Development of Next-Generation Probiotics by Investigating the Interrelationships between Gastrointestinal Microbiota and Diarrhea in Preruminant Holstein Calves
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Analyses of Fecal Score, Plasma Cytokines, Plasma IgG, and Haptoglobin
2.3. Microbiota Analysis
2.4. Isolation and Identification of Bifidobacterium longum subsp. longum
2.5. Antimicrobial Activity by an Agar Spot Test
2.6. Cytokine Production in RAW 264.7 Cells
2.7. Statistical Analysis
3. Results
3.1. The Fecal Microbiota in Healthy and Diarrheic Preruminant Calves
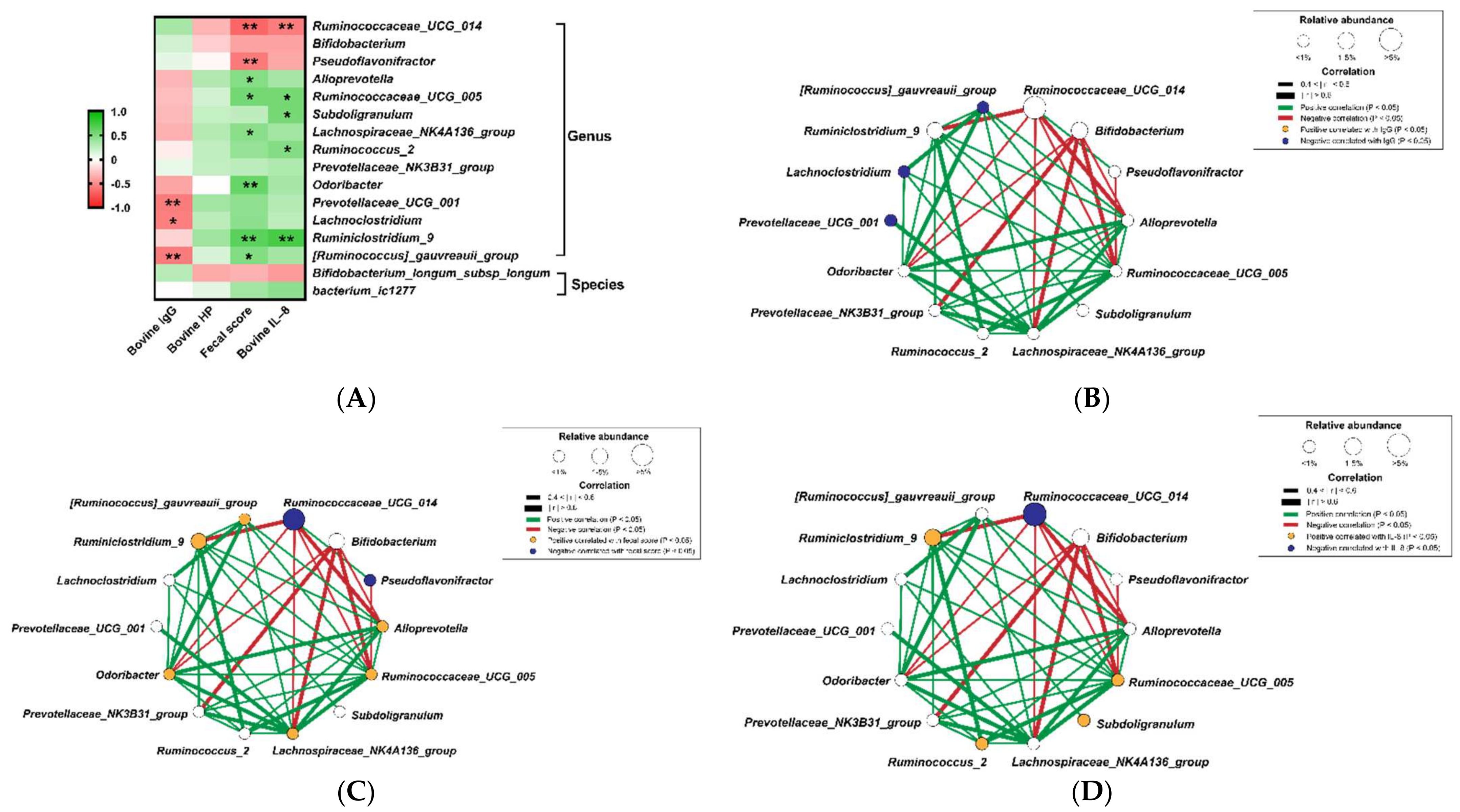
3.2. Identification of the Critical Gastrointestinal Bacterial Biomarkers
3.3. Correlation of Diarrhea and Inflammation Parameters with the Bacterial Biomarkers
3.4. The Relative Abundance of PICRUSt Functional Prediction of Fecal Microbiota in the Preruminant Calves
3.5. Isolation and Identification of Bifidobacterium longum subsp. longum from the Healthy Preruminant Calves
3.6. Determination of Antimicrobial and Immunomodulatory Effects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Millemann, Y. Diagnosis of neonatal calf diarrhoea. Rev. Médecine Vétérinaire 2009, 160, 404–409. [Google Scholar]
- Barrington, G.M.; Gay, J.M.; Evermann, J.F. Biosecurity for neonatal gastrointestinal diseases. Vet. Clin. Food Anim. Pract. 2002, 18, 7–34. [Google Scholar] [CrossRef]
- Ubeda, C.; Pamer, E.G. Antibiotics, microbiota, and immune defense. Trends Immunol. 2012, 33, 459–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.S.; Whon, T.W.; Sung, H.; Jeong, Y.S.; Jung, E.S.; Shin, N.R.; Bae, J.W. Longitudinal evaluation of fecal microbiota transplantation for ameliorating calf diarrhea and improving growth performance. Nat. Commun. 2021, 12, 161. [Google Scholar] [CrossRef]
- Maldonado, N.C.; de Ruiz, C.S.; Otero, M.C.; Sesma, F.; Nader-Macías, M.E. Lactic acid bacteria isolated from young calves–characterization and potential as probiotics. Res. Vet. Sci. 2012, 92, 342–349. [Google Scholar] [CrossRef]
- Roodposhti, P.M.; Dabiri, N. Effects of probiotic and prebiotic on average daily gain, fecal shedding of Escherichia coli, and immune system status in newborn female calves. Asian-Australas. J. Anim. Sci. 2012, 25, 1255. [Google Scholar] [CrossRef] [Green Version]
- Jatkauskas, J.; Vrotniakienė, V. Effects of encapsulated probiotic Enterococcus faecium strain on diarrhoea patterns and performance of early weaned calves. Vet. IR Zootech. 2014, 67, 47–52. [Google Scholar]
- Fernández, S.; Fraga, M.; Silveyra, E.; Trombert, A.N.; Rabaza, A.; Pla, M. Probiotic properties of native Lactobacillus spp. strains for dairy calves. Benef. Microbes 2018, 9, 613–624. [Google Scholar] [CrossRef] [Green Version]
- Alugongo, G.M.; Xiao, J.X.; Chung, Y.H.; Dong, S.Z.; Li, S.L.; Yoon, I. Effects of Saccharomyces cerevisiae fermentation products on dairy calves: Performance and health. J. Dairy Sci. 2018, 100, 1189–1199. [Google Scholar] [CrossRef] [Green Version]
- Uyeno, Y.; Shigemori, S.; Shimosato, T. Effect of probiotics/prebiotics on cattle health and productivity. Microbes Environ. 2015, 30, 126–132. [Google Scholar] [CrossRef] [Green Version]
- Alawneh, J.I.; Barreto, M.O.; Moore, R.J.; Soust, M.; Al-Harbi, H.; James, A.S. Systematic review of an intervention: The use of probiotics to improve health and productivity of calves. Prev. Vet. Med. 2020, 183, 105147. [Google Scholar] [CrossRef] [PubMed]
- McGuirk, S.; Ollivett, T.L. Calf Health Scorer Application (iOS). UWMadison School of Veterinary Medicine. Available online: https://www.vetmed.wisc.edu/fapm/svm-dairy-apps/calf-healthscorer-chs/ (accessed on 17 January 2022).
- Huang, D.B.; DuPont, H.L.; Jiang, Z.D.; Carlin, L.; Okhuysen, P.C. Interleukin-8 response in an intestinal HCT-8 cell line infected with enteroaggregative and enterotoxigenic Escherichia coli. Clin. Diagn. Lab. Immunol. 2014, 11, 548–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Tanaka, R.Y.; Masahiko, M. Improved medium for selective isolation and enumeration of Bifidobacterium. Appl. Environ. Microbiol. 1980, 40, 866–869. [Google Scholar] [CrossRef] [Green Version]
- Harrigan, W.F. Laboratory Methods in Food Microbiology; Gulf Professional Publishing: San Diego, CA, USA, 1998. [Google Scholar]
- Coico, R. Gram staining. Curr. Protoc. Microbiol. 2006, 1, A.3C.1–A.3C.2. [Google Scholar]
- Watanabe, K.; Fujimoto, J.; Sasamoto, M.; Dugersuren, J.; Tumursuh, T.; Demberel, S. Diversity of lactic acid bacteria and yeasts in Airag and Tarag, traditional fermented milk products of Mongolia. World J. Microbio. Biotechnol. 2008, 24, 1313–1325. [Google Scholar] [CrossRef]
- Miyake, T.; Watanabe, K.; Watanabe, T.; Oyaizu, H. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol. Immunol. 1998, 42, 661–667. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Tejero-Sariñena, S.; Barlow, J.; Costabile, A.; Gibson, G.R.; Rowland, I. In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: Evidence for the effects of organic acids. Anaerobe 2012, 18, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.S.; Chen, H.C.; Chen, Y.P.; Chen, M.J. Effects of kefir supernatant and lactic acid bacteria isolated from kefir grain on cytokine production by macrophage. Int. Dairy J. 2009, 19, 244–251. [Google Scholar] [CrossRef]
- Ceciliani, F.; Ceron, J.J.; Eckersall, P.D.; Sauerwein, H. Acute phase proteins in ruminants. J. Proteom. 2012, 75, 4207–4231. [Google Scholar] [CrossRef]
- Skinner, J.G.; Brown, R.A.; Roberts, L. Bovine haptoglobin response in clinically defined field conditions. Vet. Rec. 1991, 128, 147–149. [Google Scholar] [CrossRef]
- Höfner, M.C.; Fosbery, M.W.; Eckersall, P.D.; Donaldson, A.I. Haptoglobin response of cattle infected with foot-and-mouth disease virus. Res. Vet. Sci. 1994, 57, 125–128. [Google Scholar] [CrossRef]
- Ulfman, L.H.; Leusen, J.H.; Savelkoul, H.F.; Warner, J.O.; Van Neerven, R.J. Effects of bovine immunoglobulins on immune function, allergy, and infection. Front. Nutr. 2018, 5, 52. [Google Scholar] [CrossRef]
- Berge, A.C.B.; Besser, T.E.; Moore, D.A.; Sischo, W.M. Evaluation of the effects of oral colostrum supplementation during the first fourteen days on the health and performance of preweaned calves. J. Dairy Sci. 2009, 92, 286–295. [Google Scholar] [CrossRef] [Green Version]
- Jawor, P.; Stefaniak, T.; Mee, J.F. Immune and inflammatory biomarkers in cases of bovine perinatal mortality with and without infection in utero. J. Dairy Sci. 2017, 100, 1408–1416. [Google Scholar] [CrossRef] [Green Version]
- Albayrak, H.; Kabu, M. Determining serum haptoglobin and cytokine concentrations in diarrheic calves. Fırat Univ. Vet. J. Health Sci. 2016, 30, 113–117. [Google Scholar]
- Oikonomou, G.; Teixeira, A.G.V.; Foditsch, C.; Bicalho, M.L.; Machado, V.S.; Bicalho, R.C. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLoS ONE 2013, 8, e63157. [Google Scholar] [CrossRef] [PubMed]
- Foditsch, C.; Pereira, R.V.V.; Ganda, E.K.; Gomez, M.S.; Marques, E.C.; Santin, T. Oral administration of Faecalibacterium prausnitzii decreased the incidence of severe diarrhea and related mortality rate and increased weight gain in preweaned dairy heifers. PLoS ONE 2015, 10, e0145485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef] [Green Version]
- Veiga, P.; Pons, N.; Agrawal, A.; Oozeer, R.; Guyonnet, D.; Brazeilles, R. Changes of the human gut microbiome induced by a fermented milk product. Sci. Rep. 2014, 4, 6328. [Google Scholar] [CrossRef] [Green Version]
- Yasui, H.; Kiyoshima, J.; Ushijima, H. Passive protection against rotavirus-induced diarrhea of mouse pups born to and nursed by dams fed Bifidobacterium breve YIT4064. J. Infect. Dis. 1995, 172, 403–409. [Google Scholar] [CrossRef]
- Perdigón, G.; Locascio, M.; Medici, M.; Holagado, A.P.D.R.; Oliver, G. Interaction of bifidobacteria with the gut and their influence in the immune function. Biocell 2003, 27, 1–9. [Google Scholar] [CrossRef]
- Gomez, D.E.; Arroyo, L.G.; Costa, M.C.; Viel, L.; Weese, J.S. Characterization of the fecal bacterial microbiota of healthy and diarrheic dairy calves. J. Vet. Intern. Med. 2017, 31, 928–939. [Google Scholar] [CrossRef]
- Liu, J.; Bian, G.; Sun, D.; Zhu, W.; Mao, S. Starter feeding supplementation alters colonic mucosal bacterial communities and modulates mucosal immune homeostasis in newborn lambs. Front. Microbiol. 2017, 8, 429. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.R.; Chen, X.D.; Li, J.L.; Yang, Y.T.; Cui, Z.H.; Yao, J.H. Effect of alfalfa hay and starter feed supplementation on caecal microbiota and fermentation, growth, and health of yak calves. Animal 2021, 15, 100019. [Google Scholar] [CrossRef]
- Giri, R.; Hoedt, E.C.; Shamsunnahar, K.; McGuckin, M.A.; Morrison, M.; Capon, R.J. Secreted microbial metabolites modulate gut immunity and inflammatory tone. BioRxiv 2019. [Google Scholar] [CrossRef]
- Kwon, M.S.; Jo, H.E.; Lee, J.; Choi, K.S.; Yu, D.; Oh, Y.S.; Park, J.; Choi, H.-J. Alteration of the gut microbiota in post-weaned calves following recovery from bovine coronavirus-mediated diarrhea. J. Anim. Sci. 2021, 63, 125. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Cao, M.; Zhou, X.; Li, B.; Zhang, J. Epidemic characterization and molecular genotyping of Shigella flexneri isolated from calves with diarrhea in Northwest China. Antimicrob. Resist. Infect. Control 2017, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Fomenky, B.E.; Do, D.N.; Talbot, G.; Chiquette, J.; Bissonnette, N.; Chouinard, Y.P.; Lessard, M.; Ibeagha-Awemu, E.M. Direct-fed microbial supplementation influences the bacteria community composition of the gastrointestinal tract of pre-and post-weaned calves. Sci. Rep. 2018, 8, 14147. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Xue, M.Y.; Sun, H.Z.; Valencak, T.G.; Guan, L.L.; Liu, J. Rumen and hindgut bacteria are potential indicators for mastitis of mid-lactating Holstein dairy cows. Microorganisms 2020, 8, 2042. [Google Scholar] [CrossRef]
- Wan, J.; Hu, S.; Jacoby, J.J.; Liu, J.; Zhang, Y.; Yu, L.L. The impact of dietary sn-2 palmitic triacylglycerols in combination with docosahexaenoic acid or arachidonic acid on lipid metabolism and host faecal microbiota composition in Sprague Dawley rats. Food Funct. 2017, 8, 1793–1802. [Google Scholar] [CrossRef]
- Bach, A.; López-García, A.; González-Recio, O.; Elcoso, G.; Fàbregas, F.; Chaucheyras-Durand, F.; Castex, M. Changes in the rumen and colon microbiota and effects of live yeast dietary supplementation during the transition from the dry period to lactation of dairy cows. J. Dairy Sci. 2019, 102, 6180–6198. [Google Scholar] [CrossRef] [Green Version]
- Hudault, S.; Guignot, J.; Servin, A.L. Escherichia coli strains colonising the gastrointestinal tract protect germfree mice against Salmonella typhimurium infection. Gut 2001, 49, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Satomura, T.; Shimura, D.; Asai, K.; Sadaie, Y.; Hirooka, K.; Fujita, Y. Enhancement of glutamine utilization in Bacillus subtilis through the GlnK-GlnL two-component regulatory system. J. Bacteriol. 2005, 187, 4813–4821. [Google Scholar] [CrossRef] [Green Version]
- Ji, L.; Faponle, A.S.; Quesne, M.G.; Sainna, M.A.; Zhang, J.; Franke, A.; Kumar, D.; van Eldik, R.; Liu, W.; de Visser, S.P. Drug metabolism by cytochrome P450 enzymes: What distinguishes the pathways leading to substrate hydroxylation over desaturation? Chem. Eur. J. 2015, 21, 9083–9092. [Google Scholar] [CrossRef]
- Delcaru, C.; Alexandru, I.; Podgoreanu, P.; Cristea, V.C.; Bleotu, C.; Chifiriuc, M.C.; Bezirtzoglou, E.; Lazar, V. Antagonistic activities of some Bifidobacterium sp. strains isolated from resident infant gastrointestinal microbiota on Gram-negative enteric pathogens. Anaerobe 2016, 39, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Inturri, R.; Trovato, L.; Volti, G.L.; Oliveri, S.; Blandino, G. In vitro inhibitory activity of Bifidobacterium longum BB536 and Lactobacillus rhamnosus HN001 alone or in combination against bacterial and Candida reference strains and clinical isolates. Heliyon 2019, 5, e02891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaud, D.L.; Kelton, D.F.; Weese, J.S.; Noble, C.; Duffield, T.F. Evaluation of a multispecies probiotic as a supportive treatment for diarrhea in dairy calves: A randomized clinical trial. J. Dairy Sci. 2019, 102, 4498–4505. [Google Scholar] [CrossRef] [PubMed]
- Vlková, E.; Grmanová, M.; Killer, J.; Mrázek, J.; Kopečný, J.; Bunešová, V.; Rada, V. Survival of bifidobacteria administered to calves. Folia Microbiol. 2010, 55, 390–392. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Dharmavaram, S.R.; Seo, C.W.; Shahbazi, G. Antimicrobial activity of Bifidobacterium longum (NCFB 2259) as influenced by spices. Int. Nat. J. Food. 2003, 2, 6–8. [Google Scholar]



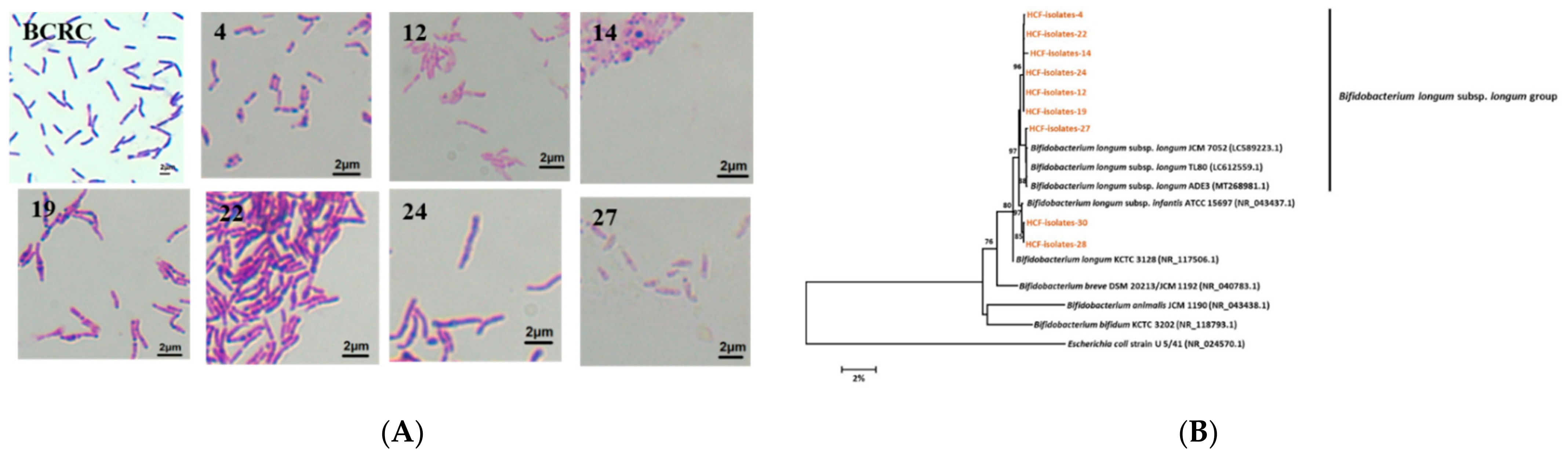

| Isolate | The Nearest Matched Species from GenBank | Similarity (%) |
|---|---|---|
| HCF-4 | B. longum subsp. longum | 97 |
| HCF-12 | B. longum subsp. longum | 97 |
| HCF-14 | B. longum subsp. longum | 97 |
| HCF-19 | B. longum subsp. longum | 97 |
| HCF-22 | B. longum subsp. longum | 97 |
| HCF-24 | B. longum subsp. longum | 97 |
| HCF-27 | B. longum subsp. longum | 97 |
| HCF-28 | B. longum subsp. infantis | 97 |
| HCF-30 | B. longum subsp. infantis | 97 |
| (A) | |||||
|---|---|---|---|---|---|
| Inhibition zone of antimicrobial activity (mm) | |||||
| Isolated B. longum subsp. longum Strain | Pathogens | ||||
| Salmonella enterica | Bacillus cereus | Escherichia coli | Staphylococcus aureus | Vibrio parahaemolyticus | |
| HCF-4 | 0.00 ± 0.00 c | 9.83 ± 0.29 a | 0.00 ± 0.00 b | 12.33 ± 0.58 b | 10.00 ± 1.00 b |
| HCF-12 | 0.00 ± 0.00 c | 10.00 ± 1.00 a | 0.00 ± 0.00 b | 12.83 ± 0.76 b | 10.83 ± 0.76 b |
| HCF-14 | 10.37 ± 0.64 b | 10.67 ± 0.58 a | 0.00 ± 0.00 b | 0.00 ± 0.00 c | 0.00 ± 0.00 c |
| HCF-19 | 10.73 ± 1.62 b | 10.33 ± 0.67 a | 0.00 ± 0.00 b | 11.67 ± 1.53 b | 10.07 ± 1.10 b |
| HCF-22 | 11.83 ± 0.76 b | 9.37 ± 1.10 a | 0.00 ± 0.00 b | 12.50 ± 0.50 b | 10.53 ± 0.84 b |
| HCF-24 | 11.50 ± 0.87 b | 10.73 ± 0.64 a | 0.00 ± 0.00 b | 12.00 ± 0.00 b | 9.33 ± 1.15 b |
| HCF-27 | 0.00 ± 0.00 c | 10.33 ± 1.15 a | 0.00 ± 0.00 b | 13.00 ± 1.00 b | 10.00 ± 1.00 b |
| PC (penicillin) | 35.00 ± 1.70 a | 10.13 ± 1.22 a | 8.15 ± 1.24 a | 15.57 ± 1.52 a | 39.59 ± 2.88 a |
| NC (broth) | 0.00 ± 0.00 c | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 c | 0.00 ± 0.00 c |
| (B) | |||||
| Inhibition zone of antimicrobial activity (mm) | |||||
| Isolated B. longum subsp. longum Strain | Pathogens | ||||
| Salmonella enterica | Bacillus cereus | Escherichia coli | Staphylococcus aureus | Vibrio parahaemolyticus | |
| HCF-4 | – | + | – | ++ | + |
| HCF-12 | – | + | – | ++ | ++ |
| HCF-14 | ++ | ++ | – | – | – |
| HCF-19 | ++ | ++ | – | ++ | ++ |
| HCF-22 | ++ | + | – | ++ | ++ |
| HCF-24 | ++ | ++ | – | ++ | + |
| HCF-27 | – | ++ | – | ++ | ++ |
| PC (penicillin) | +++ | ++ | + | ++ | +++ |
| NC (broth) | – | – | – | – | – |
| (A) | |||||
|---|---|---|---|---|---|
| Inhibition zone of antimicrobial activity (mm) | |||||
| Supernatant of isolated B. longum subsp. longum | Pathogens | ||||
| Salmonella enterica | Bacillus cereus | Escherichia coli | Staphylococcus aureus | Vibrio parahaemolyticus | |
| HCF-4 | 0.00 ± 0.00 c | 10.50 ± 1.32 ab | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| HCF-12 | 0.00 ± 0.00 c | 8.97 ± 0.06 ab | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| HCF-14 | 11.63 ± 1.10 b | 10.67 ± 0.58 ab | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| HCF-19 | 11.37 ± 0.55 b | 9.43 ± 1.01 ab | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| HCF-22 | 0.00 ± 0.00 c | 10.83 ± 1.89 ab | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| HCF-24 | 0.00 ± 0.00 c | 11.30 ± 1.13 a | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| HCF-27 | 0.00 ± 0.00 c | 8.50 ± 0.50 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| PC (penicillin) | 35.00 ± 1.70 a | 10.13 ± 1.22 ab | 8.15 ± 1.24 a | 15.57 ± 1.52 a | 39.59 ± 2.88 a |
| NC (broth) | 0.00 ± 0.00 c | 0.00 ± 0.00 c | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| (B) | |||||
| Inhibition zone of antimicrobial activity (mm) | |||||
| Supernatant of isolated B. longum subsp. longum | Pathogens | ||||
| Salmonella enterica | Bacillus cereus | Escherichia coli | Staphylococcus aureus | Vibrio parahaemolyticus | |
| HCF-4 | – | ++ | – | – | – |
| HCF-12 | – | + | – | – | – |
| HCF-14 | ++ | ++ | – | – | – |
| HCF-19 | ++ | + | – | – | – |
| HCF-22 | – | ++ | – | – | – |
| HCF-24 | – | ++ | – | – | – |
| HCF-27 | – | + | – | – | – |
| PC (penicillin) | ++ | ++ | + | ++ | +++ |
| NC (broth) | – | – | – | – | – |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuang, S.-T.; Chen, C.-T.; Hsieh, J.-C.; Li, K.-Y.; Ho, S.-T.; Chen, M.-J. Development of Next-Generation Probiotics by Investigating the Interrelationships between Gastrointestinal Microbiota and Diarrhea in Preruminant Holstein Calves. Animals 2022, 12, 695. https://doi.org/10.3390/ani12060695
Chuang S-T, Chen C-T, Hsieh J-C, Li K-Y, Ho S-T, Chen M-J. Development of Next-Generation Probiotics by Investigating the Interrelationships between Gastrointestinal Microbiota and Diarrhea in Preruminant Holstein Calves. Animals. 2022; 12(6):695. https://doi.org/10.3390/ani12060695
Chicago/Turabian StyleChuang, Shih-Te, Chien-Ting Chen, Jui-Chun Hsieh, Kuan-Yi Li, Shang-Tse Ho, and Ming-Ju Chen. 2022. "Development of Next-Generation Probiotics by Investigating the Interrelationships between Gastrointestinal Microbiota and Diarrhea in Preruminant Holstein Calves" Animals 12, no. 6: 695. https://doi.org/10.3390/ani12060695
APA StyleChuang, S.-T., Chen, C.-T., Hsieh, J.-C., Li, K.-Y., Ho, S.-T., & Chen, M.-J. (2022). Development of Next-Generation Probiotics by Investigating the Interrelationships between Gastrointestinal Microbiota and Diarrhea in Preruminant Holstein Calves. Animals, 12(6), 695. https://doi.org/10.3390/ani12060695







