoder, M.C.; Price, F.W. Alterations in the aqueous humor proteome in patients with Fuchs endothelial corneal dystrophy. Mol. Vis. 2010, 11, 2376–2383. [Google Scholar]
- Kuot, A.; Ronci, M.; Mills, R.; Klebe, S.; Snibson, G.; Wiffen, S.; Loh, R.; Corbett, M.; Zhou, T.; Chataway, T.; et al. Reduced expression of apolipoprotein E and immunoglobulin heavy constant gamma 1 proteins in Fuchs endothelial corneal dystrophy. Clin. Exp. Ophthalmol. 2019, 47, 1028–1042. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Okumura, N.; Ikegawa, M.; Toyama, Y.; Nirasawa, T.; Mascarelli, F.; Vaitinadapoule, H.; Aouimeur, I.; He, Z.; Gain, P.; et al. Shotgun proteomics identification of proteins expressed in the Descemet’ s membrane of patients with Fuchs endothelial corneal dystrophy. Sci. Rep. 2023, 13, 10401. [Google Scholar] [CrossRef] [PubMed]
- Cabrerizo, J.; Urcola, J.A.; Vecino, E.; Melles, G. Changes in lipidomic profile of aqueous humour in Fuchs endothelial dystrophy. Acta Ophthalmol. 2017, 95, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, W.N.; Blitzer, M.; Ph, D.; Insler, M.S. Aqueous amino acid levels in Fuchs’ corneal dystrophy. Am. J. Ophthalmol. 1986, 102, 570–574. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Muir, K. Awareness and attitudes toward corneal donation: Challenges and opportunities. Clin. Ophthalmol. 2018, 7, 1049–1059. [Google Scholar]
- Numa, K.; Imai, K.; Ueno, M.; Kitazawa, K.; Tanaka, H.; Bush, J.D.; Teramukai, S.; Okumura, N.; Koizumi, N.; Hamuro, J.; et al. Five-year follow-up of first 11 patients undergoing injection of cultured corneal endothelial cells for corneal endothelial failure. Ophthalmology 2021, 128, 504–514. [Google Scholar] [CrossRef]
- Kim, E.C.; Meng, H.; Jun, A.S. N-Acetylcysteine increases corneal endothelial cell survival in a mouse model of Fuchs endothelial corneal dystrophy. Exp. Eye Res. 2014, 127, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Angelbello, A.J.; Benhamou, R.I.; Rzuczek, S.G.; Tang, Z.; Chen, J.L.; Roy, M.; Wang, K.W.; Jun, A.S.; Thornton, C.A.; Disney, M.D. A small molecule that binds an RNA repeat expansion stimulates its decay via the exosome complex. Cell Chem. Biol. 2021, 28, 34–45.e6. [Google Scholar] [CrossRef] [PubMed]
- Chau, V.Q.; Hu, J.; Gong, X.; Hulleman, J.D.; Ufret-vincenty, R.L.; Rigo, F.; Prakash, T.P.; Corey, D.R.; Mootha, V.V. Delivery of antisense oligonucleotides to the cornea. Nucleic Acid Ther. 2020, 30, 207–214. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prašnikar, E.; Stunf Pukl, S. How “Omics” Studies Contribute to a Better Understanding of Fuchs’ Endothelial Corneal Dystrophy. Curr. Issues Mol. Biol. 2025, 47, 135. https://doi.org/10.3390/cimb47030135
Prašnikar E, Stunf Pukl S. How “Omics” Studies Contribute to a Better Understanding of Fuchs’ Endothelial Corneal Dystrophy. Current Issues in Molecular Biology. 2025; 47(3):135. https://doi.org/10.3390/cimb47030135
Chicago/Turabian StylePrašnikar, Erika, and Spela Stunf Pukl. 2025. "How “Omics” Studies Contribute to a Better Understanding of Fuchs’ Endothelial Corneal Dystrophy" Current Issues in Molecular Biology 47, no. 3: 135. https://doi.org/10.3390/cimb47030135
APA StylePrašnikar, E., & Stunf Pukl, S. (2025). How “Omics” Studies Contribute to a Better Understanding of Fuchs’ Endothelial Corneal Dystrophy. Current Issues in Molecular Biology, 47(3), 135. https://doi.org/10.3390/cimb47030135







