Pericentromeric Non-Coding DNA Transcription Is Associated with Niche Impairment in Patients with Ineffective or Partially Effective Multiple Myeloma Treatment
Abstract
:1. Introduction
2. Results
2.1. Hematopoietic Niche Retains Cancer-Associated Attributes after Treatment
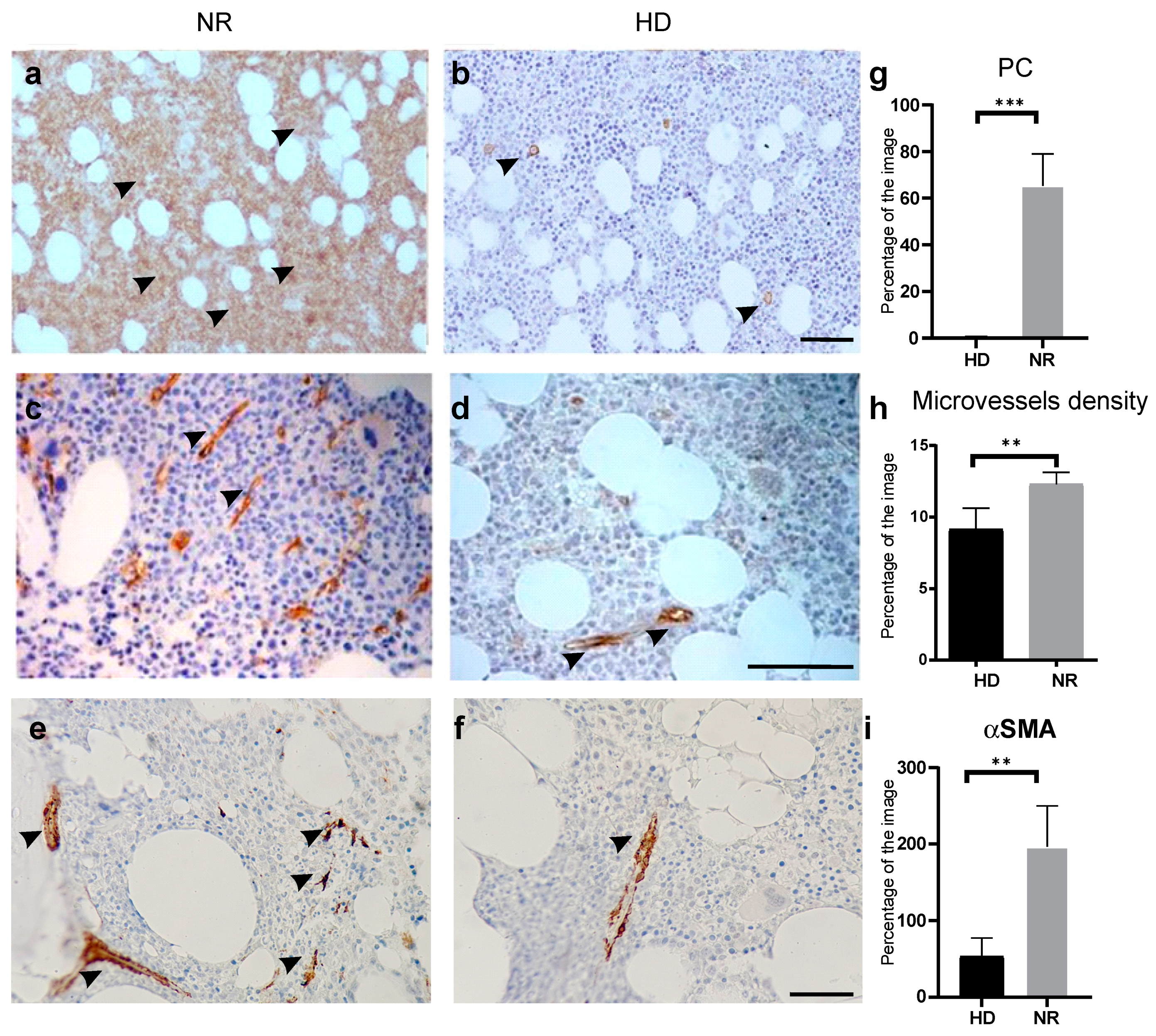
2.1.1. Histological Analysis of MM TME
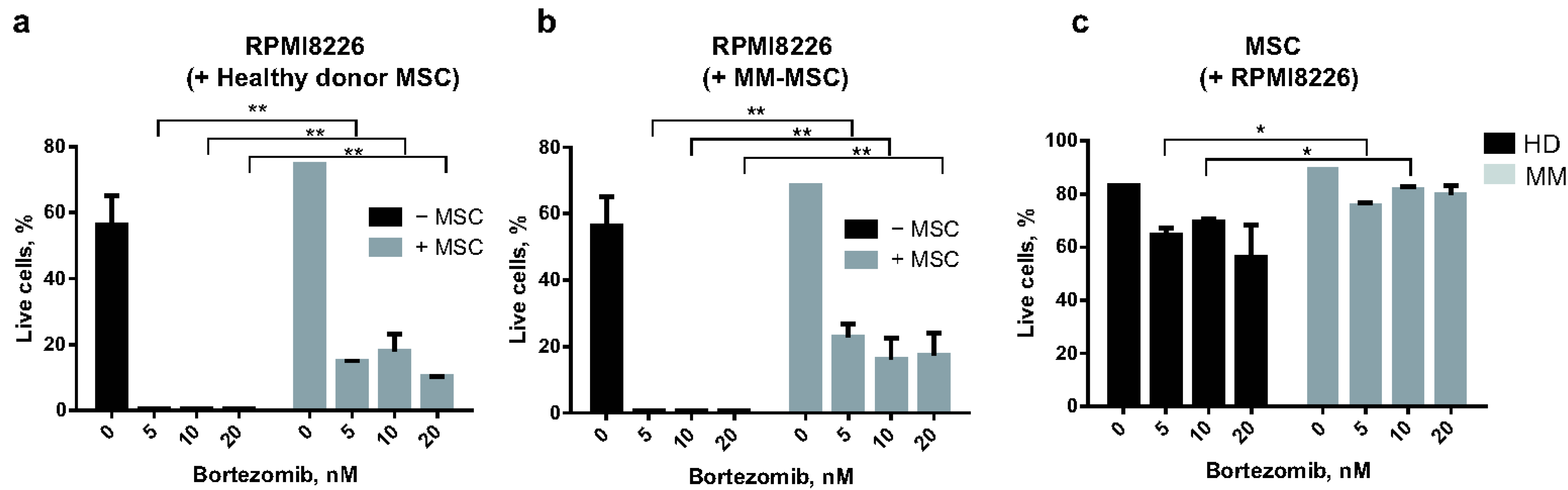
2.1.2. MSC and RPMI8226 Viability in the Presence of Bortezomib
2.1.3. HD and MM-MSC: Comparison of Functional Properties
- Surface markers of RPMI8226 and MSC.
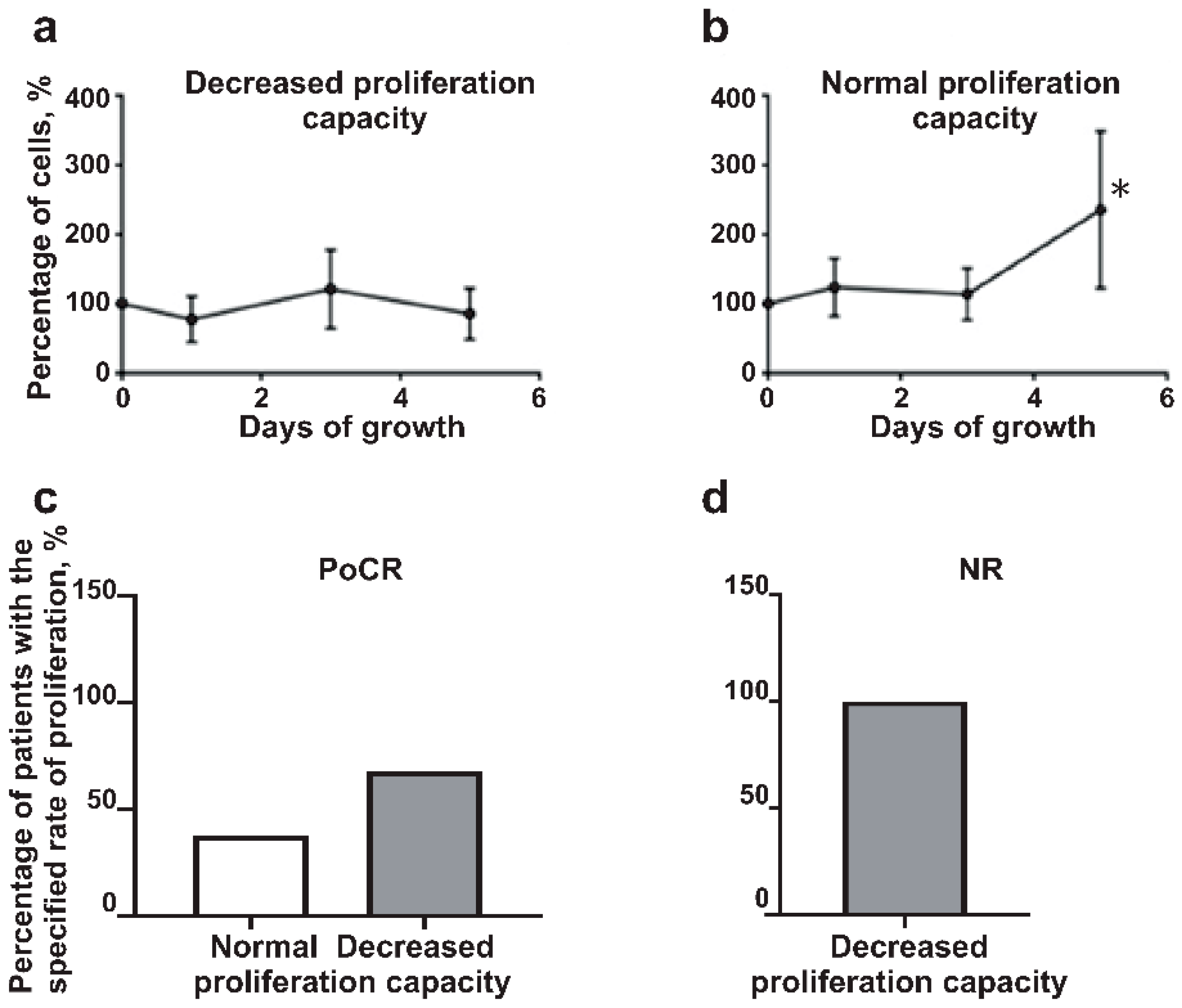
- The proliferation rate of MM-MSC
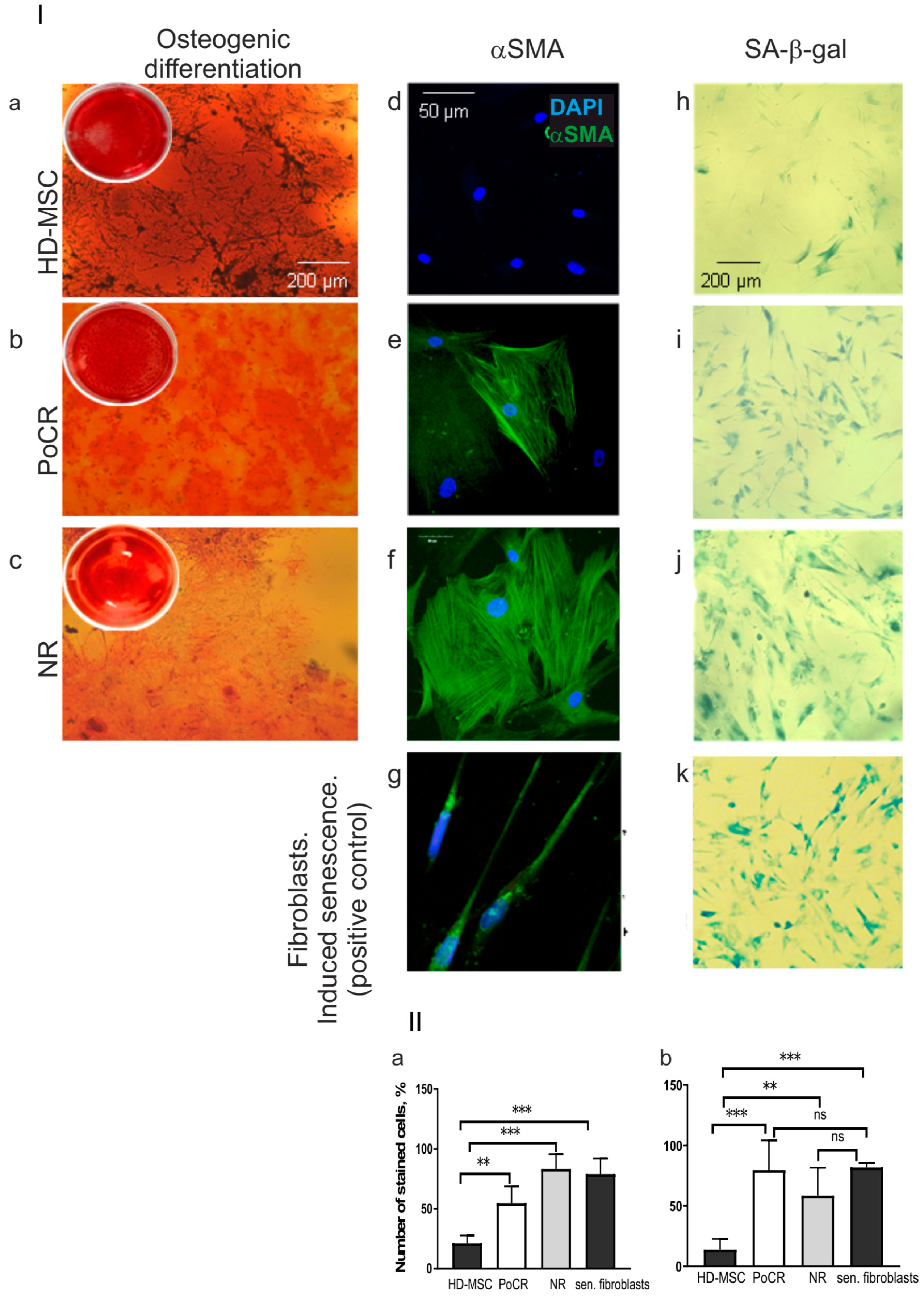
- The osteogenic differentiation of MSC from PoCR and NR patients
- Cancer-associated phenotype of MM-MSC
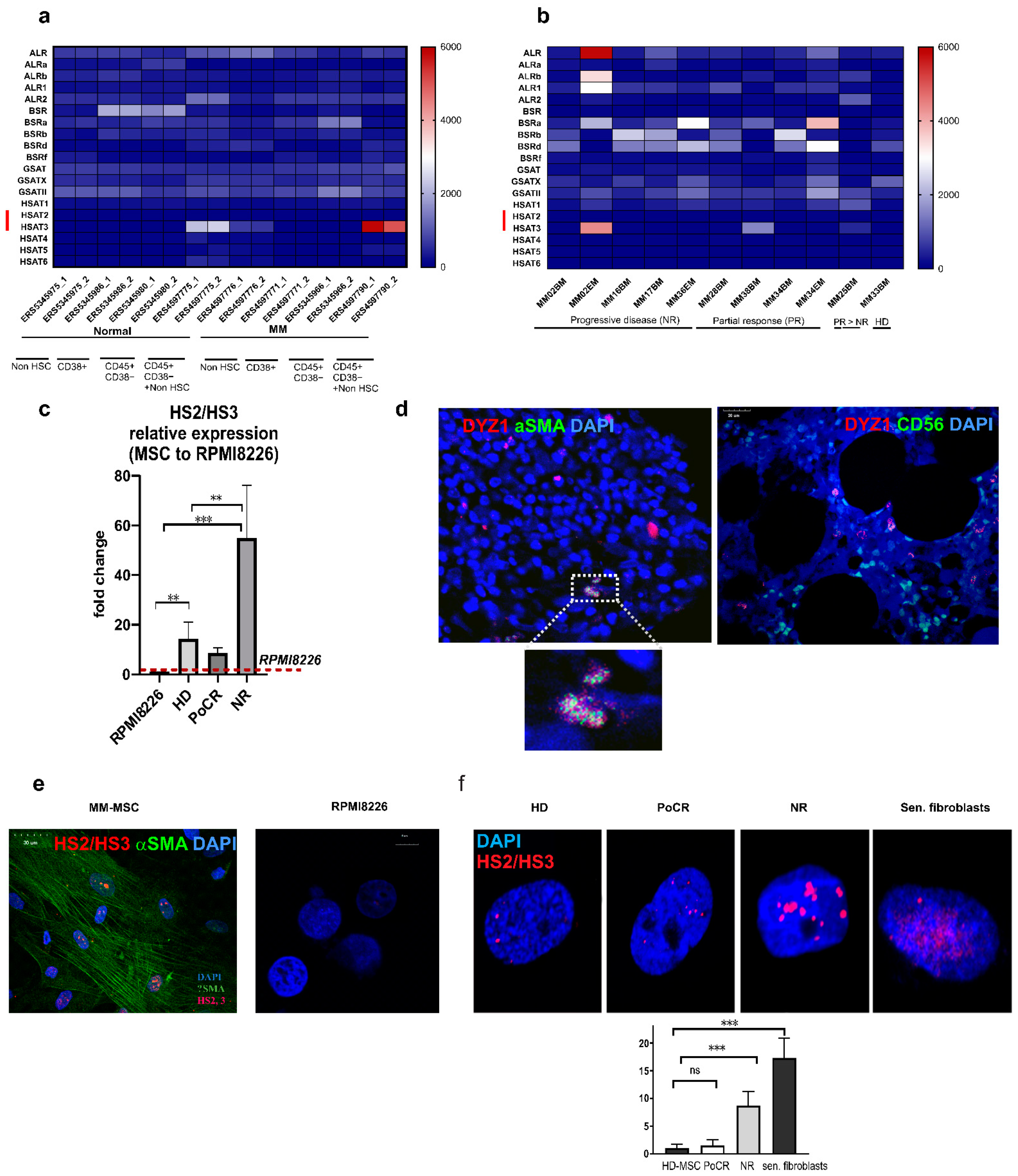
2.2. Transcription of Tandemly Repeated DNA Is Increased in MM-MSC
2.3. Pericentromeric Satellite DNA Transcription in MM-MSC of Patients with Different Response to Treatment
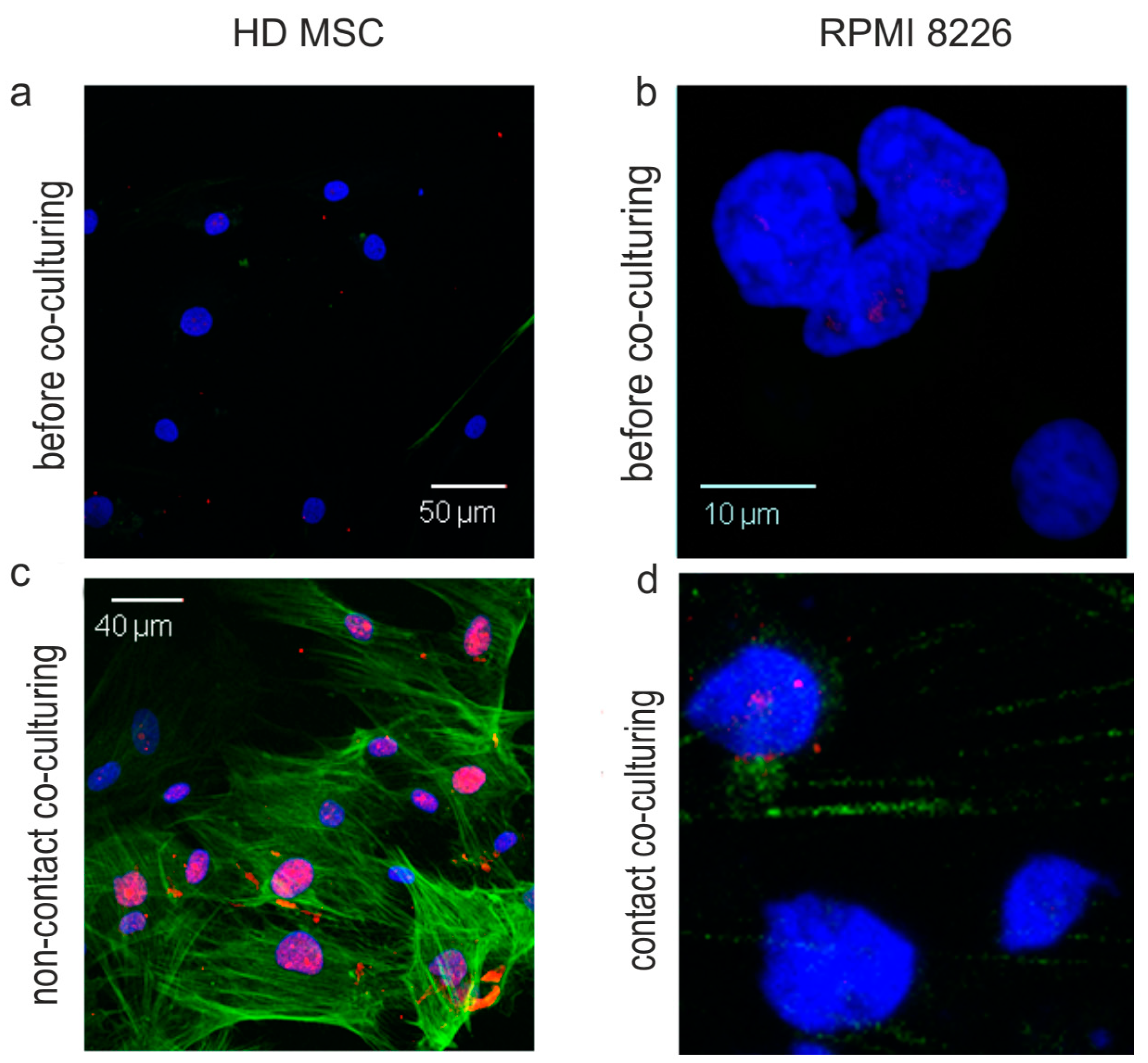
2.4. HS2/HS3 Transcription Is Induced in MM-MSC during Co-Cultivation with RPMI 8226
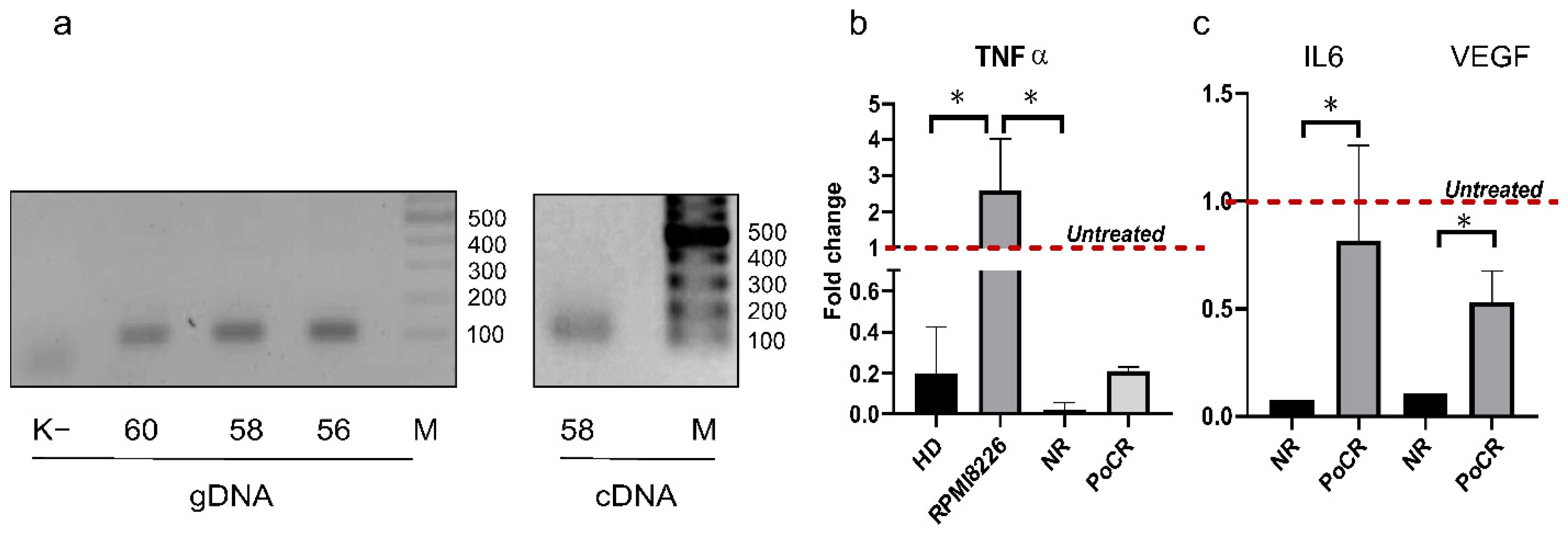
2.5. HS2/HS3 Transcription Is Downregulated in MM-MSC but Upregulated in RPMI8226 after Treatment with TNF-a
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Patients
4.3. Histopathological Examination
4.4. Bioinformatics
4.5. Cell Cultures
4.6. Co-Culturing with RPMI 8226
4.7. RNA Isolation and Quantitative PCR
4.8. Proliferation Rate Assay
4.9. Osteogenic Differentiation Assay
4.10. Galactosidase Staining
4.11. Immunofluorescence Staining
4.12. Flow Cytometry
4.13. DNA-FISH, DNA–RNA FISH, and immunoFISH
4.14. Microscopy
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, S.; De Veirman, K.; De Becker, A.; Vanderkerken, K.; Van Riet, I. Mesenchymal stem cells in multiple myeloma: A therapeutical tool or target? Leukemia 2018, 32, 1500–1514. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Anderson, K. Multiple Myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bessmeltsev, S.S. Multiple myeloma (pathogenesis, clinical features, diagnosis, differential diagnosis). Part I. Clin. Oncohematol. 2013, 6, 237–257. [Google Scholar]
- Wang, H.; Wang, L.; Chi, P.; Wang, W.; Chen, X.; Geng, Q.; Xia, Z.; Lu, Y. High level of interleukin-10 in serum predicts poor prognosis in multiple myeloma. Br. J. Cancer 2016, 114, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Aksenova, A.Y.; Zhuk, A.S.; Lada, A.G.; Zotova, I.V.; Stepchenkova, E.I.; Kostroma, I.I.; Gritsaev, S.V.; Pavlov, Y.I. Genome Instability in Multiple Myeloma: Facts and Factors. Cancers 2021, 13, 5949. [Google Scholar] [CrossRef]
- Bessmeltsev, S.S.; Abdulkadyrov, K.M. Multiple Myeloma. A Guide for Physicians; MK: Moscow, Russia, 2016. [Google Scholar]
- Greenbaum, A.; Hsu, Y.M.S.; Day, R.B.; Schuettpelz, L.G.; Christopher, M.J.; Borgerding, J.N.; Nagasawa, T.; Link, D.C. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 2013, 495, 227–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semenova, N.; Bessmel’tsev, S.; Artyukhina, Z.; Rugal’, V. Changes of hematopoietic stem cell niche in patients with multiple myeloma. Clin. Oncohematol. 2017, 10, 577–578. [Google Scholar]
- Semenova, N.Y.; Bessmel’tsev, S.S.; Rugal’, V.I. Biology of hematopoietic stem cell niche. Clin. Oncohematol. 2014, 7, 501–510. [Google Scholar]
- Romano, A.; Conticello, C.; Cavalli, M.; Vetro, C.; La Fauci, A.; Parrinello, N.L.; Di Raimondo, F. Immunological dysregulation in multiple myeloma microenvironment. Biomed Res. Int. 2014, 2014, 198539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jong, M.M.E.; Kellermayer, Z.; Papazian, N.; Tahri, S.; Hofste op Bruinink, D.; Hoogenboezem, R.; Sanders, M.A.; van de Woestijne, P.C.; Bos, P.K.; Khandanpour, C.; et al. The multiple myeloma microenvironment is defined by an inflammatory stromal cell landscape. Nat. Immunol. 2021, 22, 769–780. [Google Scholar] [CrossRef]
- Fröbel, J.; Landspersky, T.; Percin, G.; Schreck, C.; Rahmig, S.; Ori, A.; Nowak, D.; Essers, M.; Waskow, C.; Oostendorp, R.A.J. The Hematopoietic Bone Marrow Niche Ecosystem. Front. Cell Dev. Biol. 2021, 9, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Butova, R.; Vychytilova-Faltejskova, P.; Souckova, A.; Sevcikova, S.; Hajek, R. Long non-coding RNAs in multiple myeloma. Non-Coding RNA 2019, 5, 13. [Google Scholar] [CrossRef] [Green Version]
- Podgornaya, O.I.; Ostromyshenskii, D.I.; Enukashvily, N.I. Who Needs This Junk, or Genomic Dark Matter. Biochemistry 2018, 83, 450–466. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef]
- Ting, D.T.; Lipson, D.; Paul, S.; Brannigan, B.W.; Akhavanfard, S.; Coffman, E.J.; Contino, G.; Deshpande, V.; Iafrate, A.J.; Letovsky, S.; et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 2011, 331, 593–596. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; Hoong, N.; Aslanian, A.; Hara, T.; Benner, C.; Heinz, S.; Miga, K.H.; Ke, E.; Verma, S.; Soroczynski, J.; et al. Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer. Mol. Cell 2018, 70, 842–853.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enukashvily, N.I.; Donev, R.; Waisertreiger, I.S.-R.; Podgornaya, O.I. Human chromosome 1 satellite 3 DNA is decondensed, demethylated and transcribed in senescent cells and in A431 epithelial carcinoma cells. Cytogenet. Genome Res. 2007, 118, 42–54. [Google Scholar] [CrossRef]
- Wilson, A.S.; Power, B.E.; Molloy, P.L. DNA hypomethylation and human diseases. Biochim. Biophys. Acta Rev. Cancer 2007, 1775, 138–162. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Fabris, S.; Pegoraro, V.; Ronchetti, D.; Mosca, L.; Deliliers, G.L.; Motta, V.; Bertazzi, P.A.; Baccarelli, A.; Neri, A. Differential repetitive DNA methylation in multiple myeloma molecular subgroups. Carcinogenesis 2009, 30, 1330–1335. [Google Scholar] [CrossRef] [Green Version]
- Shabaneh, T.B.; Downey, S.L.; Goddard, A.L.; Screen, M.; Lucas, M.M.; Eastman, A.; Kisselev, A.F. Molecular Basis of Differential Sensitivity of Myeloma Cells to Clinically Relevant Bolus Treatment with Bortezomib. PLoS ONE 2013, 8, 10–13. [Google Scholar] [CrossRef]
- Garderet, L.; Mazurier, C.; Chapel, A.; Ernou, I.; Boutin, L.; Holy, X.; Gorin, N.C.; Lopez, M.; Doucet, C.; Lataillade, J.-J. Mesenchymal stem cell abnormalities in patients with multiple myeloma. Leuk. Lymphoma 2007, 48, 2032–2041. [Google Scholar] [CrossRef] [PubMed]
- Schmitt-Gräff, A.; Skalli, O.; Gabbiani, G. Alpha-smooth muscle actin is expressed in a subset of bone marrow stromal cells in normal and pathological conditions. Virchows Arch. B. Cell Pathol. Incl. Mol. Pathol. 1989, 57, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cheng, Q.; Li, X.; Liu, J.; Ye, Q.; Chen, Y.; Tan, S. Multiple Myeloma-Derived Exosomes Regulate the Functions of Mesenchymal Stem Cells Partially via Modulating miR-21 and miR-146a. Stem Cells Int. 2017, 2017, 9012152. [Google Scholar] [CrossRef]
- De Veirman, K.; Rao, L.; De Bruyne, E.; Menu, E.; Van Valckenborgh, E.; Van Riet, I.; Frassanito, M.A.; Di Marzo, L.; Vacca, A.; Vanderkerken, K. Cancer associated fibroblasts and tumor growth: Focus on multiple myeloma. Cancers 2014, 6, 1363–1381. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, Y.; Fei, C.; Zhao, S.; Zheng, Q.; Su, J.; Wu, D.; Li, X.; Chang, C. Dicer1 downregulation by multiple myeloma cells promotes the senescence and tumor-supporting capacity and decreases the differentiation potential of mesenchymal stem cells article. Cell Death Dis. 2018, 9, 512. [Google Scholar] [CrossRef]
- Hall, L.L.; Byron, M.; Carone, D.M.; Whitfield, T.W.; Pouliot, G.P.; Fischer, A.; Jones, P.; Lawrence, J.B. Demethylated HSATII DNA and HSATII RNA Foci Sequester PRC1 and MeCP2 into Cancer-Specific Nuclear Bodies. Cell Rep. 2017, 18, 2943–2956. [Google Scholar] [CrossRef] [PubMed]
- Ponomartsev, N.V.; Brichkina, A.I.; Enukashvily, N.I. Pericentromeric tandem DNA transcription in malignant cells and tumour microenvironment in mice NSLC model. Biopolym. Cell 2019, 35, 189–190. [Google Scholar] [CrossRef]
- Ryu, D.; Kim, S.J.; Hong, Y.; Jo, A.; Kim, N.; Kim, H.J.; Lee, H.O.; Kim, K.; Park, W.Y. Alterations in the transcriptional programs of myeloma cells and the microenvironment during extramedullary progression affect proliferation and immune evasion. Clin. Cancer Res. 2020, 26, 935–944. [Google Scholar] [CrossRef] [Green Version]
- Evdokimova, V.; Ruzanov, P.; Gassmann, H.; Zaidi, S.H.; Peltekova, V.; Heisler, L.E.; McPherson, J.D.; Orlic-Milacic, M.; Specht, K.; Steiger, K.; et al. Exosomes transmit retroelement RNAs to drive inflammation and immunosuppression in Ewing Sarcoma. bioRxiv 2019, 806851. [Google Scholar] [CrossRef] [Green Version]
- Solovyov, A.; Vabret, N.; Arora, K.S.; Snyder, A.; Funt, S.A.; Bajorin, D.F.; Rosenberg, J.E.; Bhardwaj, N.; Ting, D.T.; Greenbaum, B.D. Global Cancer Transcriptome Quantifies Repeat Element Polarization between Immunotherapy Responsive and T Cell Suppressive Classes. Cell Rep. 2018, 23, 512–521. [Google Scholar] [CrossRef]
- Ho, X.D.; Nguyen, H.G.; Trinh, L.H.; Reimann, E.; Prans, E.; Kõks, G.; Maasalu, K.; Le, V.Q.; Nguyen, V.H.; Le, N.T.N.; et al. Analysis of the expression of repetitive DNA elements in osteosarcoma. Front. Genet. 2017, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musolino, C.; Allegra, A.; Innao, V.; Allegra, A.G.; Pioggia, G.; Gangemi, S. Inflammatory and Anti-Inflammatory Equilibrium, Proliferative and Antiproliferative Balance: The Role of Cytokines in Multiple Myeloma. Mediators Inflamm. 2017, 2017, 1852517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobrynin, M.A.; Korchagina, N.M.; Prjibelski, A.D.; Shafranskaya, D. Human pericentromeric tandemly repeated DNA is transcribed at the end of oocyte maturation and is associated with membraneless mitochondria–Associated structures. Sci. Rep. 2020, 10, 19634. [Google Scholar] [CrossRef]
- Valgardsdottir, R.; Chiodi, I.; Giordano, M.; Cobianchi, F.; Riva, S.; Biamonti, G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol. Biol. Cell 2005, 16, 2597–2604. [Google Scholar] [CrossRef] [Green Version]
- Ichii, M.; Hosen, N. Current understanding of myelomatous mesenchymal stromal cells extended through advances in experimental methods. Cancers 2021, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Mosteo, L.; Storer, J.; Batta, K.; Searle, E.J.; Duarte, D.; Wiseman, D.H. The Dynamic Interface Between the Bone Marrow Vascular Niche and Hematopoietic Stem Cells in Myeloid Malignancy. Front. Cell Dev. Biol. 2021, 9, 1–24. [Google Scholar] [CrossRef]
- Kunisaki, Y.; Bruns, I.; Scheiermann, C.; Ahmed, J.; Pinho, S.; Zhang, D.; Mizoguchi, T.; Wei, Q.; Lucas, D.; Ito, K.; et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature 2013, 502, 637–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussong, J.W.; Rodgers, G.M.; Shami, P.J. Evidence of increased angiogenesis in patients with acute myeloid leukemia. Blood 2000, 95, 309–313. [Google Scholar] [CrossRef]
- Giuliani, N.; Colla, S.; Morandi, F.; Lazzaretti, M.; Sala, R.; Bonomini, S.; Grano, M.; Colucci, S.; Svaldi, M.; Rizzoli, V. Myeloma cells block RUNX2/CBFA1 activity in human bone marrow osteoblast progenitors and inhibit osteoblast formation and differentiation. Blood 2005, 106, 2472–2483. [Google Scholar] [CrossRef] [PubMed]
- Tian, E.; Zhan, F.; Walker, R.; Rasmussen, E.; Ma, Y.; Barlogie, B.; Shaughnessy, J.D. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N. Engl. J. Med. 2003, 349, 2483–2494. [Google Scholar] [CrossRef] [PubMed]
- Phinney, D.G. Functional Heterogeneity of Mesenchymal Stem Cells: Implications for Cell Therapy. J. Cell. Biochem. 2012, 113, 2806–2812. [Google Scholar] [CrossRef] [PubMed]
- André, T.; Meuleman, N.; Stamatopoulos, B.; De Bruyn, C.; Pieters, K.; Bron, D.; Lagneaux, L. Evidences of Early Senescence in Multiple Myeloma Bone Marrow Mesenchymal Stromal Cells. PLoS ONE 2013, 8, e59756. [Google Scholar] [CrossRef] [PubMed]
- Talele, N.P.; Fradette, J.; Davies, J.E.; Kapus, A.; Hinz, B. Expression of α-Smooth Muscle Actin Determines the Fate of Mesenchymal Stromal Cells. Stem Cell Rep. 2015, 4, 1016–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiang, Y.W.; Hu, B.; Chen, Y.; Zhong, Y.; Shi, B.; Barlogie, B.; Shaughnessy, J.D. Bortezomib induces osteoblast differentiation via Wnt-independent activation of β-catenin/TCF signaling. Blood 2009, 113, 4319–4330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Fan, R.; Lei, L.; Lei, L.; Wang, Y.; Lv, N.; Chen, P.; Williamson, R.A.; Wang, B.; Hu, J. Cell cycle exit during bortezomib-induced osteogenic differentiation of mesenchymal stem cells was mediated by Xbp1s-upregulated p21 Cip1 and p27 Kip1. J. Cell. Mol. Med. 2020, 24, 9428–9438. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, H.; Mundel, T.M.; Kieran, M.W.; Kalluri, R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol. Ther. 2006, 5, 1640–1646. [Google Scholar] [CrossRef] [Green Version]
- Frassanito, M.A.; De Veirman, K.; Desantis, V.; Di Marzo, L.; Vergara, D.; Ruggieri, S.; Annese, T.; Nico, B.; Menu, E.; Catacchio, I.; et al. Halting pro-survival autophagy by TGFβ inhibition in bone marrow fibroblasts overcomes bortezomib resistance in multiple myeloma patients. Leukemia 2016, 30, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.H. Transposable elements in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Younger, S.T.; Rinn, J.L. Silent pericentromeric repeats speak out. Proc. Natl. Acad. Sci. USA 2015, 112, 15008–15009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanne, A.; Muniz, L.R.; Puzio-Kuter, A.; Leonova, K.I.; Gudkov, A.V.; Ting, D.T.; Monasson, R.; Cocco, S.; Levine, A.J.; Bhardwaj, N.; et al. Distinguishing the immunostimulatory properties of noncoding RNAs expressed in cancer cells. Proc. Natl. Acad. Sci. USA 2015, 112, 15154–15159. [Google Scholar] [CrossRef] [Green Version]
- Bronkhorst, A.J.; Wentzel, J.F.; Ungerer, V.; Peters, D.L.; Aucamp, J.; de Villiers, E.P.; Holdenrieder, S.; Pretorius, P.J. Sequence analysis of cell-free DNA derived from cultured human bone osteosarcoma (143B) cells. Tumor Biol. 2018, 40, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hideshima, T.; Chauhan, D.; Schlossman, R.; Richardson, P.; Anderson, K.C. The role of tumor necrosis factor α in the pathophysiology of human multiple myeloma: Therapeutic applications. Oncogene 2001, 20, 4519–4527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamik, J.; Jin, S.; Sun, Q.; Zhang, P.; Weiss, K.R.; Anderson, J.L.; Silbermann, R.; Roodman, G.D.; Galson, D.L. EZH2 or HDAC1 inhibition reverses multiple myeloma-induced epigenetic suppression of osteoblast differentiation. Mol. Cancer Res. 2017, 15, 405–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Medical Association Declaration of Helsinki, Ethical Principles for Scientific Requirements and Research Protocols. Bull. World Health Organ. 2013, 79, 373.
- Mendeleeva, L.P.; Votyakova, O.M.; Pokrovskaya, O.S.; Rekhtina, I.G.; Darskaya, E.I.; Galtseva, I.V.; Kaplanov, K.D.; Motorin, D.V.; Samoylova, O.S.; Semochkin, S.V.; et al. National clinical recommendations on diagnosis and treatment of multiple myeloma. Hematol. Transfusiol. 2016, 61, 1–24. [Google Scholar] [CrossRef]
- Lee, N.; Moon, S.Y.; Lee, J.H.; Park, H.K.; Kong, S.Y.; Bang, S.M.; Lee, J.H.; Yoon, S.S.; Lee, D.S. Discrepancies between the percentage of plasma cells in bone marrow aspiration and BM biopsy: Impact on the revised IMWG diagnostic criteria of multiple myeloma. Blood Cancer J. 2017, 7, e530. [Google Scholar] [CrossRef]
- Bessmeltsev, S.S. Multiple myeloma (management of newly diagnosed patients): Literature review and our own data. Part II. Clin. Oncohematol. 2013, 6, 379–414. [Google Scholar]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Ivanovic, Z. Hypoxia or in situ normoxia: The stem cell paradigm. J. Cell. Physiol. 2009, 219, 271–275. [Google Scholar] [CrossRef]
- Buravkova, L.B.; Andreeva, E.R.; Gogvadze, V.; Zhivotovsky, B. Mesenchymal stem cells and hypoxia: Where are we? Mitochondrion 2014, 19, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Hannah, S.S.; McFadden, S.; McNeilly, A.; McClean, C. “Take My Bone Away?” Hypoxia and bone: A narrative review. J. Cell. Physiol. 2021, 236, 721–740. [Google Scholar] [CrossRef]
- Moore, G.E.; Kitamura, H. Cell line derived from patient with myeloma. N. Y. State J. Med. 1968, 68, 2054–2060. [Google Scholar] [PubMed]
- Eccles, M.; Li, C. Senescence Associated β-galactosidase Staining. Bio-protocol 2012, 2, e247. [Google Scholar] [CrossRef]
- Kozhukharova, I.; Zemelko, V.; Kovaleva, Z.; Alekseenko, L.; Lyublinskaya, O.; Nikolsky, N. Therapeutic doses of doxorubicin induce premature senescence of human mesenchymal stem cells derived from menstrual blood, bone marrow and adipose tissue. Int. J. Hematol. 2018, 107, 286–296. [Google Scholar] [CrossRef]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J.; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K.; et al. Landscape of somatic retrotransposition in human cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| PC < 10% (n = 9) | Type of Response | Diagnosis | Type of M Protein | BM Infiltration | Microvessel Density of BM | Myelogram, Percentage of PC | M-Component in Serum (in Urine for Bence-Jones Protein), g/L | Karyotype | MRD | Treatment |
| SD | non-secretory MM | − | 10% | 7.20% | 6% | 0 | No abnormalities | + | 4 VCD courses | |
| CR | IIA | Ig G κ pa | 10% | 8.90% | 3% | 0 | No abnormalities | + | 6 VCD courses, aHSCT | |
| PR | IIIA | Ig A κ | 5% | 10.20% | 2.40% | 4 | No abnormalities | + | 5 VCD courses | |
| CR | IIIB | Ig G λ | 1–2% | 7.40% | 1% | 0 | No abnormalities | + | 5 VCD courses, aHSCT | |
| VGPR | IIIA | Ig G κ | 3% | 8.50% | 3.60% | 10.7 | No abnormalities | - | 2 VRD courses + 2 KRd courses, aHSCT | |
| VGPR | IIIA | Ig G κ | 30% | 11.70% | 2.20% | 7.7 | 13q14 and 13q34 deletions | + | 3 VCD courses, aHSCT | |
| PR | IIIA | Ig G κ | 1–2% | 10.60% | 1.40% | 15 | No abnormalities | + | 5 VCD courses, aHSCT | |
| CR | Bence-Jones MM IIIA | Ig G κ | 1–2% | 8.60% | 2.60% | 0 | No abnormalities | - | 6 VD courses | |
| VGPR | IIA | Ig G λ | 1–2% | 9.20% | 2.80% | 0 | No abnormalities | - | VRD, aHSCT | |
| Non-responders (n = 3) | SD | IIIA | Ig G κ | 50% | 13.00% | 47% | 0.47 | 14q32 monosomy, 13q14 and 13q34 deletions, TP53/17p13 deletions or monosomy | + | 6 VD course, aHSCT |
| SD | IIIA | Ig A κ | 50% | 12.50% | 14.40% | 6 | No abnormalities | + | 3 VCD course, aHSCT | |
| SD | IIIA | Ig G λ | 90% | 11.40% | 82.40% | 0 | Y chromosome loss | + | 2 VCD course |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enukashvily, N.I.; Semenova, N.; Chubar, A.V.; Ostromyshenskii, D.I.; Gushcha, E.A.; Gritsaev, S.; Bessmeltsev, S.S.; Rugal, V.I.; Prikhodko, E.M.; Kostroma, I.; et al. Pericentromeric Non-Coding DNA Transcription Is Associated with Niche Impairment in Patients with Ineffective or Partially Effective Multiple Myeloma Treatment. Int. J. Mol. Sci. 2022, 23, 3359. https://doi.org/10.3390/ijms23063359
Enukashvily NI, Semenova N, Chubar AV, Ostromyshenskii DI, Gushcha EA, Gritsaev S, Bessmeltsev SS, Rugal VI, Prikhodko EM, Kostroma I, et al. Pericentromeric Non-Coding DNA Transcription Is Associated with Niche Impairment in Patients with Ineffective or Partially Effective Multiple Myeloma Treatment. International Journal of Molecular Sciences. 2022; 23(6):3359. https://doi.org/10.3390/ijms23063359
Chicago/Turabian StyleEnukashvily, Natella I., Natalia Semenova, Anna V. Chubar, Dmitry I. Ostromyshenskii, Ekaterina A. Gushcha, Sergei Gritsaev, Stanislav S. Bessmeltsev, Viktor I. Rugal, Egor M. Prikhodko, Ivan Kostroma, and et al. 2022. "Pericentromeric Non-Coding DNA Transcription Is Associated with Niche Impairment in Patients with Ineffective or Partially Effective Multiple Myeloma Treatment" International Journal of Molecular Sciences 23, no. 6: 3359. https://doi.org/10.3390/ijms23063359
APA StyleEnukashvily, N. I., Semenova, N., Chubar, A. V., Ostromyshenskii, D. I., Gushcha, E. A., Gritsaev, S., Bessmeltsev, S. S., Rugal, V. I., Prikhodko, E. M., Kostroma, I., Zherniakova, A., Kotova, A. V., Belik, L. A., Shumeev, A., Maslennikova, I. I., & Ivolgin, D. I. (2022). Pericentromeric Non-Coding DNA Transcription Is Associated with Niche Impairment in Patients with Ineffective or Partially Effective Multiple Myeloma Treatment. International Journal of Molecular Sciences, 23(6), 3359. https://doi.org/10.3390/ijms23063359







