Signaling Mechanism of Budding, Proliferation, and Tissue Regeneration in Cnidaria
Abstract
1. Introduction
1.1. Cnidarian Life Cycle Overview
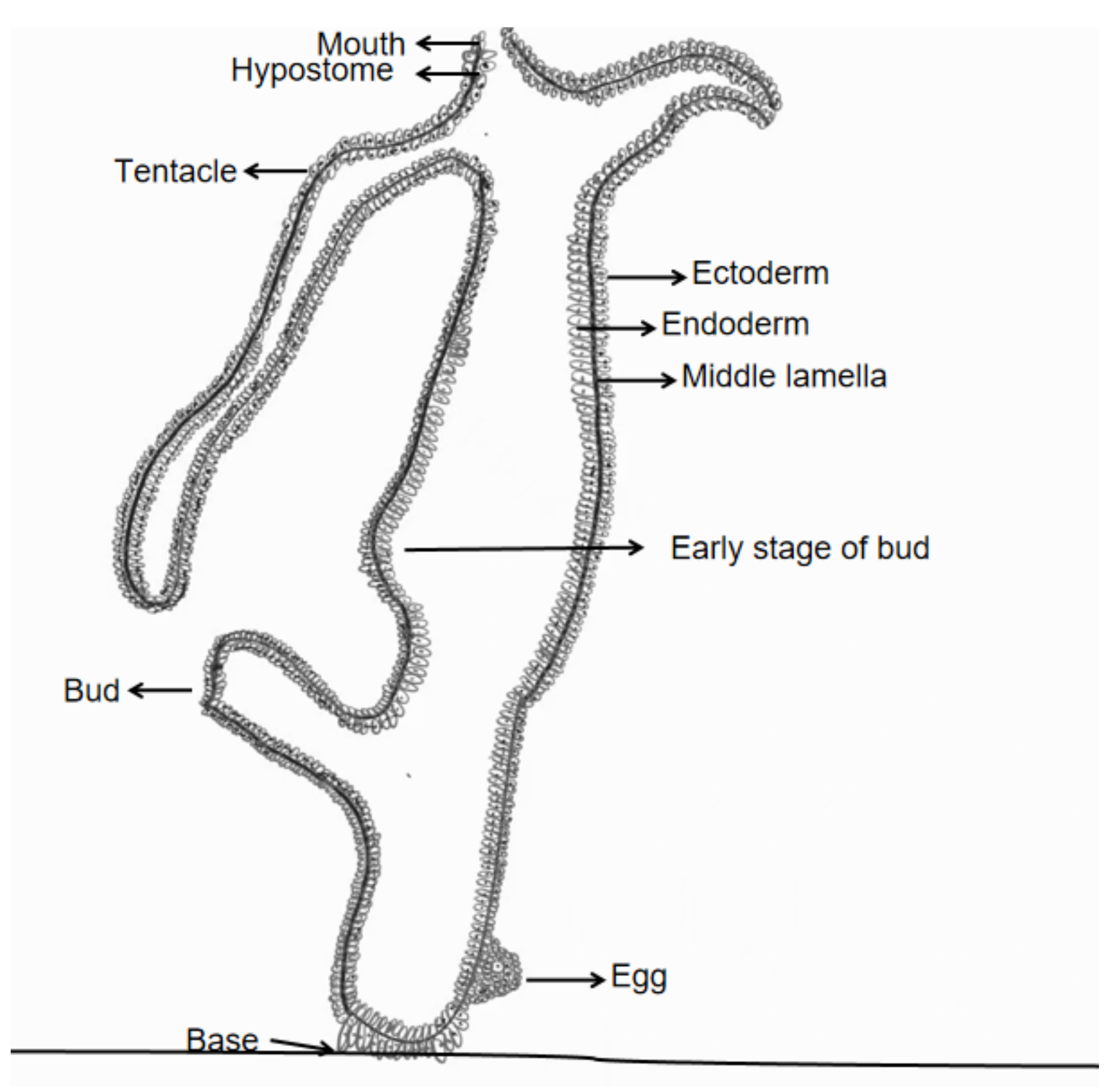
1.2. Budding Mechanisms in Hydrozoans
1.3. Species-Specific Variations in Budding
1.4. Genomic and Evolutionary Insights
1.5. Ecological Impacts of Cnidarian Proliferation
1.6. Signaling Pathways in Budding and Regeneration
1.7. Hydra as a Model Organism
2. Common Methods for Studying Budding and Tissue Regeneration in Cnidaria
3. Influence Factors on the Budding and Proliferation
3.1. Feeding Level
3.2. Temperature
3.3. Salinity
3.4. Other Factors
4. Cell Types and Their Roles Involved in Budding and Proliferation of Prickly Cell Animals
4.1. Epithelial Muscle Cells
4.2. Cnidoblasts
4.3. Neural Cells
4.4. Germ Cells
4.5. Glandular Cells
5. Signaling Pathways and Their Effects on the Proliferation and Sprouting of Prickly Cell Animals
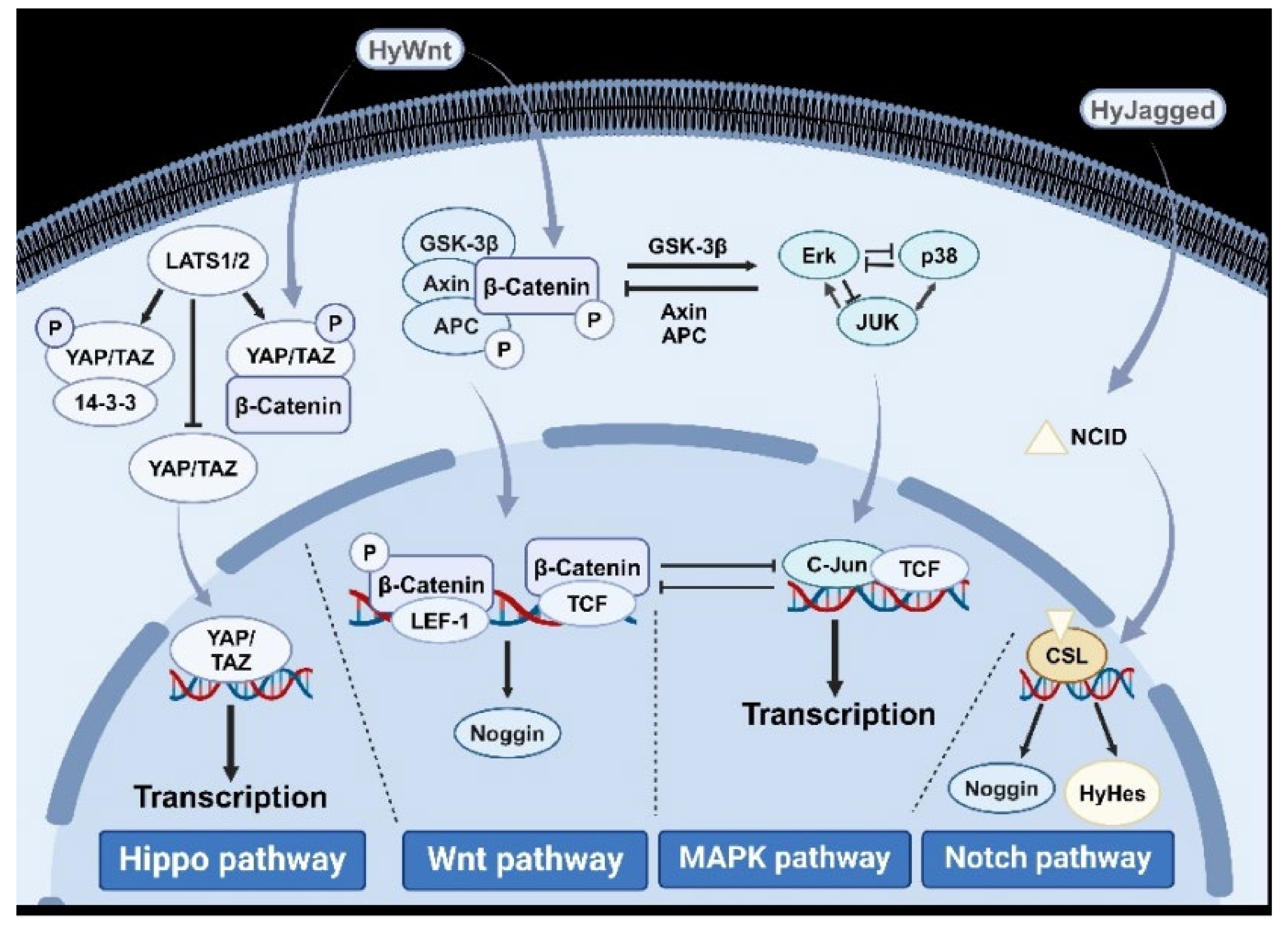
5.1. Wnt/β-Catenin Signaling Pathway
5.2. Hippo Signaling Pathway
5.3. Mitogen-Activated Protein Kinase (MAPK) Signaling Pathway
5.4. PI3K Signaling Pathway
5.5. JNK Signaling Pathway
5.6. ERK Signaling Pathway
5.7. Protein Kinase C (PKC) Signaling Pathway
5.8. FGFR Signaling Pathway
5.9. Bone Morphogenetic Protein (BMP) Signaling Pathway
5.10. Notch Signaling Pathway
5.11. Vascular Endothelial Growth Factor (VEGF)/FGF Signaling Pathway
6. Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Feng, H.; Lv, S.; Li, R.; Shi, J.; Wang, J.; Cao, P. Mitochondrial genome comparison reveals the evolution of cnidarians. Ecol. Evol. 2023, 13, e10157. [Google Scholar]
- Skokan, T.D.; Hobmayer, B.; McKinley, K.L.; Vale, R.D. Mechanical stretch regulates macropinocytosis in Hydra vulgaris. Mol. Biol. Cell 2024, 35, br9. [Google Scholar]
- Holstein, T.W. The Hydra stem cell system—Revisited. Cells Dev. 2023, 174, 203846. [Google Scholar] [PubMed]
- MacWilliams, H.K. Hydra transplantation phenomena and the mechanism of Hydra head regeneration. II. Properties of the head activation. Dev. Biol. 1983, 96, 239–257. [Google Scholar] [PubMed]
- Schnedler-Meyer, N.A.; Kiørboe, T.; Mariani, P. Boom and Bust: Life History, Environmental Noise, and the (un)Predictability of Jellyfish Blooms. Front. Mar. Sci. 2018, 5, 257. [Google Scholar]
- Tang, C.; Sun, S.; Zhang, F. Intraguild predation by polyps of three scyphozoan jellyfish: Nemopilema nomurai, Aurelia coerulea, and Rhopilema esculentum. J. Oceanol. Limnol. 2019, 38, 1755–1761. [Google Scholar]
- Zhu, J.; Wang, J.; Zhen, Y.; Mi, T. Analyzed the effects of different food levels on asexual reproduction of jellyfish polyps based on transcriptome. Mar. Environ. Sci. 2023, 42, 169–175. [Google Scholar]
- Ghaskadbi, S. Cell signaling molecules in hydra: Insights into evolutionarily ancient functions of signaling pathways. Int. J. Dev. Biol. 2020, 64, 141–149. [Google Scholar]
- Kayal, E.; Bentlage, B.; Sabrina, M.; Ohdera, A.; Ryan, J. Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol. Biol. 2018, 18, 68. [Google Scholar]
- Fu, Y.; Dong, Z.; Liu, D. Factors influencing the asexual reproduction of moon jellyfish Aurelia aurita. Ecol. Sci. 2012, 31, 335–339. [Google Scholar]
- Travert, M.; Boohar, R.; Sanders, S.; Boosten, M.; Leclère, L.; Steel, R.; Cartwright, P. Coevolution of the Tlx homeobox gene with medusa development (Cnidaria: Medusozoa). Commun. Biol. 2022, 6, 709. [Google Scholar] [CrossRef] [PubMed]
- Leclère, L.; Horin, C.; Chevalier, S.; Lapébie, P.; Dru, P.; Peron, S.; Jager, M.; Condamine, T.; Pottin, K.; Romano, S.; et al. The genome of the jellyfish Clytia hemisphaerica and the evolution of the cnidarian life-cycle. Nat. Ecol. Evol. 2019, 3, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Leclere, L.; Copley, R.; Momose, T.; Houliston, E. Hydrozoan insights in animal development and evolution. Curr. Opin. Genet. Dev. 2016, 39, 157–167. [Google Scholar] [CrossRef]
- Turing, A.M. The chemical basis of morphogenesis. Bull. Math. Biol. 1990, 52, 153–197. [Google Scholar] [CrossRef]
- Bosch, T.C.; Anton-Erxleben, F.; Hemmrich, G.; Khalturin, K. The Hydra polyp: Nothing but an active stem cell community. Dev. Growth Differ. 2010, 52, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Purcell, J.E.; Uye, S.; Lo, W. Anthropogenic causes of jellyfish blooms and their direct consequences for humans: A review. Mar. Ecol.-Prog. Ser. 2007, 350, 153–174. [Google Scholar] [CrossRef]
- Chiaverano, L.M.; Robinson, K.L.; Tam, J.; Ruzicka, J.J.; Quiones, J.; Aleksa, K.; Hernandez, F.; Brodeur, R.; Uye, R.L. Evaluating the role of large jellyfish and forage fishes as energy pathways, and their interplay with fisheries, in the Northern Humboldt Current System. Prog. Oceanogr. 2018, 164, 28–36. [Google Scholar] [CrossRef]
- Fu, Z.; Shibata, M.; Makabe, R.; Ikeda, H.; Uye, S. Body size reduction under starvation, and the point of no return, in ephyrae of the moon jellyfish Aurelia aurita. Mar. Ecol.-Prog. Ser. 2014, 510, 255–263. [Google Scholar] [CrossRef]
- Dong, Z.; Liu, D.; Keesing, J.K. Jellyfish blooms in China: Dominant species, causes and consequences. Mar. Pollut. Bull. 2010, 60, 954–963. [Google Scholar] [CrossRef]
- Chapman, J.A.; Kirkness, E.F.; Simakov, O.; Hampson, S.E.; Mitros, T.; Weinmaier, T.; Rattei, T.; Balasubramanian, P.G.; Borman, J.; Busam, D.; et al. The dynamic genome of Hydra. Nature 2010, 464, 592–596. [Google Scholar] [CrossRef]
- Lengfeld, T.; Watanabe, H.; Simakow, O.; Lindgens, D.; Gee, L.; Law, L.; Schmidt, H.A.; Özbek, S.; Bode, H.; Holstein, T.W. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev. Biol. 2009, 330, 186–199. [Google Scholar] [PubMed]
- Manuel, G.C.; Reynoso, R.; Gee, L.; Salgado, L.M.; Bode, H.R. PI3K and ERK 1-2 regulate early stages during head regeneration in hydra. Dev. Growth Differ. 2006, 48, 129–138. [Google Scholar] [PubMed]
- Meinhardt, H.; Gierer, A. Generation and regeneration of sequence of structures during morphogenesis. J. Theor. Biol. 1980, 85, 429–450. [Google Scholar] [PubMed]
- Wenger, Y.; Buzgariu, W.; Perruchoud, C.; Loichot, G.; Galliot, B. Generic and context-dependent gene modulations during Hydra whole body regeneration. bioRxiv 2019, 6, 587147. [Google Scholar]
- Sehwag, V.; Wang, S.; Mittal, P.; Jana, S. HYDRA: Pruning Adversarially Robust Neural Networks. NeurIPS 2020, 33, 19655–19666. [Google Scholar]
- Chera, S.; Ghila, L.; Wenger, Y.; Galliot, B. Injury-induced activation of the MAPK/CREB pathway triggers apoptosis-induced compensatory proliferation in hydra head regeneration. Dev. Growth Differ. 2011, 53, 186–201. [Google Scholar] [CrossRef]
- Johnson, S.E.; Ilagan, M.X.G.; Kopan, R.; Barrick, D. Thermodynamic Analysis of the CSL·Notch Interaction. J. Biol. Chem. 2010, 285, 6681–6692. [Google Scholar]
- Böttger, A.; Alexandrova, O.; Cikala, M.; Schade, M.; Herold, M.; David, C.N. GFP expression in Hydra: Lessons from the particle gun. Dev. Genes Evol. 2002, 212, 302–305. [Google Scholar]
- Khalturin, K.; Anton-Erxleben, F.; Milde, S.; Plötz, C.; Wittlieb, J.; Hemmrich, G.; Bosch, T.C. Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev. Biol. 2007, 309, 32–44. [Google Scholar] [CrossRef]
- Berking, S. Bud formation inHydra: Inhibition by an endogenous morphogen. Wilehm Roux Arch. Dev. Biol. 1977, 181, 215–225. [Google Scholar] [CrossRef]
- Beermann, A.; Aranda, M.; Schröder, R. The Sp8 zinc-finger transcription factor is involved in allometric growth of the limbs in the beetle Tribolium castaneum. Development 2004, 131, 733–742. [Google Scholar] [PubMed]
- Camara, P.G. Methods and challenges in the analysis of single-cell RNA-sequencing data. Curr. Opin. Syst. Biol. 2018, 7, 47–53. [Google Scholar]
- Perween, N.; Pekhale, K.; Haval, G.; Mittal, S.; Ghaskadbi, S.; Ghaskadbi, S.S. Cloning and characterization of Thioredoxin 1 from the Cnidarian Hydra. J. Biochem. 2022, 171, 41–51. [Google Scholar] [PubMed]
- Purcell, J.E.; Bondyale-Juez, D.R.; Romero-Kutzner, V.; Martínez, I.; Caprioli, R.; Tames-Espinosa, M.; Almunia, J.; Alonso, E.; Packard, T.T.; Gómez, M. Food supply effects on the asexual reproduction and respiratory metabolism of Aurelia aurita polyps. Hydrobiologia 2019, 846, 135–146. [Google Scholar]
- Shostak, S. Origin of Asexual Reproduction in Hydra. Biomed. J. Sci. Tech. Res. 2018, 10, 7867–7871. [Google Scholar]
- Fuchs, B.; Wang, W.; Graspeuntner, S.; Li, Y.; Insua, S.; Herbst, E.; Dirksen, P.; Böhm, A.; Hemmrich, G.; Sommer, F.; et al. Regulation of polyp-to-jellyfish transition in Aurelia aurita. Curr. Biol. 2014, 24, 263–273. [Google Scholar]
- Xing, Y.Z.; Zhang, M.; Zhen, Y.; Tiezhu, M. Effects of salinity on the survival and asexual reproduction of Aurelia coerulea polyps at different temperatures. Ying Yong Sheng Tai Xue Bao 2019, 30, 2087–2092. [Google Scholar]
- Chuard, P.J.C.; Johnson, M.D.; Guichard, F. Ocean acidification causes mortality in the medusa stage of the cubozoan Carybdea xaymacana. Sci. Rep. 2019, 9, 5611. [Google Scholar]
- Yuan, S.; Huang, J.Y.; Qian, W.; Zhu, X.S.; Wang, S.H.; Jiang, X. Are Physical Sunscreens Safe for Marine Life? A Study on a Coral-Zooxanthellae Symbiotic System. Environ. Sci. Technol. 2023, 57, 15846–15857. [Google Scholar]
- Muscatine, L.; Porter, J.W. Reef Corals: Mutualistic Symbioses Adapted to Nutrient-Poor Environments. BioScience 1977, 27, 454–460. [Google Scholar]
- Anthony, K.R.N.; Hoegh-Guldberg, O. Kinetics of photoacclimation in corals. Oecologia 2003, 134, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.; Esko, J.; Freeze, H.H.; Stanley, P.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [PubMed]
- LaJeunesse, T.C.; Parkinson, J.E.; Gabrielson, P.W.; Jeong, H.J.; Reimer, J.D.; Voolstra, C.R.; Santos, S.R. Systematic Revision of Symbiodiniaceae Highlights the Antiquity and Diversity of Coral Endosymbionts. Curr. Biol. 2018, 28, 2570–2580. [Google Scholar] [PubMed]
- Eom, H.-J.; Lee, N.; Yum, S. Effects of extremely high concentrations of polystyrene microplastics on asexual reproduction and nematocyst discharge in the jellyfish Sand eria malayensis. Sci. Total Environ. 2022, 807, 150988. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Q.; Xiao, L.; Zhang, L. Intervention effects of five cations and their correction on hemolytic activity of tentacle extract from the jellyfish Cyanea capillata. PeerJ 2017, 5, 3338. [Google Scholar] [CrossRef]
- Fabila, Y.; Navarro, L.; Fujisawa, T.; Bode, H.R.; Salgado, L.M. Selective inhibition of protein kinases blocks the formation of a new axis, the beginning of budding, in Hydra. Mech. Dev. 2002, 119, 157–164. [Google Scholar] [CrossRef]
- Brooun, M.; Salvenmoser, W.; Dana, C.; Sudol, M.; Steele, R.; Hobmayer, B.; McNeill, H. The Hippo pathway regulates axis formation and morphogenesis in Hydra. Proc. Natl. Acad. Sci. USA 2022, 119, e2203257119. [Google Scholar] [CrossRef]
- Wang, R.; Steele, R.E.; Collins, E.S. Wnt signaling determines body axis polarity in regenerating Hydra tissue fragments. Dev. Biol. 2020, 467, 88–94. [Google Scholar] [CrossRef]
- Piccolo, S.; Dupont, M.; Cordenonsi, M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 2014, 94, 1287–1312. [Google Scholar] [CrossRef]
- Wang, C.; Cheng, F.Y.; Han, Z.; Yan, P.; Yin, Z.; Ge, X.; Li, D.; Zhong, R.; Liu, Q.; Chen, F.L.; et al. Human-induced pluripotent stem cell–derived neural stem cell exosomes improve blood–brain barrier function after intracerebral hemorrhage by activating astrocytes via PI3K/AKT/MCP-1 axis. Neural Regen. Res. 2024, 20, 518–532. [Google Scholar] [CrossRef]
- Bosch, T.C. Hydra and the evolution of stem cells. Bioessays 2009, 31, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Siebert, S.; Farrell, J.A.; Cazet, J.F.; Abeykoon, Y.; Juliano, C. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 2019, 365, eaav9314. [Google Scholar] [CrossRef] [PubMed]
- Bosch, T.C. Symmetry breaking in stem cells of the basal metazoan Hydra. Prog. Mol. Subcell. Biol. 2007, 45, 61–78. [Google Scholar]
- Vogg, M.C.; Buzgariu, W.; Suknovic, N.S.; Galliot, B. Cellular, Metabolic, and Developmental Dimensions of Whole-Body Regene ration in hydra. CSH Perspect. Biol. 2021, 13, a040725. [Google Scholar]
- Unni, M.; Reddy, P.C.; Pal, M.; Sagi, I.; Galande, S. Identification of Components of the Hippo Pathway in Hydra and potential role of YAP in cell division and differentiation. Front. Genet. 2021, 12, 676182. [Google Scholar] [CrossRef]
- He, J.; Bosch, T.C.G. Hydra’s Lasting Partnership with Microbes: The Key for Escaping Senescence? Microorganisms 2022, 10, 774. [Google Scholar] [CrossRef]
- Rathbun, L.I.; Everett, C.A.; Bergstralh, D. Emerging Cnidarian Models for the Study of Epithelial Polarity. Front. Cell Dev. Biol. 2022, 10, 854373. [Google Scholar] [CrossRef]
- Babonis, L.S.; Enjolras, C.; Ryan, J.F.; Martindale, M.Q. A novel regulatory gene promotes novel cell fate by suppressing ancestral fate in the sea anemone Nematostella vectensis. Proc. Natl. Acad. Sci. USA 2022, 119, e2113701119. [Google Scholar] [CrossRef]
- Zhang, R.; Jin, L.; Zhang, N.; Petridis, A.K.; Eckert, T.; Scheiner-Bobis, G.; Bergmann, M.; Scheidig, A.; Schauer, R.; Yan, M.; et al. The Sialic Acid-Dependent Nematocyst Discharge Process in Relation to Its Physical-Chemical Properties Is a Role Model for Nanomedical Diagnostic and Therapeutic Tools. Mar. Drugs 2019, 17, 469. [Google Scholar] [CrossRef]
- Bode, H.R.; David, C.N. Regulation of a multipotent stem cell, the interstitial cell of hydra. Prog. Biophys. Mol. Biol. 1978, 33, 189–206. [Google Scholar] [CrossRef]
- Sebestyén, F.; Barta, Z.; Tökölyi, J. Reproductive mode, stem cells and regeneration in a freshwater cnidarian with post-repro ductive senescence. Funct. Ecol. 2018, 32, 2497–2508. [Google Scholar]
- Wittlieb, J.; Khalturin, K.; Lohmann, J.U.; Anton-Erxleben, F.; Bosch, T. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 6208–6211. [Google Scholar] [CrossRef] [PubMed]
- Columbus-Shenkar, Y.Y.; Sachkova, M.Y.; Macrander, J.; Fridrich, A.; Modepalli, V.; Reitzel, A.M.; Sunagar, K.; Moran, Y. Dynamics of venom composition across a complex life cycle. Elife 2018, 7, e35014. [Google Scholar] [CrossRef]
- Yap, W.Y.; Tan, K.; Hwang, J.S. Expansion of Hydra actinoporin-like toxin (HALT) gene family: Expression divergence and functional convergence evolved through gene duplication. Toxicon 2019, 170, 10–20. [Google Scholar] [CrossRef]
- Bosch, T.C.; David, C.N. Male and female stem cells and sex reversal in Hydra polyps. Proc. Natl. Acad. Sci. USA 1986, 83, 9478–9482. [Google Scholar] [CrossRef]
- Arvizu, F.; Aguilera, A.; Salgado, L.M. Activities of the protein kinases STK, PI3K, MEK, and ERK are required for the development of the head organizer in Hydra magnipapillata. Differentiation 2006, 74, 305–312. [Google Scholar]
- Brooun, M.; Manoukian, A.; Shimizu, H.; Bode, H.R.; Mcneill, H. Organizer formation in Hydra is disrupted by thalidomide treatment. Dev. Biol. 2013, 378, 51–63. [Google Scholar]
- Münder, S.; Käsbauer, T.; Prexl, A.; Aufschnaiter, R.; Zhang, X.; Towb, P.; Böttger, A. Notch signalling defines critical boundary during budding in Hydra. Dev. Biol. 2010, 344, 331–345. [Google Scholar] [CrossRef]
- Gufler, S.; Artes, B.; Bielen, H.; Krainer, I.; Eder, M.K.; Falschlunger, J.; Bollmann, A.; Ostermann, T.; Valovka, T.; Hartl, M.; et al. beta-Catenin acts in a position-independent regeneration response in the simple eumetazoan Hydra. Dev. Biol. 2018, 433, 310–323. [Google Scholar]
- Qiu, L.; Sun, Y.; Ning, H.; Chen, G.; Zhao, W.; Gao, Y. The scaffold protein AXIN1: Gene ontology, signal network, and physiological function. Cell Commun. Signal. 2024, 22, 77. [Google Scholar]
- Tursch, A.; Bartsch, N.; Mercker, M.; Schlüter, J.; Lommel, M.; Marciniak, C.A.; Özbek, S.; Holstein, T.W. Injury-induced MAPK activation triggers body axis formation in hydra by default Wnt signaling. Proc. Natl. Acad. Sci. USA 2022, 119, e2204122119. [Google Scholar] [PubMed]
- Zhao, Y. Chronic stress dysregulates the Hippo/YAP/14-3-3η pathway and induces mitochondrial damage in basolateral amygdala in a mouse model of depression. Theranostics 2024, 14, 3653–3673. [Google Scholar] [PubMed]
- Shao, Q.; Duong, T.N.; Park, I.; Nomura, D.K. Covalent 14-3-3 Molecular Glues and Heterobifunctional Molecules Against Nuclear Transcription Factors and Regulators. bioRxiv 2023, 11, 565850. [Google Scholar]
- Namoto, K.; Baader, C.; Orsini, V.; Landshammer, A.; Breuer, E.; Dinh, K.T.; Ungricht, R.; Pikiolek, M.; Laurent, S.; Lu, B.; et al. NIBR-LTSi is a selective LATS kinase inhibitor activating YAP signaling and expanding tissue stem cells in vitro and in vivo. Cell Stem Cell 2024, 31, 554–569. [Google Scholar]
- Taouktsi, E.; Kyriakou, E.; Voulgaraki, E.; Verganelakis, D.; Krokou, S.; Rigas, S.; Voutsinas, G.E.; Syntichaki, P. Mitochondrial p38 Mitogen-Activated Protein Kinase: Insights into Its Regulation of and Role in LONP1-Deficient Nematodes. Int. J. Mol. Sci. 2023, 24, 17209. [Google Scholar] [CrossRef]
- Philipp, I.; Holstein, T.W.; Hobmayer, B. HvJNK, a Hydra member of the c-Jun NH2-terminal kinase gene family, is expressed during nematocyte differentiation. Gene Expr. Patterns 2005, 5, 397–402. [Google Scholar] [CrossRef]
- Tursch, A.; Bartsch, N.; Holstein, T.W. MAPK signaling links the injury response to Wnt-regulated patterning in Hydra regen eration. bioRxiv 2020, 6, 189795. [Google Scholar]
- Sewing, J.; Klinger, M.; Notbohm, H. Jellyfish collagen matrices conserve the chondrogenic phenotype in two-and three-dimensional collagen matrices. J. Tissue Eng. Regen. Med. 2017, 11, 916–925. [Google Scholar]
- Hasse, C.; Holz, O.; Lange, E.; Pisowodzki, L.; Rebscher, N.; Eder, M.C.; Hobmayer, B.; Hassel, M. FGFR-ERK signaling is an essential component of tissue separation. Dev. Biol. 2014, 395, 154–166. [Google Scholar]
- DuBuc, T.Q.; Traylor-Knowles, N.; Martindale, M.Q. Initiating a regenerative response; cellular and molecular features of wound healing in the cnidarian Nematostella vectensis. BMC Biol. 2014, 12, 24. [Google Scholar] [CrossRef]
- Holz, O.; Apel, D.; Steinmetz, P.; Lange, E.; Hopfenmuller, S.; Ohler, K.; Sudhop, S.; Hassel, M. Bud detachment in hydra requires activation of fibroblast growth factor receptor and a Rho-ROCK-myosin II signaling pathway to ensure formation of a basal constriction. Dev. Dyn. 2017, 246, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Prexl, A.; Münder, S.; Loy, B.; Kremmer, E.; Tischer, S.; Böttger, A. The putative Notch ligand HyJagged is a transmembrane protein present in all cell types of adult Hydra and upregulated at the boundary between bud and parent. BMC Cell Biol. 2021, 12, 38. [Google Scholar]
- Iommelli, F.; De Rosa, V.; Terlizzi, C.; Fonti, R.; Camerlingo, R.; Stoppelli, M.P.; Stewart, C.A.; Byers, L.A.; Piwnica-Worms, D.; Del Vecchio, S. A Reversible Shift of Driver Dependence from EGFR to Notch1 in Non-Small Cell Lung Cancer as a Cause of Resistance to Tyrosine Kinase Inhibitors. Cancers 2021, 13, 2022. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, H. DLL4/Notch blockade disrupts mandibular advancement-induced condylar osteogenesis by inhibiting H-type angiogenesis. J. Oral Rehabil. 2024, 51, 754–761. [Google Scholar]
- Cardenas, M.M.; Salgado, L.M. STK, the src homologue, is responsible for the initial commitment to develop head structures in Hydra. Dev. Biol. 2003, 264, 495–505. [Google Scholar]
- Turwankar, A.; Ghaskadbi, S. VEGF and FGF signaling during head regeneration in hydra. Gene 2019, 717, 144047. [Google Scholar]
- Fields, C.; Levin, M. Why isn’t sex optional? Stem-cell competition, loss of regenerative capacity, and cancer in meta zoan evolution. Commun. Integr. Biol. 2020, 13, 170–183. [Google Scholar]
| Number | Technology | Function | Advantage | Disadvantage | Author | Ref. |
|---|---|---|---|---|---|---|
| 1 | In situ hybridization | A gene probe was introduced into Hydra to locate the expression results and expression amount of key genes and related proteins, gaining insight into the role of signal pathways in bud proliferation. | The spatial distribution of RNA in tissues or cells is directly mapped to retain morphological information. No need to rely on genetically modified animals, suitable for non-model organisms. The cost is relatively low, and the technology is mature. | Sensitivity is limited, and low abundance RNAs may not be detectable. The steps (fixation, slicing, probe design) are complicated and take a long time. It is not possible to dynamically observe real-time changes in gene expression. | Perween et al. | [33] |
| 2 | Fluorescence tracing in vivo | Introduction of fluorescent protein in vivo and imaging tracing | Real-time dynamic observation of cell migration, differentiation, or molecular activity (such as calcium signaling). It is suitable for long-term tracking of live samples to reduce the interference of sample processing. High specificity can be achieved in combination with transgenic fluorescent labeling. | Depending on the fluorescent labeling technology, phototoxicity or photobleaching effects may be introduced. Imaging depth is limited and is not suitable for thick tissue or large samples. Expensive equipment (such as confocal microscopes) and complex data analysis are required. | Böttger et al.; Khalturin et al. | [28,29] |
| 3 | Signal pathway inhibitor | Specific inhibition of signal pathway to regulate bud proliferation | Rapid and reversible blocking of specific pathways to study short-term effects. Simple operation (such as drug immersion or local injection), and low cost. It is suitable for screening key regulatory pathways. | There may be off-target effects that interfere with other pathways. Inhibitor concentration and duration of action need to be strictly optimized; otherwise, false positives/negatives may occur. It is not possible to distinguish the specific functions of different components in the same pathway. | Berking et al. | [30] |
| 4 | Knockdown/ Overexpression | Inactivate or overexpress the target gene | Stable genetic strains can be constructed to support long-term research. Rapid verification of gene function gain effect, suitable for phenotypic screening. The spatiotemporal specificity of expression can be controlled by inducible promoters. | Genetic redundancy may result in phenotypic insensitivity (especially in species with high regenerative capacity). The construction time is long (such as stable mutant screening), and the cost is high. Overexpression may exceed physiological levels, leading to artificial illusions. | Beermann et al. | [31] |
| 5 | Single Nuclei RNA Sequencing snRNA-seq | Comments on cell differentiation, signal pathways and key molecules | Cell heterogeneity is resolved to identify rare cell types or states. Comprehensive mapping of gene expression to reveal new regulatory factors or pathways. Data can be integrated into multiple omics analyses (e.g., spatial transcriptome). | High cost (sample preparation, sequencing, computing resources). Loss of spatial position information (combined with in situ hybridization verification). Sensitive to sample quality (high requirements for cell activity and RNA integrity). | Camara et al. | [32] |
| Number | Impact Factor | Function | Author | Ref. |
|---|---|---|---|---|
| 1 | Food | Less food supply promotes asexual proliferation | Purcell et al.; Shostak et al. | [34,35] |
| 2 | Temperature | 15 °C is the optimal temperature for sprouting and proliferation | Jianbin et al. | [7] |
| 3 | Salinity | 20–32‰ is the optimal salinity for sprouting and proliferation | Xing et al. | [37] |
| 4 | O2 | Low/extremely low oxygen reduces the germination efficiency | Fu et al.; Ishii et al. | [18,37] |
| 5 | PH | The optimal pH is 7 | Perween et al.; Chuard et al. | [33,38] |
| 6 | Microplastics | Obstruct the sprouting and proliferation of polyps | Eom et al. | [44] |
| 7 | Heavy metal | Lower the sprouting and proliferation | Zhang et al. | [45] |
| Number | Cell Type | Function | Author | Ref. |
|---|---|---|---|---|
| 1 | Epithelial muscle cell | The types of gastric epidermal cells proliferate to promote the digestive function of polyps and promote budding. | Siebert et al.; Bosch et al.; Vogg et al. | [52,53,54] |
| 2 | Prickle cell | Prey, attack, defense | Babonis et al.; Bode et al. | [58,60] |
| 3 | Nerve cell | At various stages of budding, proliferation and tissue regeneration of Hydra | Wittlieb, J.; Columbus-Shenkar, Y.Y. | [62,63] |
| 4 | Germ cell | Ensure the occurrence of sexual reproduction | Unni, M.; He, J.;Sebestyén et al.; | [55,56,61] |
| 5 | Glandular cell | Help polyps attach, prey, and digest | Columbus-Shenkar et al.; Yap et al. | [63,64] |
| Number | Impact Factor | Regulation | Author | Ref. |
|---|---|---|---|---|
| 1 | Wnt | Coding polyp mouth-exit axis body | Lengfeld et al.; Wang et al. | [21,48] |
| 2 | β-catenin | Promote the regeneration of head and feet | Gufler et al. | [69] |
| 3 | Hippo | Regulation and induction of the formation of new body axis of bud | Brooun et al. | [47] |
| 4 | MAPK | Promote head and foot regeneration and bud proliferation | Fabila et al.; Cardenas et al. | [46,85] |
| 5 | PI3K | Promote the growth of the head and bud of polyps | Fabila et al. | [46] |
| 6 | JNK | Regulating the differentiation and proliferation of prickle cells; regulating compensatory proliferation | Chera et al.; Philipp et al. | [26,76] |
| 7 | ERK | Promote polyp sprouting and regulate head development | Sewing, J.; Hasse, C. | [78,79] |
| 8 | PKC | Promote head regeneration | Fabila et al. | [46] |
| 9 | FGFR | Promote the separation of polyps and buds | DuBuc, T.Q.; Holz, O | [80,81] |
| 10 | BMP | Participate in the growth of columnar gland cells | Brooun et al. | [67] |
| 11 | Notch | Promote bud differentiation | Iommelli, F. | [83] |
| 12 | VEGF/FGF | Promote the regeneration of the head and tentacles | Hu, Y. | [84] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, J.; Chen, J.; Li, L.; Geng, X.; Li, B.; Wang, M.; Yang, J. Signaling Mechanism of Budding, Proliferation, and Tissue Regeneration in Cnidaria. Curr. Issues Mol. Biol. 2025, 47, 219. https://doi.org/10.3390/cimb47040219
Lv J, Chen J, Li L, Geng X, Li B, Wang M, Yang J. Signaling Mechanism of Budding, Proliferation, and Tissue Regeneration in Cnidaria. Current Issues in Molecular Biology. 2025; 47(4):219. https://doi.org/10.3390/cimb47040219
Chicago/Turabian StyleLv, Jie, Jinhong Chen, Liangzhi Li, Xiaoyu Geng, Bingbing Li, Mingke Wang, and Jishun Yang. 2025. "Signaling Mechanism of Budding, Proliferation, and Tissue Regeneration in Cnidaria" Current Issues in Molecular Biology 47, no. 4: 219. https://doi.org/10.3390/cimb47040219
APA StyleLv, J., Chen, J., Li, L., Geng, X., Li, B., Wang, M., & Yang, J. (2025). Signaling Mechanism of Budding, Proliferation, and Tissue Regeneration in Cnidaria. Current Issues in Molecular Biology, 47(4), 219. https://doi.org/10.3390/cimb47040219


_Kim.png)




