Comprehensive Analysis of Differentially Expressed Profiles of mRNA 5-Methylcytosine Modification in Metabolic Dysfunction-Associated Steatotic Liver Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Studies and Sample Collection
2.2. Liver Lipid Analysis and Biochemical Assessment
2.3. RNA Isolation and Preparation
2.4. RNA-BS-Seq, RNA-Seq, and Data Analysis
2.5. RT-qPCR Validation
2.6. Statistical Analysis
3. Results
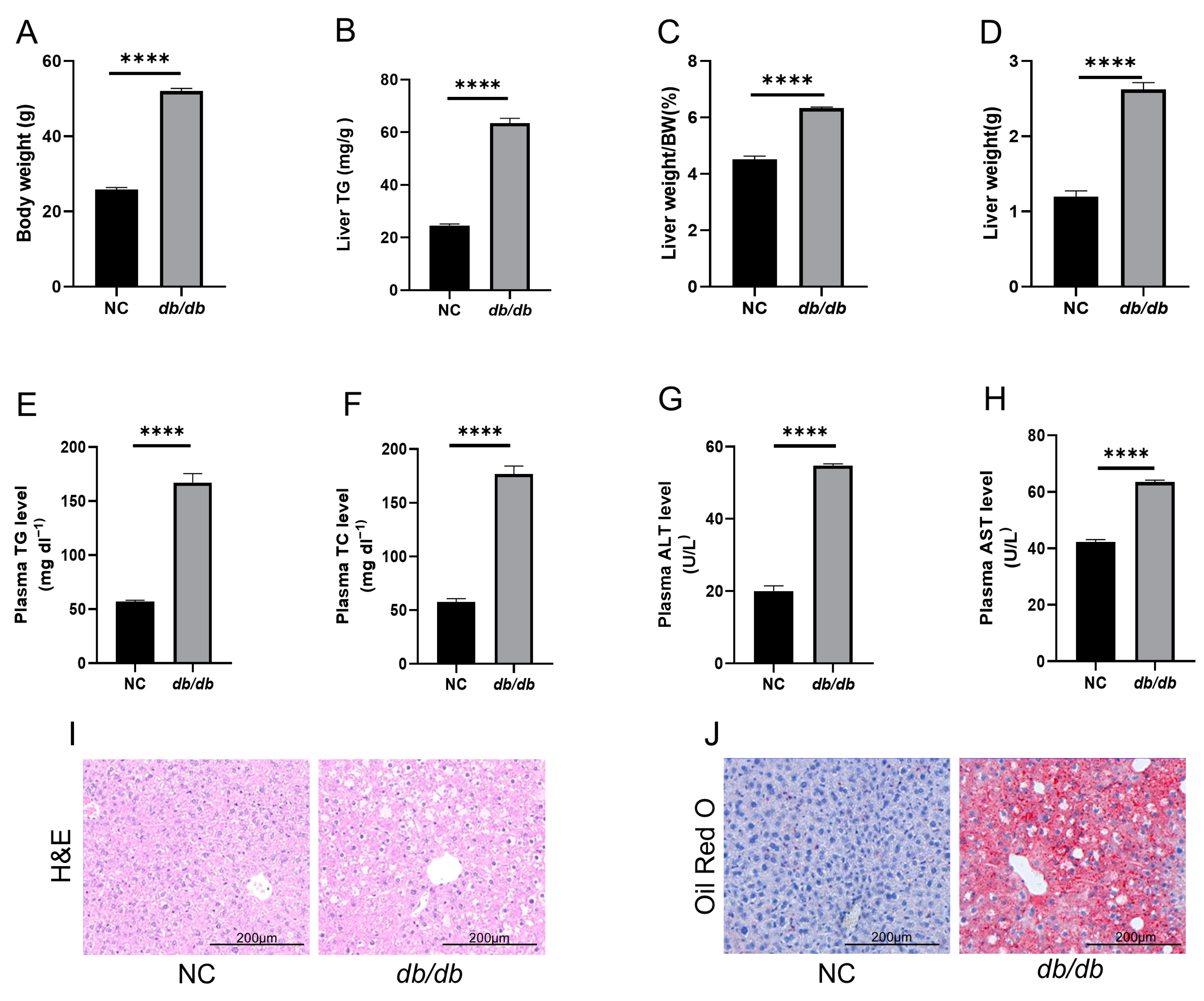
3.1. db/db Mice as a Model of MASLD
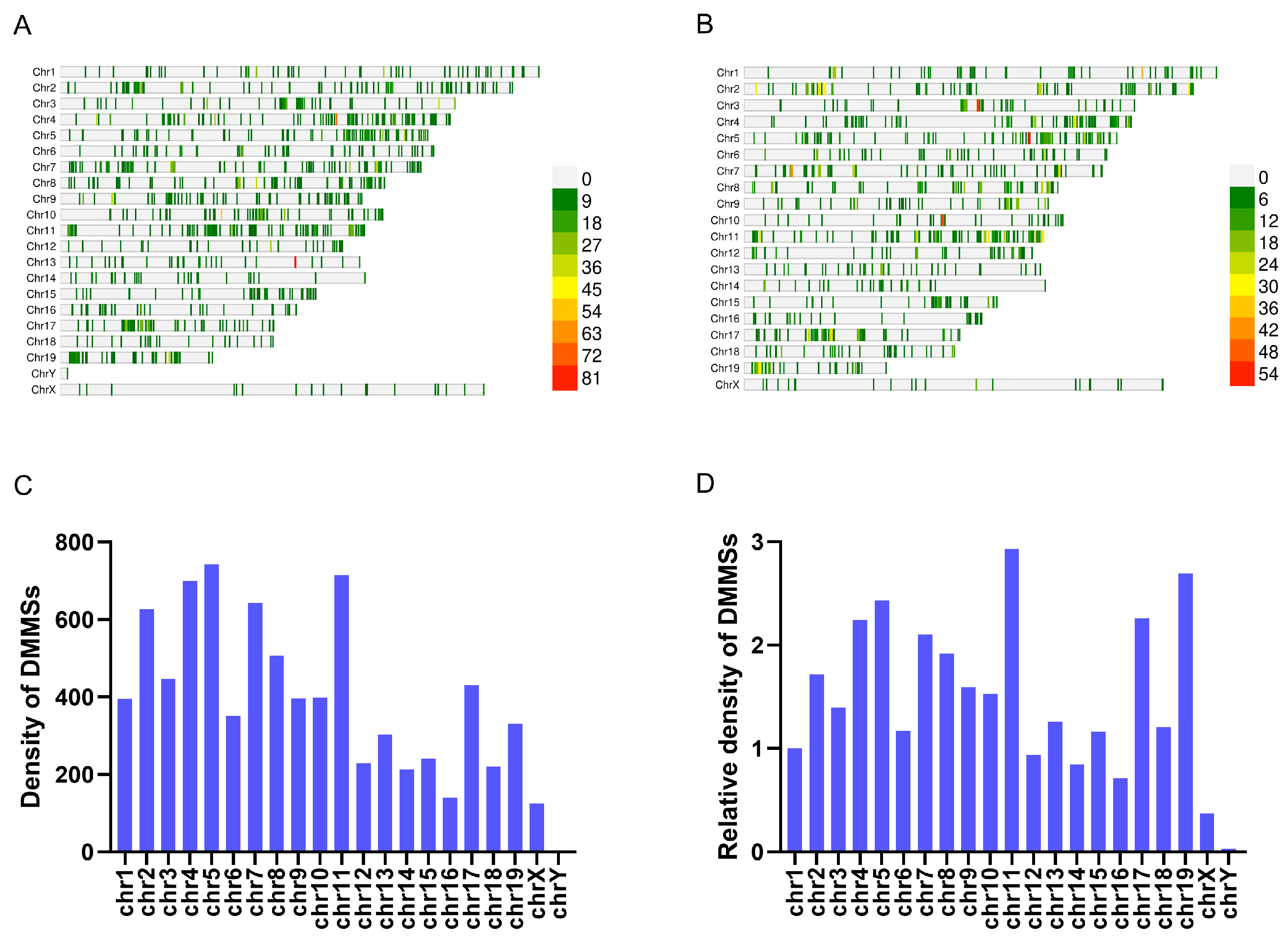
3.2. Overview of m5C Methylation in the Livers of db/db and NC Mice
3.3. Distribution and Biological Function of Different m5C Methylations in db/db and NC Mice
3.3.1. The Analysis of Differentially Methylated m5C Sites (DMMSs) in db/db and NC Mice
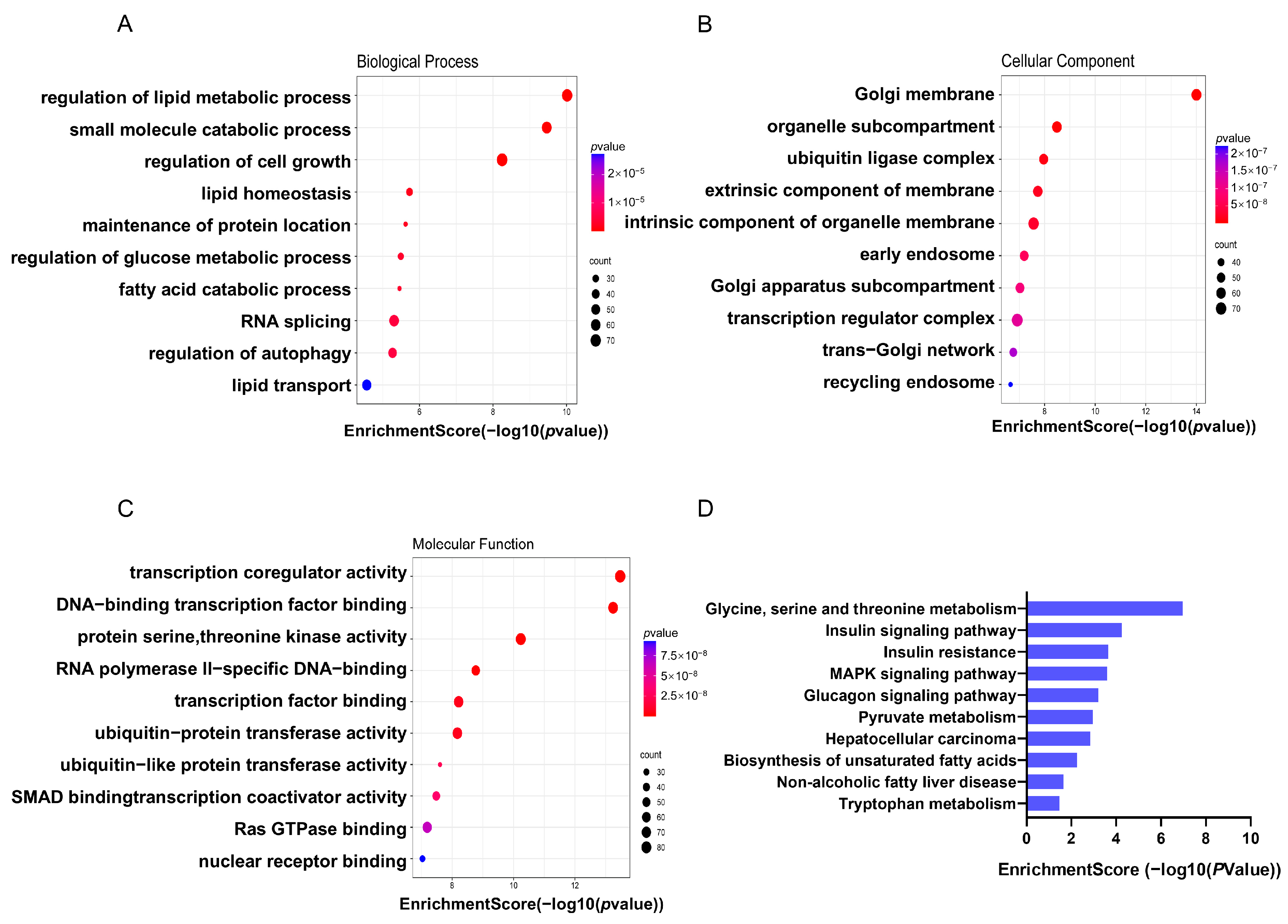
3.3.2. Biological Function of Differentially Methylated m5C Genes (DMMGs) in db/db and NC Mice
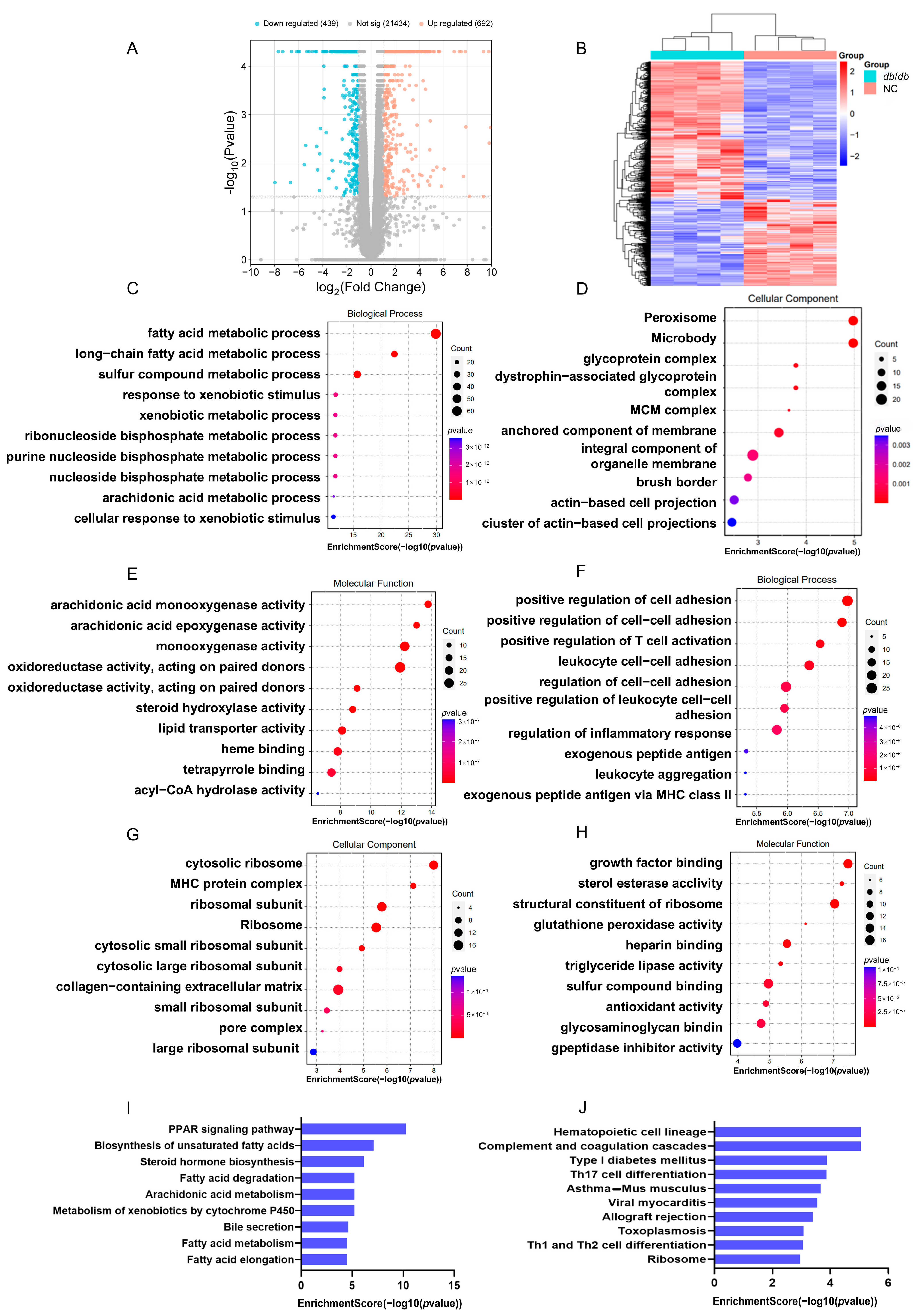
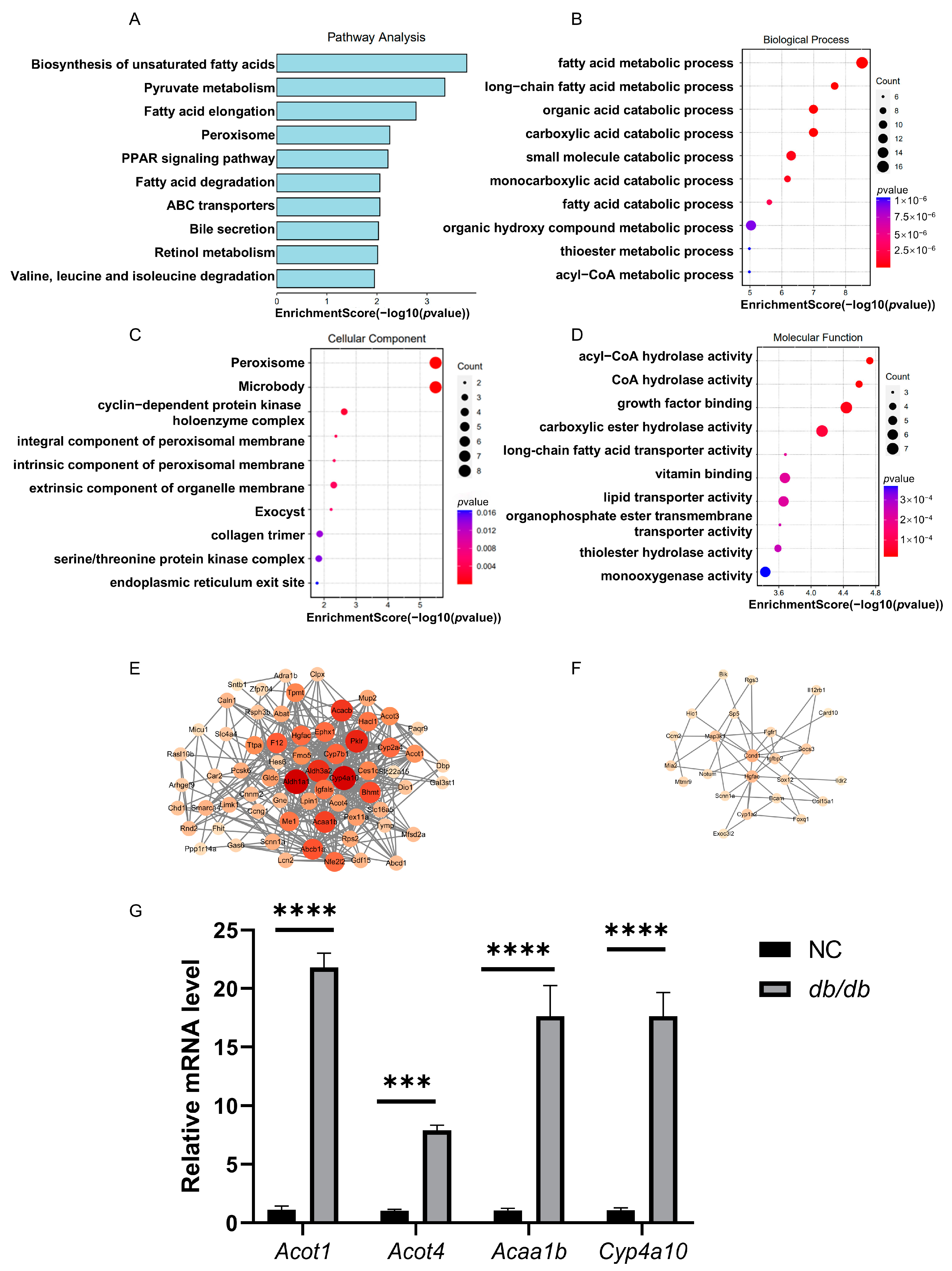
3.4. Detection of RNA-Seq-Based Gene Expression Variations in db/db Mice
3.5. Integrated Analysis of RNA-BS-Seq and RNA-Seq Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DMMGs | Differentially methylated m5C genes |
| DMMSs | Differentially methylated m5C sites |
| HCC | Hepatocellular carcinoma |
| MASH | Metabolic dysfunction-associated steatohepatitis |
| MASL | Metabolic dysfunction-associated steatotic liver |
| RNA-BS-Seq | RNA bisulfite sequencing |
| RNA-Seq | RNA transcriptome sequencing |
| m5C | 5-methylcytosine |
| MASLD | Metabolic dysfunction-associated steatotic liver disease |
| H&E | Hematoxylin and eosin |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| DEGs | Differentially expressed genes |
References
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology 2023, 78, 1966–1986. [Google Scholar] [CrossRef]
- Targher, G.; Byrne, C.D.; Tilg, H. MASLD: A systemic metabolic disorder with cardiovascular and malignant complications. Gut 2024, 73, 691–702. [Google Scholar] [CrossRef]
- Jiang, J.; Gao, Y.; Wang, J.; Huang, Y.; Yang, R.; Zhang, Y.; Ma, Y.; Wen, Y.; Luo, G.; Zhang, S.; et al. Hepatic sphingomyelin phosphodiesterase 3 promotes steatohepatitis by disrupting membrane sphingolipid metabolism. Cell Metab. 2025, 37, 1–18. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, X.J.; Li, H. Role of Innate Immune Signaling in Non-Alcoholic Fatty Liver Disease. Trends Endocrinol. Metab. 2018, 29, 712–722. [Google Scholar] [CrossRef]
- Yu, Y.; Cai, J.; She, Z.; Li, H. Insights into the Epidemiology, Pathogenesis, and Therapeutics of Nonalcoholic Fatty Liver Diseases. Adv. Sci. 2019, 6, 1801585. [Google Scholar] [CrossRef]
- Li, Y.; Yang, P.; Ye, J.; Xu, Q.; Wu, J.; Wang, Y. Updated mechanisms of MASLD pathogenesis. Lipids Health Dis. 2024, 23, 117. [Google Scholar] [CrossRef]
- Zhao, Y.C.; Zhao, G.J.; Chen, Z.; She, Z.G.; Cai, J.; Li, H. Nonalcoholic Fatty Liver Disease: An Emerging Driver of Hypertension. Hypertension 2020, 75, 275–284. [Google Scholar] [CrossRef]
- Song, P.; Tayier, S.; Cai, Z.; Jia, G. RNA methylation in mammalian development and cancer. Cell Biol. Toxicol. 2021, 37, 811–831. [Google Scholar] [CrossRef]
- Chen, X.; Li, A.; Sun, B.F.; Yang, Y.; Han, Y.N.; Yuan, X.; Chen, R.X.; Wei, W.S.; Liu, Y.; Gao, C.C.; et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019, 21, 978–990. [Google Scholar] [CrossRef]
- Li, X.; Meng, Y. Expression and prognostic characteristics of m(5) C regulators in low-grade glioma. J. Cell Mol. Med. 2021, 25, 1383–1393. [Google Scholar] [CrossRef]
- Nombela, P.; Miguel-López, B.; Blanco, S. The role of m(6)A, m(5)C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol. Cancer 2021, 20, 18. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, M.; Zhang, Q.; Yu, X.; Sun, Z.; He, Y.; Guo, W. Role of Main RNA Methylation in Hepatocellular Carcinoma: N6-Methyladenosine, 5-Methylcytosine, and N1-Methyladenosine. Front. Cell Dev. Biol. 2021, 9, 767668. [Google Scholar] [CrossRef]
- Zhou, B.; Luo, Y.; Bi, H.; Zhang, N.; Ma, M.; Dong, Z.; Ji, N.; Zhang, S.; Wang, X.; Liu, Y.; et al. Amelioration of nonalcoholic fatty liver disease by inhibiting the deubiquitylating enzyme RPN11. Cell Metab. 2024, 36, 2228–2244.e2227. [Google Scholar] [CrossRef]
- Zhou, B.; Luo, Y.; Ji, N.; Mao, F.; Xiang, L.; Bian, H.; Zheng, M.H.; Hu, C.; Li, Y.; Lu, Y. Promotion of nonalcoholic steatohepatitis by RNA N(6)-methyladenosine reader IGF2BP2 in mice. Life Metab. 2022, 1, 161–174. [Google Scholar] [CrossRef]
- Zheng, L.; Duan, Y.; Li, M.; Wei, J.; Xue, C.; Chen, S.; Wei, Q.; Tang, F.; Xiong, W.; Zhou, M.; et al. Deciphering the vital roles and mechanism of m5C modification on RNA in cancers. Am. J. Cancer Res. 2023, 13, 6125–6146. [Google Scholar]
- Fang, L.; Huang, H.; Lv, J.; Chen, Z.; Lu, C.; Jiang, T.; Xu, P.; Li, Y.; Wang, S.; Li, B.; et al. m5C-methylated lncRNA NR_033928 promotes gastric cancer proliferation by stabilizing GLS mRNA to promote glutamine metabolism reprogramming. Cell Death Dis. 2023, 14, 520. [Google Scholar] [CrossRef]
- Yang, M.; Wei, R.; Zhang, S.; Hu, S.; Liang, X.; Yang, Z.; Zhang, C.; Zhang, Y.; Cai, L.; Xie, Y. NSUN2 promotes osteosarcoma progression by enhancing the stability of FABP5 mRNA via m(5)C methylation. Cell Death Dis. 2023, 14, 125. [Google Scholar] [CrossRef]
- Zuo, S.; Li, L.; Wen, X.; Gu, X.; Zhuang, A.; Li, R.; Ye, F.; Ge, S.; Fan, X.; Fan, J.; et al. NSUN2-mediated m(5) C RNA methylation dictates retinoblastoma progression through promoting PFAS mRNA stability and expression. Clin. Transl. Med. 2023, 13, e1273. [Google Scholar] [CrossRef]
- Chen, S.Y.; Chen, K.L.; Ding, L.Y.; Yu, C.H.; Wu, H.Y.; Chou, Y.Y.; Chang, C.J.; Chang, C.H.; Wu, Y.N.; Wu, S.R.; et al. RNA bisulfite sequencing reveals NSUN2-mediated suppression of epithelial differentiation in pancreatic cancer. Oncogene 2022, 41, 3162–3176. [Google Scholar] [CrossRef]
- Shinoda, S.; Kitagawa, S.; Nakagawa, S.; Wei, F.Y.; Tomizawa, K.; Araki, K.; Araki, M.; Suzuki, T.; Suzuki, T. Mammalian NSUN2 introduces 5-methylcytidines into mitochondrial tRNAs. Nucleic Acids Res. 2019, 47, 8734–8745. [Google Scholar] [CrossRef]
- Lewinska, A.; Adamczyk-Grochala, J.; Wnuk, M. TRDMT1-mediated RNA C-5 methylation as a novel target in anticancer therapy. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188964. [Google Scholar] [CrossRef]
- Li, H.; Zhu, D.; Wu, J.; Ma, Y.; Cai, C.; Chen, Y.; Qin, M.; Dai, H. New substrates and determinants for tRNA recognition of RNA methyltransferase DNMT2/TRDMT1. RNA Biol. 2021, 18, 2531–2545. [Google Scholar] [CrossRef]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef]
- Chen, Y.S.; Yang, W.L.; Zhao, Y.L.; Yang, Y.G. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip. Rev. RNA 2021, 12, e1639. [Google Scholar] [CrossRef]
- Zhang, M.; Song, J.; Yuan, W.; Zhang, W.; Sun, Z. Roles of RNA Methylation on Tumor Immunity and Clinical Implications. Front. Immunol. 2021, 12, 641507. [Google Scholar] [CrossRef]
- Ji, A.L.; Rubin, A.J.; Thrane, K.; Jiang, S.; Reynolds, D.L.; Meyers, R.M.; Guo, M.G.; George, B.M.; Mollbrink, A.; Bergenstråhle, J.; et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell 2020, 182, 497–514.e422. [Google Scholar] [CrossRef]
- Löffler, M.W.; Mohr, C.; Bichmann, L.; Freudenmann, L.K.; Walzer, M.; Schroeder, C.M.; Trautwein, N.; Hilke, F.J.; Zinser, R.S.; Mühlenbruch, L.; et al. Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma. Genome Med. 2019, 11, 28. [Google Scholar] [CrossRef]
- Xiao, Y.; Ma, D.; Zhao, S.; Suo, C.; Shi, J.; Xue, M.Z.; Ruan, M.; Wang, H.; Zhao, J.; Li, Q.; et al. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer. Clin. Cancer Res. 2019, 25, 5002–5014. [Google Scholar] [CrossRef]
- Xiang, L.; Li, X.; Luo, Y.; Zhou, B.; Liu, Y.; Li, Y.; Wu, D.; Jia, L.; Zhu, P.W.; Zheng, M.H.; et al. A multi-omic landscape of steatosis-to-NASH progression. Life Metab. 2022, 1, 242–257. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Shen, L.; Shao, N.Y.; Liu, X.; Maze, I.; Feng, J.; Nestler, E.J. diffReps: Detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE 2013, 8, e65598. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Sarin, L.P.; Leidel, S.A. Modify or die?—RNA modification defects in metazoans. RNA Biol. 2014, 11, 1555–1567. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, S.; Wu, X.; Zhou, X. m(6)A RNA Methylation in Cardiovascular Diseases. Mol. Ther. 2020, 28, 2111–2119. [Google Scholar] [CrossRef]
- Edelheit, S.; Schwartz, S.; Mumbach, M.R.; Wurtzel, O.; Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013, 9, e1003602. [Google Scholar] [CrossRef]
- Hoernes, T.P.; Clementi, N.; Faserl, K.; Glasner, H.; Breuker, K.; Lindner, H.; Hüttenhofer, A.; Erlacher, M.D. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 2016, 44, 852–862. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, C.; Nachtergaele, S.; Wunderlich, M.; Qing, Y.; Deng, X.; Wang, Y.; Weng, X.; Hu, C.; et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 2018, 172, 90–105.e123. [Google Scholar] [CrossRef]
- Zou, F.; Tu, R.; Duan, B.; Yang, Z.; Ping, Z.; Song, X.; Chen, S.; Price, A.; Li, H.; Scott, A.; et al. Drosophila YBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs. Proc. Natl. Acad. Sci. USA 2020, 117, 3603–3609. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.S.; Xu, J.W.; Lai, W.Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, Y.; Wu, R.; Chen, Y.; Chen, W.; Liu, Y.; Luo, Y.; Huang, C.; Zeng, B.; Liao, X.; et al. mRNA m5C controls adipogenesis by promoting CDKN1A mRNA export and translation. RNA Biol. 2021, 18, 711–721. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Wu, R.; Gao, C.C.; Liao, X.; Han, X.; Zeng, B.; Huang, C.; Luo, Y.; Liu, Y.; et al. mRNA m(5)C inhibits adipogenesis and promotes myogenesis by respectively facilitating YBX2 and SMO mRNA export in ALYREF-m(5)C manner. Cell Mol. Life Sci. 2022, 79, 481. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, J.; Yang, X.; Wang, K.; Sun, K.; Yang, Z.; Zhang, L.; Yang, L.; Gu, C.; Huang, X.; et al. Dysregulated m6A modification promotes lipogenesis and development of non-alcoholic fatty liver disease and hepatocellular carcinoma. Mol. Ther. 2022, 30, 2342–2353. [Google Scholar] [CrossRef]
- Guo, J.; Ren, W.; Li, A.; Ding, Y.; Guo, W.; Su, D.; Hu, C.; Xu, K.; Chen, H.; Xu, X.; et al. Fat mass and obesity-associated gene enhances oxidative stress and lipogenesis in nonalcoholic fatty liver disease. Dig. Dis. Sci. 2013, 58, 1004–1009. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, J.; Han, C.; Wang, Z.; Li, C.; Wang, X.; Liu, Z.; Wen, J.; Zheng, L. Expression and significance of fat mass and obesity associated gene and forkhead transcription factor O1 in non-alcoholic fatty liver disease. Chin. Med. J. 2014, 127, 3771–3776. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, X.; Wu, W.; Wang, X.; Wang, Y. FTO-dependent function of N6-methyladenosine is involved in the hepatoprotective effects of betaine on adolescent mice. J. Physiol. Biochem. 2015, 71, 405–413. [Google Scholar] [CrossRef]
- Dou, X.; Xia, Y.; Chen, J.; Qian, Y.; Li, S.; Zhang, X.; Song, Z. Rectification of impaired adipose tissue methylation status and lipolytic response contributes to hepatoprotective effect of betaine in a mouse model of alcoholic liver disease. Br. J. Pharmacol. 2014, 171, 4073–4086. [Google Scholar] [CrossRef]
- Feng, Q.; Wang, D.; Xue, T.; Lin, C.; Gao, Y.; Sun, L.; Jin, Y.; Liu, D. The role of RNA modification in hepatocellular carcinoma. Front. Pharmacol. 2022, 13, 984453. [Google Scholar] [CrossRef]
- Li, D.; Li, K.; Zhang, W.; Yang, K.W.; Mu, D.A.; Jiang, G.J.; Shi, R.S.; Ke, D. The m6A/m5C/m1A Regulated Gene Signature Predicts the Prognosis and Correlates with the Immune Status of Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 918140. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Chan, T.H.; Tenen, D.G.; Chen, L. RNA editome imbalance in hepatocellular carcinoma. Cancer Res. 2014, 74, 1301–1306. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Li, X.; Xiong, X.; Wang, J.; Zhou, Z.; Zhu, X.; Gu, Y.; Dominissini, D.; He, L.; et al. N(1)-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 2021, 12, 6314. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Gu, X.; Zheng, Q.; Shi, Q.; Yuan, X.; Su, Y.; Jia, J.; Jiang, J.; Lu, J.; Li, L. ALYREF mediates RNA m(5)C modification to promote hepatocellular carcinoma progression. Signal Transduct. Target. Ther. 2023, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Xue, S.; Zhang, M.; Xu, H.; Hu, X.; Chen, S.; Liu, Y.; Guo, M.; Cui, H. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene 2020, 39, 6906–6919. [Google Scholar] [CrossRef]
- Fabbrini, E.; Sullivan, S.; Klein, S. Obesity and nonalcoholic fatty liver disease: Biochemical, metabolic, and clinical implications. Hepatology 2010, 51, 679–689. [Google Scholar] [CrossRef]
- GB/T 35892–2018; Laboratory Animal—Guideline for Ethical Review of Animal Welfare. General Administration of Quality Supervision, Inspection and Quarantine: Beijing, China, 2018.

| Chromosome | RefPos | RefStrand | GeneName | Foldchange |
|---|---|---|---|---|
| 19 | 7516421 | + | Atl3 | 3.577619607 |
| 10 | 76896699 | − | Col18a1 | 3.101572825 |
| 19 | 7516425 | + | Atl3 | 3.090842199 |
| 11 | 118302035 | − | Cant1 | 3.044066762 |
| 19 | 7516426 | + | Atl3 | 2.946495372 |
| 10 | 75773545 | + | Chchd10 | 2.677710514 |
| 11 | 118302029 | − | Cant1 | 2.591881931 |
| 2 | 144112449 | − | Snx5 | 2.581124981 |
| 19 | 38111275 | − | Rbp4 | 2.529759085 |
| 10 | 30076483 | − | Cenpw | 2.507066041 |
| Chromosome | RefPos | RefStrand | GeneName | Foldchange |
|---|---|---|---|---|
| 4 | 148543216 | + | Mtor | 3.739900791 |
| 6 | 94676492 | − | Lrig1 | 3.61000291 |
| 19 | 47568459 | + | Slk | 3.462946262 |
| 12 | 51737918 | + | Ap4s1 | 3.441410522 |
| 3 | 108191651 | + | Sort1 | 3.301221634 |
| 8 | 123876449 | − | Sult5a1 | 3.264812185 |
| 7 | 127159861 | + | Gm42715 | 3.23104311 |
| 1 | 9818350 | − | Vcpip1 | 3.21986455 |
| 14 | 70526697 | − | Ppp3cc | 3.197623321 |
| 17 | 31519274 | + | Slc37a1 | 3.164549205 |
| Gene Name | Chrome | Strand | Regulation | Fold Change | p-Value |
|---|---|---|---|---|---|
| Sult3a1 | chr10:33863934–33879475 | + | Up | 980.08 | 0.00185 |
| Cyp2b13 | chr7:26061494–26096196 | + | Up | 901.21 | 0.0039 |
| Sult1e1 | chr5:87575967–87591611 | − | Up | 875.72 | 0.00005 |
| Cyp2b9 | chr7:26173410–26210661 | + | Up | 649.61 | 0.00005 |
| Sult2a5 | chr7:13623966–13670801 | + | Up | 647.37 | 0.0499 |
| Hao2 | chr3:98874576–98893239 | − | Up | 287.06 | 0.0491 |
| Cyp3a41a | chr5:145694059–145720136 | − | Up | 235.47 | 0.0018 |
| Sult2a7 | chr7:14465158–14492926 | − | Up | 226.97 | 0.00005 |
| A4gnt | chr9:99612501–99622367 | + | Up | 204.94 | 0.01895 |
| Ly6d | chr15:74762055–74763567 | − | Up | 179.17 | 0.00005 |
| Serpina9 | chr12:103980242–104013755 | − | Down | 252.27 | 0.0255 |
| Rpl21 | chr5:146832889–146837032 | + | Down | 207.83 | 0.00005 |
| Camk2b | chr11:5969643–6066362 | − | Down | 159.94 | 0.00005 |
| Hsd3b5 | chr3:98382480–98763128 | − | Down | 102.79 | 0.02555 |
| Serpina1e | chr12:103946930–103958975 | − | Down | 97.10 | 0.00005 |
| Adh6-ps1 | chr3:138374120–138388291 | + | Down | 70.99 | 0.00005 |
| Enho | chr4:41569774–41641416 | − | Down | 64.92 | 0.00005 |
| BC048546 | chr6:128539821–128581606 | − | Down | 62.24 | 0.00005 |
| Cib3 | chr8:72204334–72212837 | − | Down | 46.44 | 0.0108 |
| Gm3839 | chr14:11280734–11356726 | − | Down | 42.66 | 0.00435 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Wang, E.; Zhou, B.; Lu, Y.; Ding, X.; Li, Y. Comprehensive Analysis of Differentially Expressed Profiles of mRNA 5-Methylcytosine Modification in Metabolic Dysfunction-Associated Steatotic Liver Disease. Curr. Issues Mol. Biol. 2025, 47, 305. https://doi.org/10.3390/cimb47050305
Yang Y, Wang E, Zhou B, Lu Y, Ding X, Li Y. Comprehensive Analysis of Differentially Expressed Profiles of mRNA 5-Methylcytosine Modification in Metabolic Dysfunction-Associated Steatotic Liver Disease. Current Issues in Molecular Biology. 2025; 47(5):305. https://doi.org/10.3390/cimb47050305
Chicago/Turabian StyleYang, Yueying, E Wang, Bing Zhou, Yan Lu, Xiaoying Ding, and Yao Li. 2025. "Comprehensive Analysis of Differentially Expressed Profiles of mRNA 5-Methylcytosine Modification in Metabolic Dysfunction-Associated Steatotic Liver Disease" Current Issues in Molecular Biology 47, no. 5: 305. https://doi.org/10.3390/cimb47050305
APA StyleYang, Y., Wang, E., Zhou, B., Lu, Y., Ding, X., & Li, Y. (2025). Comprehensive Analysis of Differentially Expressed Profiles of mRNA 5-Methylcytosine Modification in Metabolic Dysfunction-Associated Steatotic Liver Disease. Current Issues in Molecular Biology, 47(5), 305. https://doi.org/10.3390/cimb47050305






