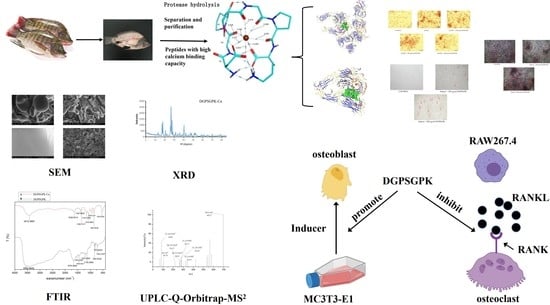
Purification and Characterization of a Novel Calcium-Binding Heptapeptide from the Hydrolysate of Tilapia Bone with Its Osteogenic Activity
Abstract
:1. Introduction
2. Results and Discussion
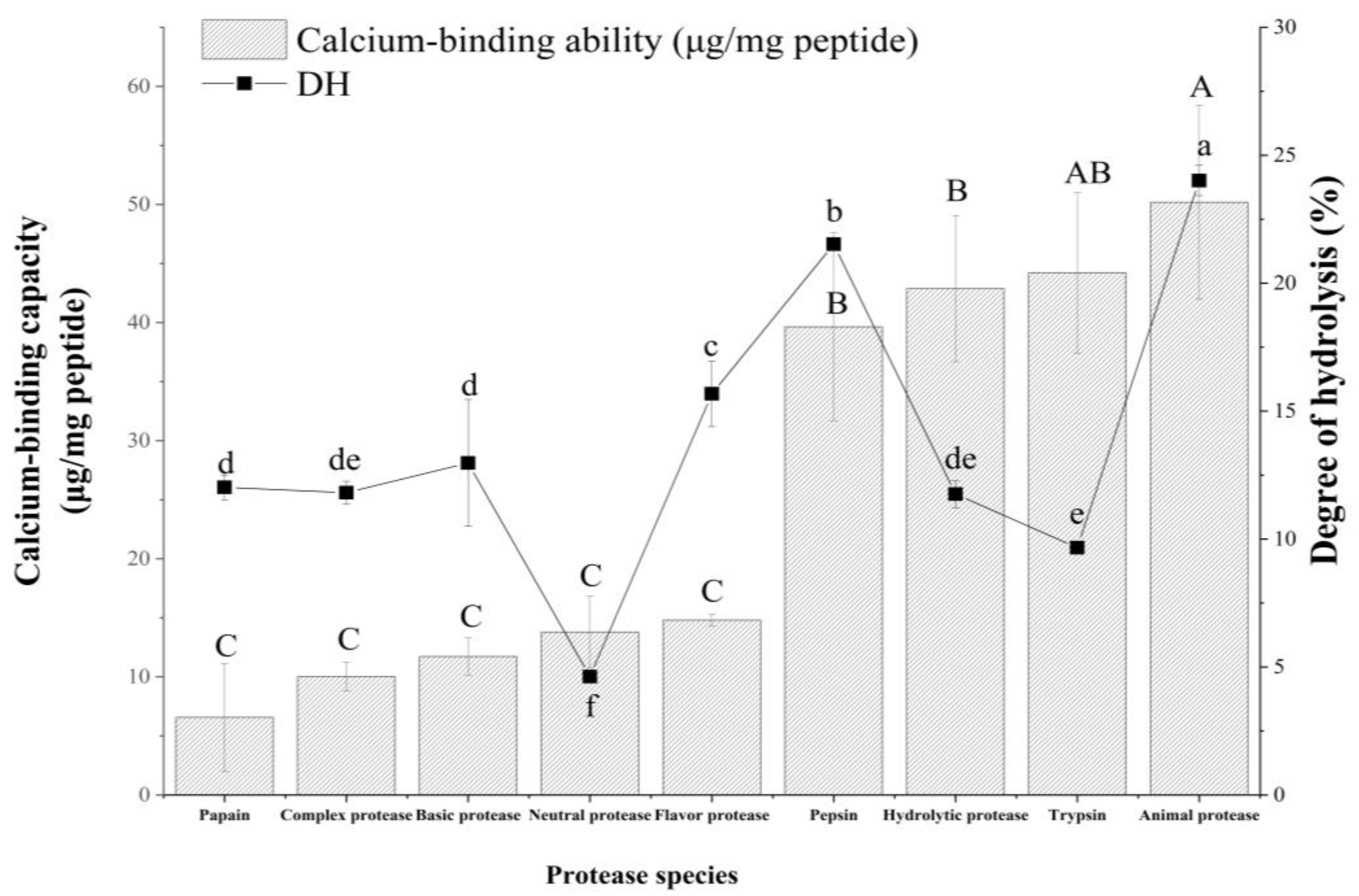
2.1. Different Degrees of Hydrolysis (DH) and Calcium Binding Capacities of Different Hydrolysates
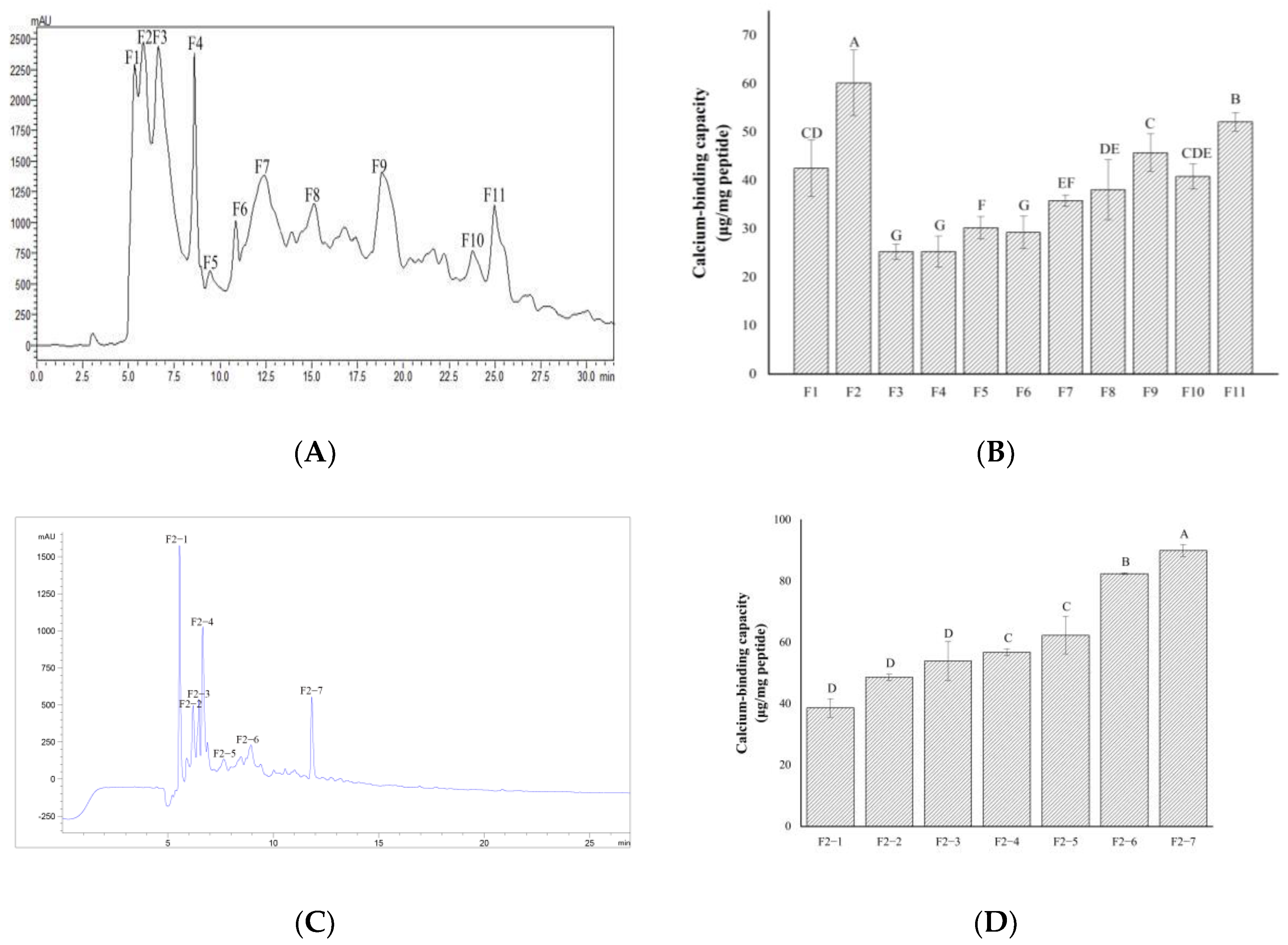
2.2. Separation and Purification of Calcium-Chelating Peptides
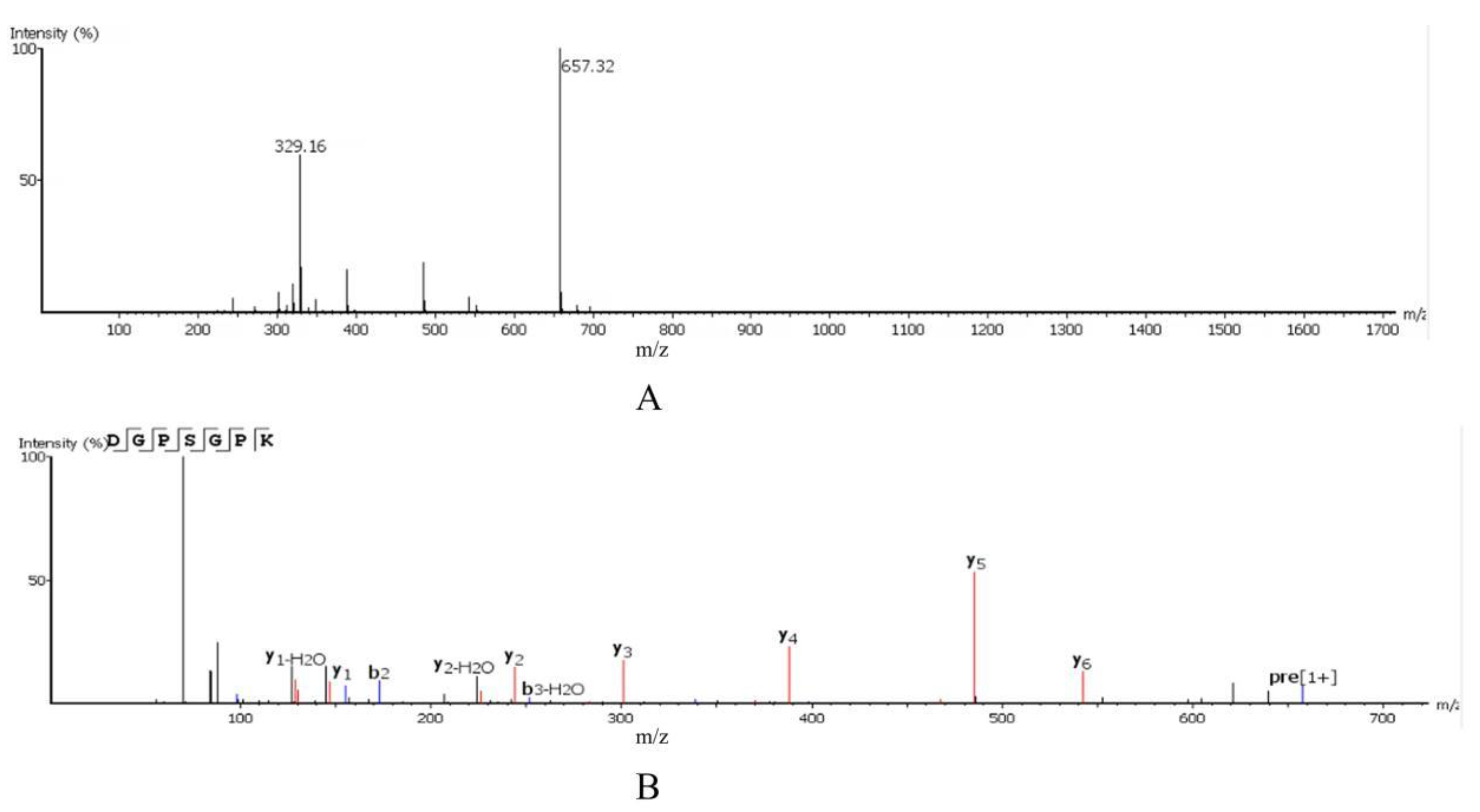
2.3. Amino Acid Sequence Identification
2.4. Structural Characterization
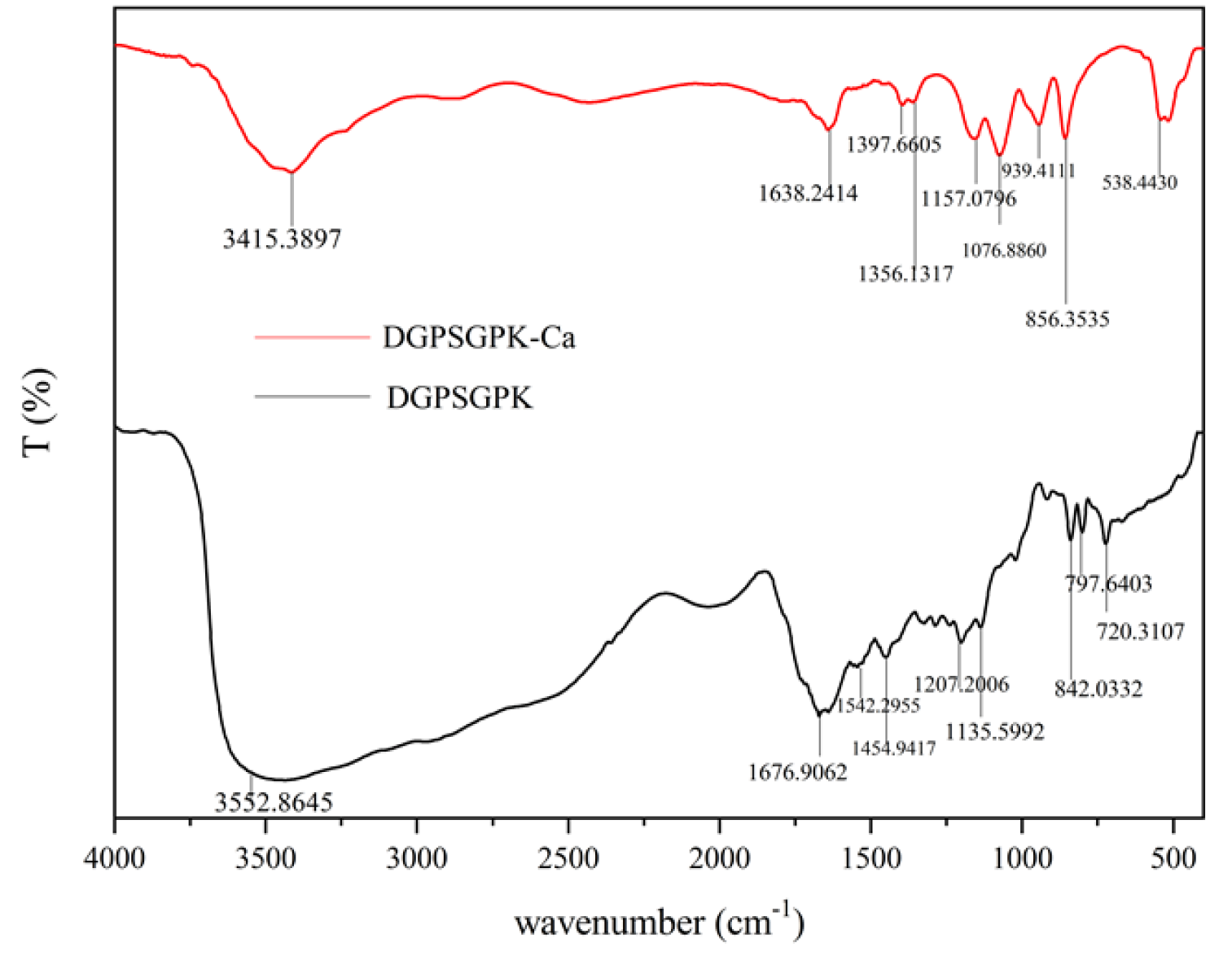
2.4.1. FTIR
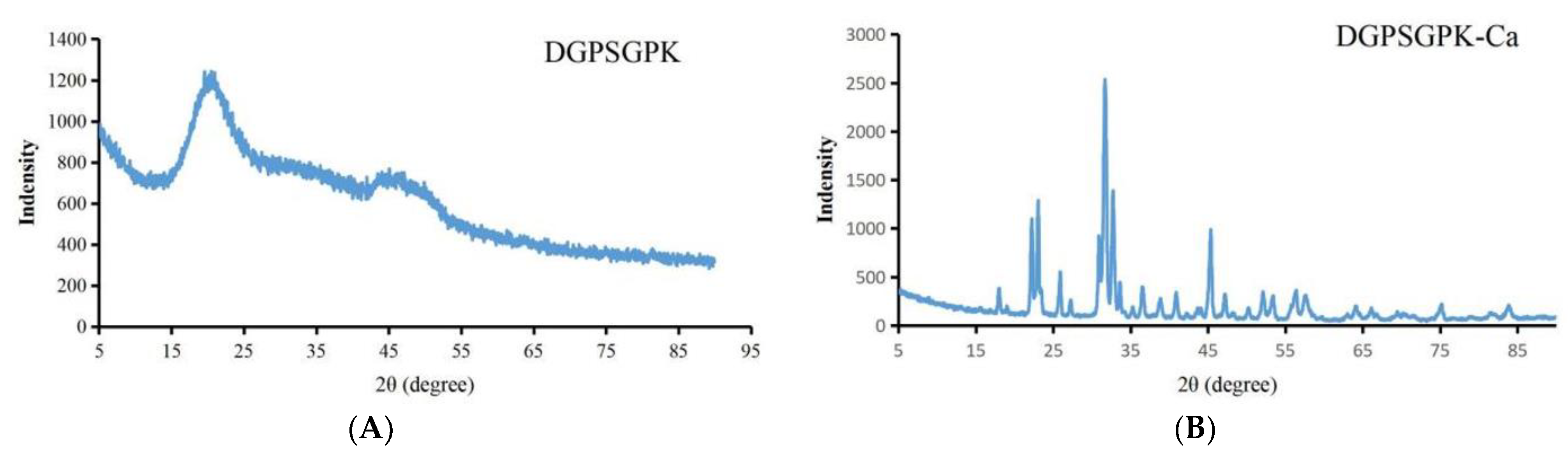
2.4.2. XRD
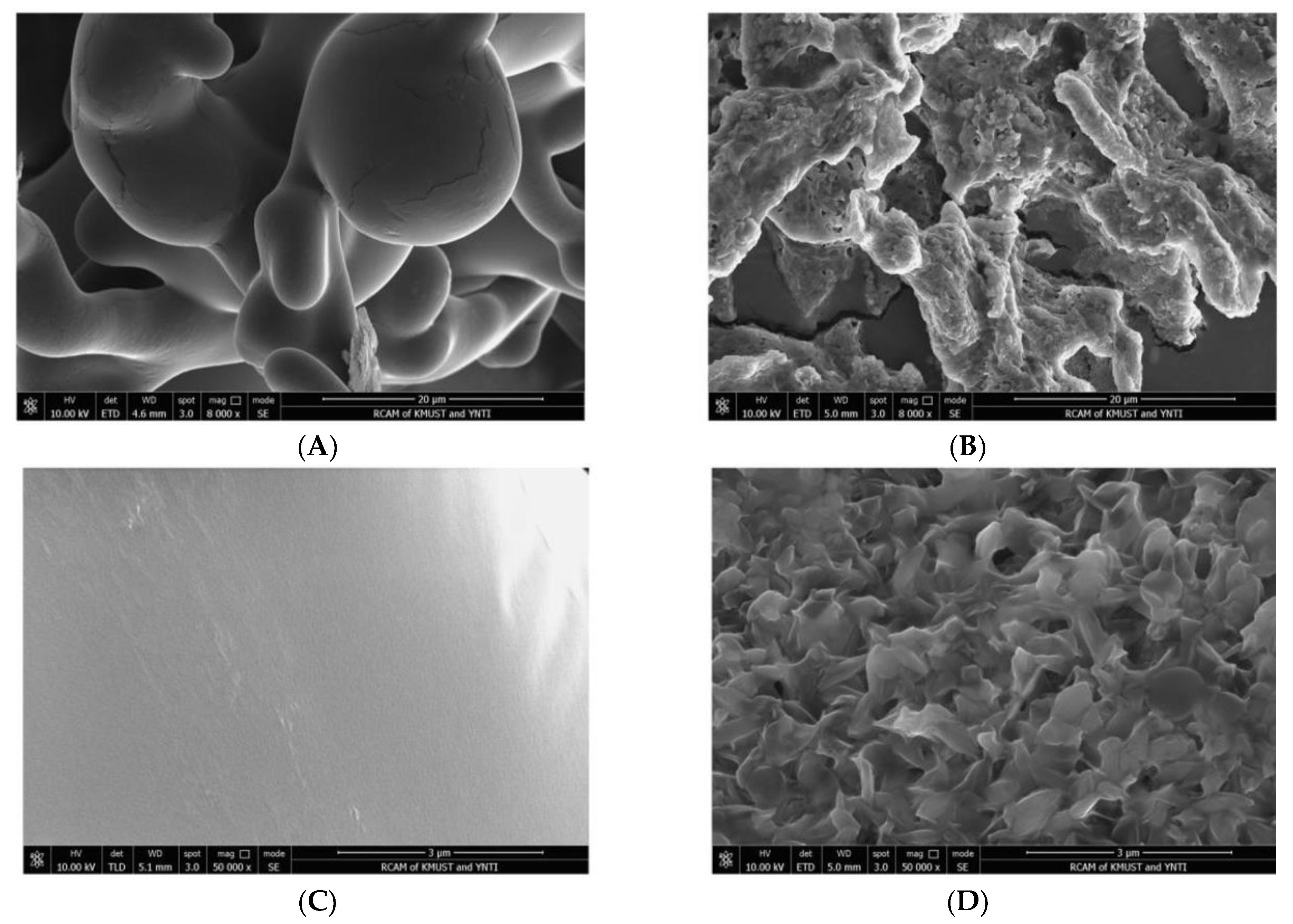
2.4.3. SEM
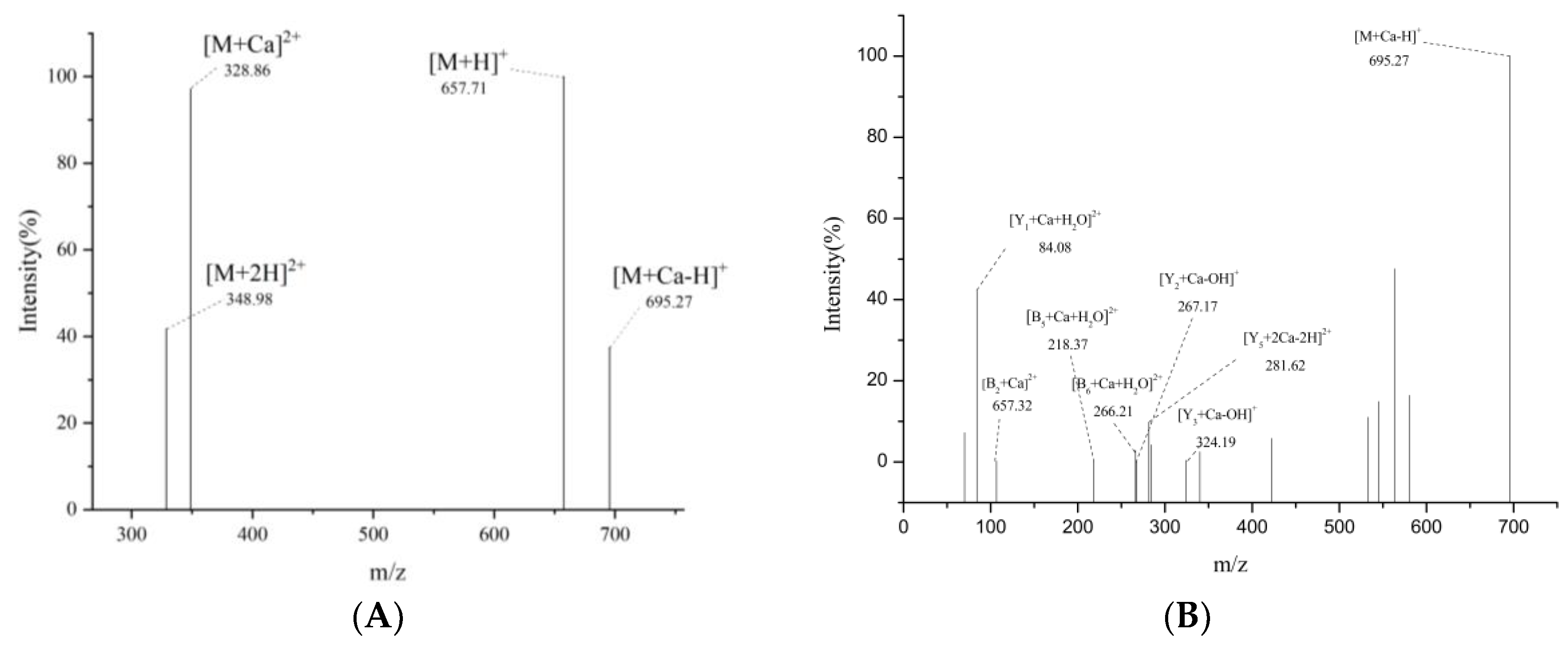
2.4.4. UPLC-Q-Orbitrap-MS2
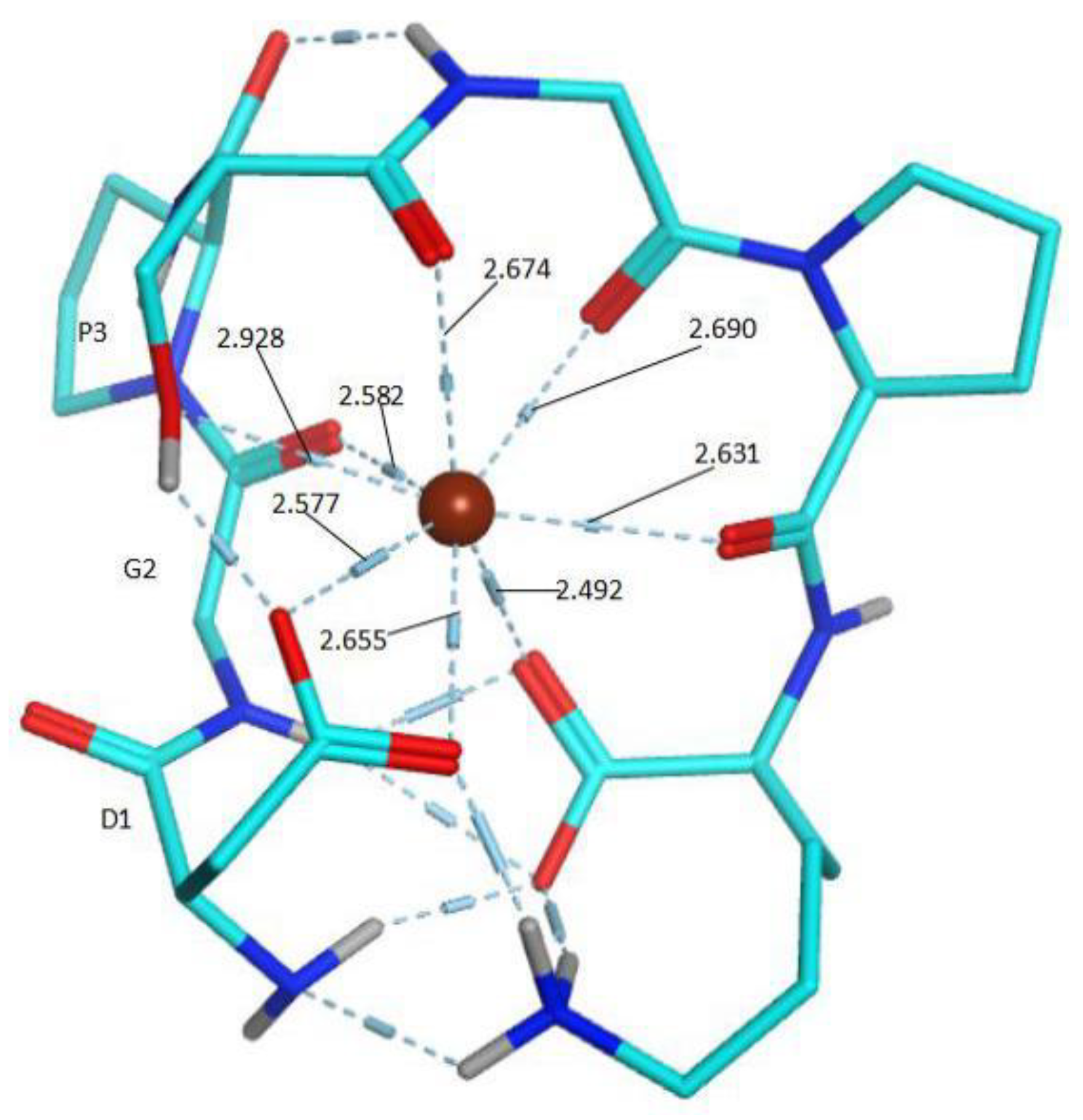
2.4.5. Construction of the Possible Molecular Modes
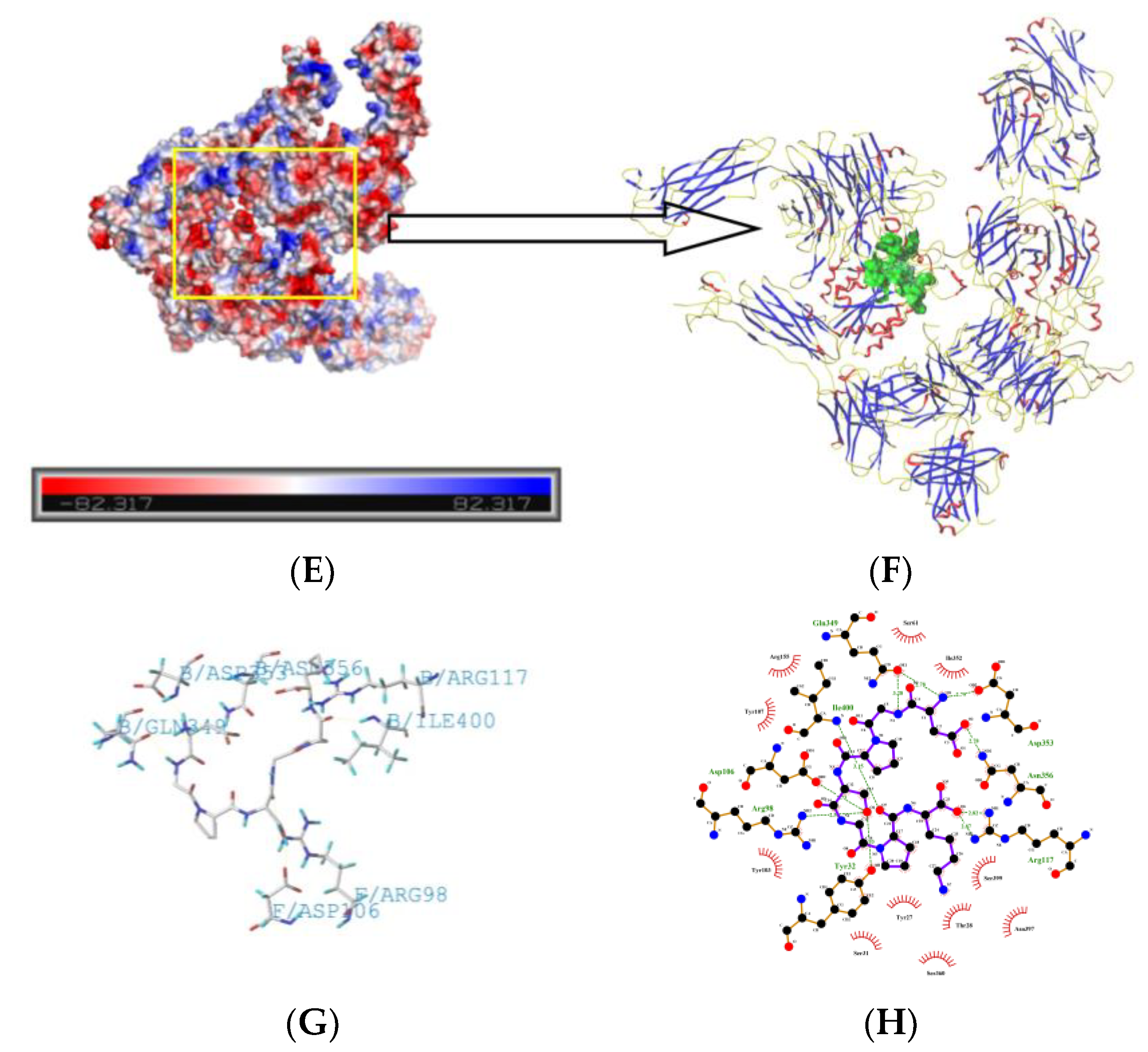
2.5. Molecule Docking
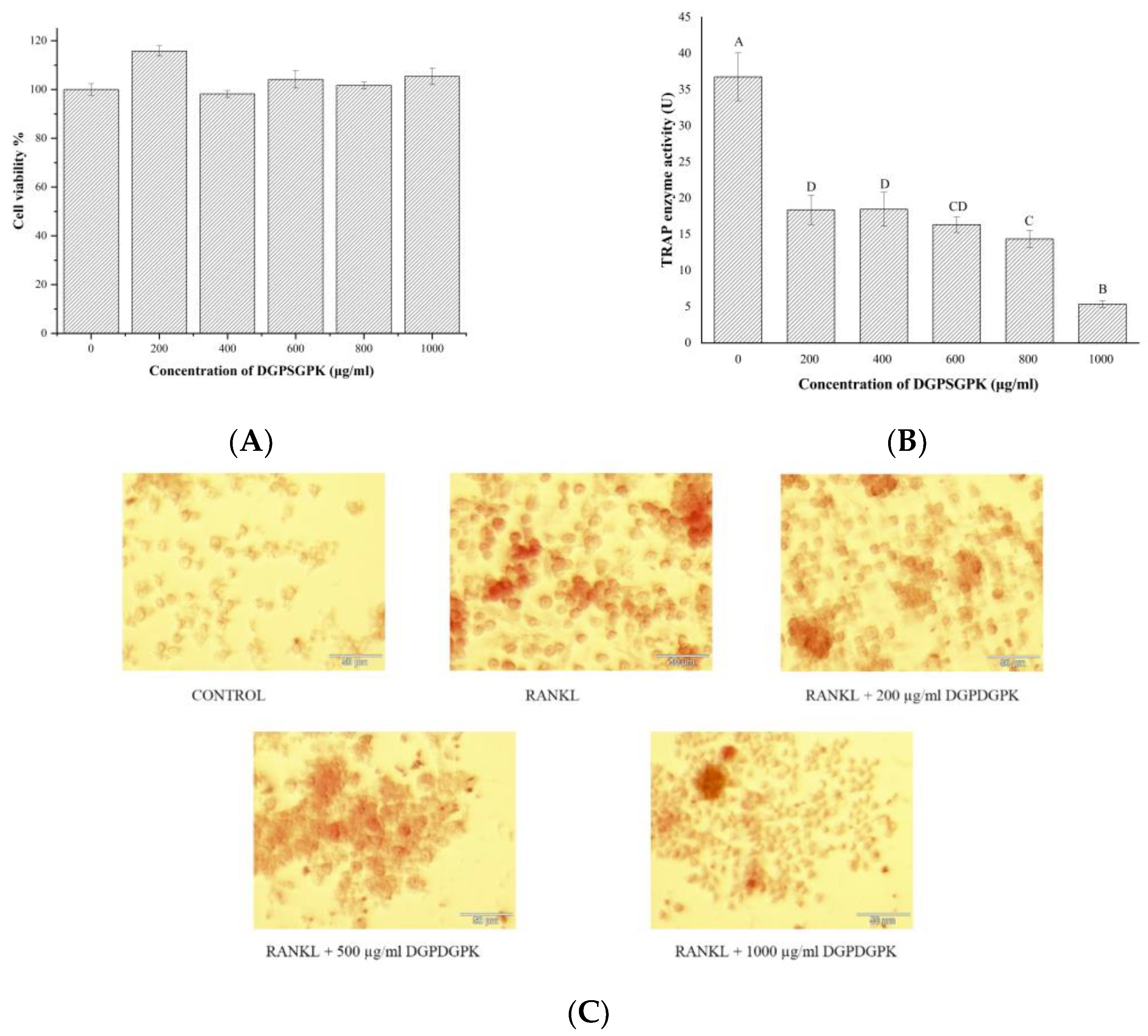
2.6. Effects of DGPSGPK on the RANKL-Induced Osteoclasts
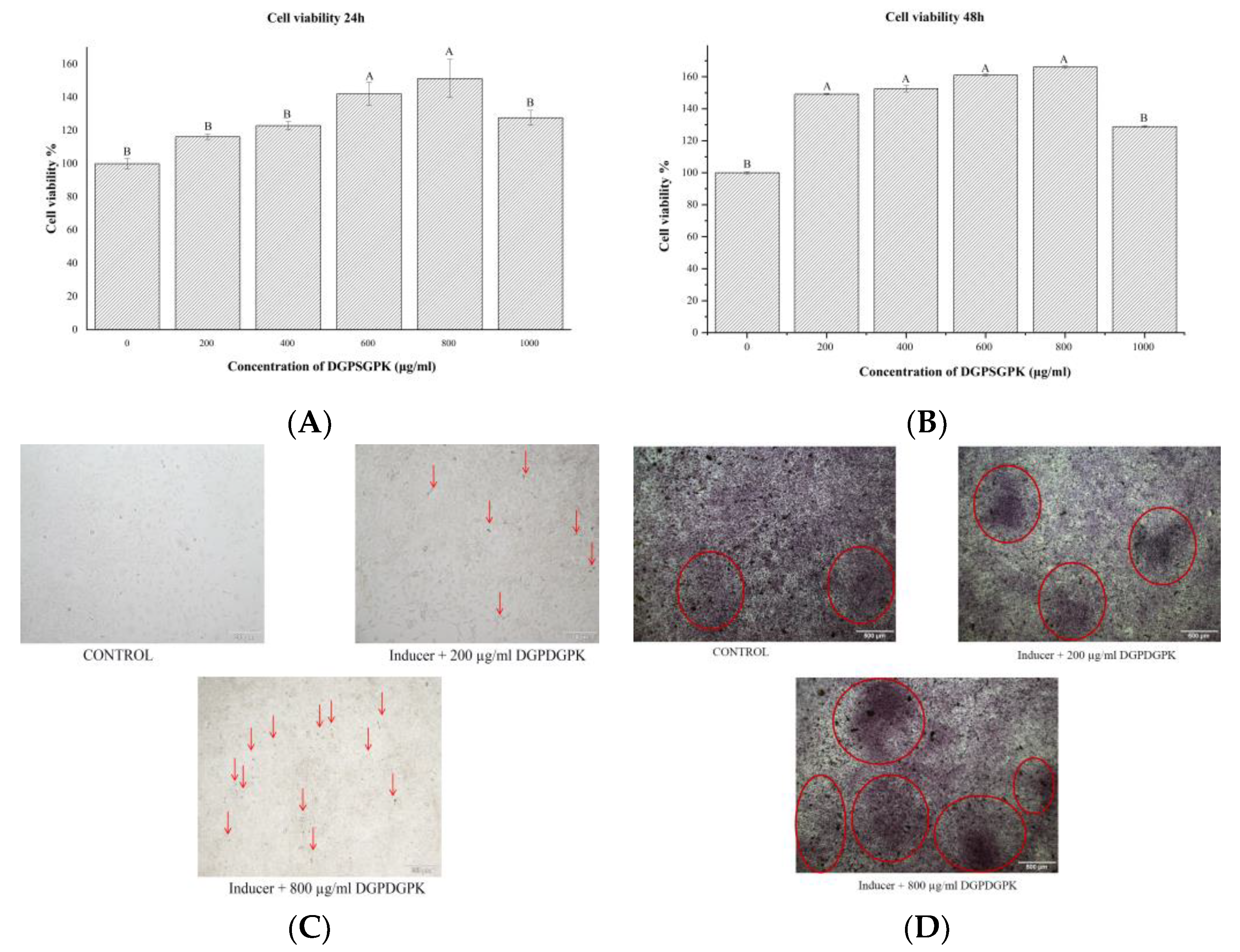
2.7. Effects of DGPSGPK on the Proliferation, Differentiation, and Mineralization of MC3T3-E1 Cells
3. Materials and Methods
3.1. Materials and Reagents
3.2. Preparation of Different Tilapia Bone Enzymatic Hydrolysates
3.3. Determination of DH
3.4. Calcium-Binding Capacity Assay
3.5. Calcium-Binding Peptides
3.6. Identification of Amino Acid Sequence
3.7. Preparation of Calcium–Peptide Chelate
3.8. Structure Characterization of the Calcium–Peptide Chelate
3.8.1. FTIR
3.8.2. XRD
3.8.3. UPLC-Q-Orbitrap-MS2
3.8.4. Construction of the Possible Molecular Modes
3.9. Molecular Docking
3.10. RAW264.7 Cell Culture
3.10.1. TRAP Activity Assay
3.10.2. TRAP Staining
3.11. MC3T3-E1 Cell Culture and Staining
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hou, T.; Wang, C.; Ma, Z.; Lui, W.; He, H. Desalted duck egg white peptides: Promotion of calcium uptake and structure characterization. J. Agric. Food Chem. 2015, 63, 8170–8176. [Google Scholar] [CrossRef]
- Shin, H.L.; Jung, I.Y.; Suck, M.H.; Dae, H.H.; Shin, Y.L.; Il, H.K.; Sang, Y.C. Phosphorylation of peptides derived from isolated soybean protein: Effects on calcium binding, solubility and influx into Caco-2 cells. BioFactors 2005, 23, 121–128. [Google Scholar]
- Caroline, M.G. Calcium-sensing receptor signaling-How human disease informs biology. Curr. Opin. Endocr. Metab. Res. 2021, 16, 10–18. [Google Scholar]
- Giovianni, B.; Jonathan, O.R.; Alan, L.K.; James, A.O. Physicochemical and bulk handling properties of micronised calcium salts and their application in calcium fortification of whey protein-based solutions. J. Food Eng. 2020, 293, 110213. [Google Scholar]
- Guo, L.; Harnedy, P.A.; Li, B.; Hou, H.; Zhang, Z.; Zhao, X.; Fitzgerald, R.J. Food protein-derived chelating peptides: Biofunctional ingredients for dietary mineral bioavailability enhancement. Trends Food Sci. Technol. 2014, 37, 92–105. [Google Scholar] [CrossRef]
- Martina, V.; Leif, H.S. Calcium nutrition. bioavailability and fortification. LWT Food Sci. Technol. 2014, 59, 1198–1204. [Google Scholar]
- Zhang, X.W.; Qi, J.; Li, M.Y.; Liu, H.P.; Wang, Q.; Wu, Y.R.; Niu, L.L.; Liu, Z.T. Isolation of a novel calcium-binding peptide from phosvitin hydrolysates and the study of its calcium chelation mechanism. Food Res. Int. 2021, 141, 110169. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; He, L.; Li, C.; Zhao, S.; Liang, Y.; Yang, F.; Zhang, M.; Jing, G.; Ma, M. Phosphorylation of porcine bone collagen peptide to improve its calcium chelating capacity and its effect on promoting the proliferation, differentiation and mineralization of osteoblastic MC3T3-E1 cells. J. Funct. Foods 2019, 64, 103701. [Google Scholar] [CrossRef]
- Shunichi, Y.; Gen, M.; Tomohiro, S.; Tomoka, H.; Taku, E.; Daisuke, T.; Cai, H.; Yuan, T.; Hend, A.; Masahiko, T.; et al. Cardiotrophin Like Cytokine Factor 1 (CLCF1) alleviates bone loss in osteoporosis mouse models by suppressing osteoclast differentiation through activating interferon signaling and repressing the nuclear factor-κB signaling pathway. Bone 2021, 153, 116140. [Google Scholar]
- Wang, Q.; Shen, X.; Chen, Y.; Chen, J.; Li, Y. Osteoblasts-derived exosomes regulate osteoclast differentiation through miR-503-3p/Hpse axis. Acta Histochem. 2021, 123, 151790. [Google Scholar] [CrossRef]
- Qing, X.B.; Wen, K.; Wu, X.X.; Yao, Z. MiR-183 regulates the differentiation of osteoblasts in the development of osteoporosis by targeting Smad4. Acta Histochem. 2021, 123, 151786. [Google Scholar]
- Zu, Y.; Liang, X.D.; Du, J.; Zhou, S.; Yang, C. Binding of integrin α1 to bone morphogenetic protein receptor IA suggests a novel role of integrin α1β1 in bone morphogenetic protein 2 signalling. J. Biomech. 2015, 48, 3950–3954. [Google Scholar] [CrossRef] [PubMed]
- Zuzana, S.; Carole, L.H.; Sofia, A.; Caroline, M.; Sophie, D.N.; Pascal, S.; Pierre, J.M. Wnt/β-catenin signaling mediates osteoblast differentiation triggered by peptide-induced α5β1 integrin priming in mesenchymal skeletal cells. J. Biol. Chem. 2015, 11, 6903–6912. [Google Scholar]
- Polong, W.; Shaochen, L.; Chiachen, C.; Seiji, M.; Nobuaki, A.; Wenguey, W.; Yoshikazu, T. Non-cytotoxic cobra cardiotoxin A5 binds to αvβ3 integrin and inhibits bone resorption. J. Biol. Chem. 2006, 12, 7937–7945. [Google Scholar]
- Weilin, S.; Matsui, T. Current knowledge of intestinal absorption of bioactive peptides. Food Funct. 2017, 8, 4306–4314. [Google Scholar]
- Chen, D.; Mu, X.; Huang, H.; Nie, R.; Liu, Z.; Zeng, M. Isolation of a calcium-binding peptide from tilapia scale protein hydrolysate and its calcium bioavailability in rats. J. Funct. Foods 2014, 6, 575–584. [Google Scholar] [CrossRef]
- Charoenphun, N.; Cheirsilp, B.; Sirinupong, N.; Youravong, W. Calcium-binding peptides derived from tilapia (Oreochromis niloticus) protein hydrolysate. Eur. Food Res. Technol. 2013, 236, 57–63. [Google Scholar] [CrossRef]
- Liu, B.T.; Zhuang, Y.L.; Sun, L.P. Identification and characterization of the peptides with calcium-binding capacity from tilapia (Oreochromis niloticus) skin gelatin enzymatic hydrolysates. J. Food Sci. 2019, 85, 114–122. [Google Scholar]
- Shun, L.H.; Li, N.Z.; Xixi, C.; Shaoyun, W.; Yifan, H.; Jing, H.; Pingfan, R. Purification and characterisation of a glutamic acid-containing peptide with calcium-binding capacity from whey protein hydrolysate. J. Dairy Res. 2015, 82, 29–35. [Google Scholar]
- Xu, Z.; Chen, H.; Wang, Z.; Fan, F.; Shi, P.; Tu, M.; Du, M. Isolation and characterization of peptides from Mytilus edulis with osteogenic activity in mouse MC3T3-E1 preosteoblast cells. Agric. Food Chem. 2019, 67, 1572–1584. [Google Scholar] [CrossRef]
- Yuan, Y.; Dai, Y.; Zhang, Z.; Gong, Y.; Yuan, Y. Technical efficiency of different farm sizes for tilapia farming in China. Aquac. Res. 2020, 51, 307–315. [Google Scholar] [CrossRef]
- Lin, S.; Hu, X.; Li, L.; Yang, X.; Chen, S.; Wu, Y.; Yang, S. Preparation, purification and identification of iron-chelating peptides derived from tilapia (Oreochromis niloticus) skin collagen and characterization of the peptide-iron complexes. LWT Food Sci. Technol. 2021, 149, 111796. [Google Scholar] [CrossRef]
- Chuesiang, P.; Sanguandeekul, R. Protein hydrolysate from tilapia frame: Antioxidant and angiotensin I converting enzyme inhibitor properties. Int. J. Food Sci. Technol. 2015, 50, 1436–1444. [Google Scholar] [CrossRef]
- Liao, W.; Chen, H.; Jin, W.; Yang, Z.; Cao, Y.; Miao, J. Three newly isolated calcium-chelating peptides from tilapia bone collagen hydrolysate enhance calcium absorption activity in intestinal caco-2 cells. J. Agric. Food Chem. 2020, 68, 2091–2098. [Google Scholar] [CrossRef]
- Zhuang, Y.L.; Sun, L.P. Preparation of reactive oxygen scavenging peptides from tilapia (Oreochromis niloticus) skin gelatin: Optimization using response surface methodology. J. Food Sci. 2011, 76, C483–C489. [Google Scholar] [CrossRef]
- Sun, L.P.; Zhuang, Y.L. Antiphotoaging effect and purification of an antioxidant peptide from tilapia (Oreochromis niloticus) gelatin peptides. J. Funct. Foods 2013, 5, 154–162. [Google Scholar] [CrossRef]
- Zhao, T.; Liu, B.T.; Yuan, L.; Sun, L.P.; Zhuang, Y.L. ACE inhibitory activity in vitro and antihypertensive effect in vivo of LSGYGP and its transepithelial transport by Caco-2 cell monolayer. J. Funct. Foods 2019, 61, 103488. [Google Scholar]
- Clemente, A. Enzymatic protein hydrolysates in human nutrition. Trends Food Sci. Technol. 2001, 11, 254–262. [Google Scholar] [CrossRef]
- Beaubier, S.; Framboisier, X.; Ioannou, I.; Galet, O.; Kapel, R. Simultaneous quantification of the degree of hydrolysis, protein conversion rate and mean molar weight of peptides released in the course of enzymatic proteolysis. J. Chromatogr. B 2018, 1105, 1–9. [Google Scholar] [CrossRef]
- Sandroddin, S.G.; Georges, G. Theory of optimization of the experimental conditions of preparative elution chromatography: Optimization of the column efficiency. Anal. Chem. 2002, 61, 1368–1382. [Google Scholar]
- Yanlan, L.; Xixi, C.; Xiaoping, W.; Shengnan, L.; Shaoyun, W. Fabrication of snapper fish scales protein hydrolysate-calcium complex and the promotion in calcium cellular uptake. J. Funct. Foods 2019, 65, 103717. [Google Scholar]
- Wayne, A.H.; Jerme, K. Carp muscle calcium-binding protein. J. Biol. Chem. 1973, 248, 3313–3326. [Google Scholar]
- Wu, W.; Li, B.; Hu, H.; Zhang, H.; Xue, Z. Isolation and identification of calcium-chelating peptides from pacific cod skin gelatin and their binding properties with calcium. Food Funct. 2017, 8, 103701. [Google Scholar] [CrossRef]
- Cui, P.; Lin, S.; Jin, Z.; Zhu, B.; Liang, S.; Sun, N. In vitro digestion profile and calcium absorption studies of a sea cucumber ovum derived heptapeptide-calcium complex. Food Funct. 2018, 9, 4582–4592. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, Y.; Wang, S. Purification of algal calcium-chelating peptide and its physical chemical properties. J. Aquat. Food Prod. Technol. 2018, 27, 518–530. [Google Scholar] [CrossRef]
- Li, W.; Ding, Y.; Zhang, X.; Li, Y.; Chen, Z. Isolation of a novel calcium-binding peptide from wheat germ protein hydrolysates and the prediction for its mechanism of combination. Food Chem. 2017, 239, 416–426. [Google Scholar]
- Wu, W.; He, L.; Liang, Y.; Yue, L.; Peng, W.; Jin, G.; Ma, M. Preparation process optimization of pig bone collagen peptide-calcium chelate using response surface methodology and its structural characterization and stability analysis. Food Chem. 2019, 284, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Huang, S.; Cai, X.; Hong, J.; Wang, S. A specific peptide with calcium chelating capacity isolated from whey protein hydrolysate. J. Funct. Foods 2014, 10, 46–53. [Google Scholar] [CrossRef]
- Zhang, K.; Li, J.; Hou, H.; Zhang, H.; Li, B. Purification and characterization of a novel calcium-biding decapeptide from pacific cod (Gadus macrocephalus) bone: Molecular properties and calcium chelating modes. J. Funct. Foods 2019, 52, 670–679. [Google Scholar] [CrossRef]
- Cai, X.; Yang, Q.; Lin, J.; Fu, N.; Wang, S. A specific peptide with calcium-binding capacity from defatted Schizochytrium sp. protein hydrolysates and the molecular properties. Molecules 2017, 22, 544. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Gao, A.; Yue, C.; Zhang, X.; Li, S.; Ye, C. Preparation of cucumber seed peptide-calcium chelate by liquid state fermentation and its characterization. Food Chem. 2017, 229, 487–494. [Google Scholar]
- Sun, X.D.; Ruan, S.Y.; Zhuang, Y.L.; Sun, L.P. Anti-osteoporosis effect and purification of peptides with high calcium-binding capacity from walnut protein hydrolysate. Food Funct. 2021, 12, 8454–8466. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Z.; Xu, H.; Li, X.; Hao, X. Preparation of sheep bone collagen peptide-calcium chelate using enzymolysis-fermentation methodology and its structural characterization and stability analysis. RSC Adv. 2020, 10, 11624–11633. [Google Scholar] [CrossRef]
- Tanaka, M.; Hong, S.M.; Akiyama, S.; Hu, Q.Q.; Matsui, T. Visualized absorption of anti-atherosclerotic dipeptide, Trp-His, in Sprague-Dawley rats by LC-MS and MALDI-MS imaging analyses. Mol. Nutr. Food Res. 2015, 59, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tuffery, P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar]
- Ruan, S.Y.; Sun, L.P.; He, J.L.; Zhuang, Y.L. Novel umami peptides from tilapia lower jaw and molecular docking to the taste receptor T1R1/T1R3. Food Chem. 2021, 362, 130249. [Google Scholar]
- Tzu, L.; Rong, Y.; Huang, J.T.; Liou, H.C.; Lin, Y.M.; Woiejer, C.; Fu, W.M. Inhibition of osteoporosis by the αvβ3 integrin antagonist of rhodostomin variants. Eur. J. Pharmacol. 2017, 804, 94–101. [Google Scholar]
- Vermont, P.D.; Elvira, G.M. Lunasin potentiates the effect of oxaliplatin preventing outgrowth of colon cancer metastasis, binds to α5β1 integrin and suppresses FAK/ERK/NF-κB signaling. Cancer Lett. 2011, 313, 167–180. [Google Scholar]
- Roberta, F.; Florian, R.; Stefanie, N.; Jose, M.M.; Javier, G.; Horst, K.; Carlos, M.M. Mimicking bone extracellular matrix: Integrin-binding peptidomimetics enhance osteoblast-like cells adhesion, proliferation, and differentiation on titanium. Colloids Surf. B Biointerfaces 2015, 128, 191–200. [Google Scholar]
- Bondžić, A.M.; Senćanski, M.V.; Nikezić, A.V.V.; Kirillova, M.V.; André, V.; Kirillov, A.M.; Bondžić, B.P. Aminoalcoholate-driven tetracopper(II) cores as dual acetyl and butyrylcholinesterase inhibitors: Experimental and theoretical elucidation of mechanism of action. J. Inorg. Biochem. 2020, 205, 110990. [Google Scholar] [CrossRef]
- Dawei, J.; Min, X.; Chibuike, C.U.; Domonic, A. Physicochemical characterisation, molecular docking, and drug-likeness evaluation of hypotensive peptides encrypted in flaxseed proteome. Curr. Res. Food Sci. 2020, 3, 41–50. [Google Scholar]
- Xu, Z.; Chen, H.; Fan, F.; Shi, P.; Tu, M.; Cheng, S.; Wang, Z.; Du, M. Bone formation activity of an osteogenic dodecapeptide from blue mussels (Mytilus edulis). Food Funct. 2019, 10, 5616–5625. [Google Scholar] [CrossRef]
- Daye, L.; Wan, K.K.; Seong, J.K.; In, B.H.; Je, B.H.; Seung, H.S.; Seil, S. Inhibitory effects of gold and silver nanoparticles on the differentiation into osteoclasts in vitro. Pharmaceutics 2021, 13, 462. [Google Scholar]
- Takeshi, M.; Fumio, A.; Osamu, O.; Katsumasa, T.; Dirk, A.; Toshio, S. An adherent condition is required for formation of multinuclear osteoclasts in the presence of macrophage colony-stimulating factor and receptor activator of nuclear factor κB ligand. Blood 2000, 15, 4335. [Google Scholar]
- Lundberg, P.; Lie, A.; Bjurholm, A.; Lehenkari, P.P.; Horton, M.A.; Lerner, U.H.; Ransjo, M. Vasoactive intestinal peptide regulates osteoclast activity via specific binding sites on both osteoclasts and osteoblasts. Bone 2000, 27, 803–810. [Google Scholar] [CrossRef]
- Kaneda, T.; Yoshida, H.; Nakajima, Y.; Toishi, M.; Nugroho, A.E.; Morita, H. Cyclolinopeptides, cyclic peptides from flaxseed with osteoclast differentiation inhibitory activity. Bioorganic Med. Chem. Lett. 2016, 26, 1760–1761. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Fan, F.; Chen, H.; Xu, Z.; Cheng, S.; Lu, W.L.; Du, M. A bovine lactoferrin-derived peptide induced osteogenesis via regulation of osteoblast proliferation and differentiation. J. Dairy Sci. 2020, 103, 2950–3960. [Google Scholar] [CrossRef]
- Yuki, T.; Masashi, K.; Kiyoko, O.G.; Shunji, H.; Noriko, F. Collagen-derived X-Hyp-Gly-type tripeptides promote differentiation of MC3T3-E1 pre-osteoblasts. J. Funct. Foods 2018, 46, 456–462. [Google Scholar]
- Liu, B.T.; Zhuang, Y.L. Structural characterization of peptide calcium chelate VGLPNSR-Ca and its calcium absorption ability in Caco-2 cell monolayer. Chem. J. Chin. Univ.-Chin. 2019, 40, 1643–1648. [Google Scholar]
- Yuan, L.; Sun, L.P.; Zhuang, Y.L. Preparation and identification of novel inhibitory angiotensin-I-converting enzyme peptides from tilapia skin gelatin hydrolysates: Inhibition kinetics and molecular docking. Food Funct. 2018, 9, 5251–5259. [Google Scholar]
- Hong, S.M.; Tanaka, M.; Koyanagi, R.; Shen, W.; Matsui, T. Structural design of oligopeptides for intestinal transport model. J. Agric. Food Chem. 2016, 64, 2072–2079. [Google Scholar] [CrossRef] [PubMed]

| Types | E/S/% | Temperature/°C | pH | Time/h | Source | Type |
|---|---|---|---|---|---|---|
| Papain | 2.5 | 55 | 7.0 | 4 | Vegetal | Endopeptidase |
| Flavoring protease | 2.5 | 55 | 7.0 | 4 | Microbial | Endo and exopeptidase |
| Complex protease | 2.5 | 55 | 6.0 | 4 | Microbial | Endopeptidase |
| Neutral protease | 2.5 | 50 | 7.0 | 4 | Microbial | Endopeptidase |
| Basic protease | 2.5 | 55 | 9.0 | 4 | Microbial | Endopeptidase |
| Pepsin | 2.5 | 37 | 2.0 | 4 | Animal | Endopeptidase |
| Hydrolytic protease | 2.5 | 60 | 7.0 | 4 | Microbial | Endopeptidase |
| Trypsin | 2.5 | 37 | 8.0 | 4 | Animal | Endopeptidase |
| Animal protease | 2.5 | 50 | 7.0 | 4 | Animal | Endo and exopeptidase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Guo, H.; Zhang, M.; Wang, M.; Sun, L.; Zhuang, Y. Purification and Characterization of a Novel Calcium-Binding Heptapeptide from the Hydrolysate of Tilapia Bone with Its Osteogenic Activity. Foods 2022, 11, 468. https://doi.org/10.3390/foods11030468
He J, Guo H, Zhang M, Wang M, Sun L, Zhuang Y. Purification and Characterization of a Novel Calcium-Binding Heptapeptide from the Hydrolysate of Tilapia Bone with Its Osteogenic Activity. Foods. 2022; 11(3):468. https://doi.org/10.3390/foods11030468
Chicago/Turabian StyleHe, Jinlun, Hao Guo, Mei Zhang, Meng Wang, Liping Sun, and Yongliang Zhuang. 2022. "Purification and Characterization of a Novel Calcium-Binding Heptapeptide from the Hydrolysate of Tilapia Bone with Its Osteogenic Activity" Foods 11, no. 3: 468. https://doi.org/10.3390/foods11030468
APA StyleHe, J., Guo, H., Zhang, M., Wang, M., Sun, L., & Zhuang, Y. (2022). Purification and Characterization of a Novel Calcium-Binding Heptapeptide from the Hydrolysate of Tilapia Bone with Its Osteogenic Activity. Foods, 11(3), 468. https://doi.org/10.3390/foods11030468