In Silico Molecular Docking Analysis of Karanjin against Alzheimer’s and Parkinson’s Diseases as a Potential Natural Lead Molecule for New Drug Design, Development and Therapy
Abstract
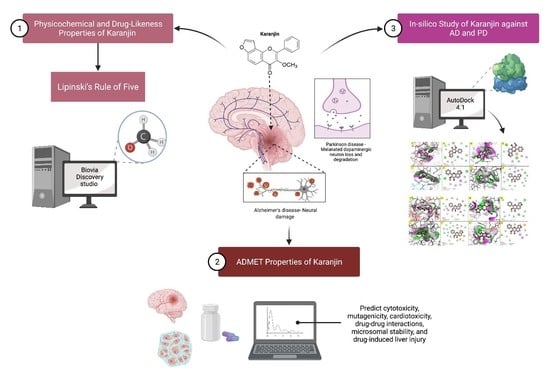
:1. Introduction
2. Results and Discussion
2.1. Physicochemical, Drug-Likeness and ADMET Properties of Karanjin
2.2. In Silico Results of Karanjin against AD and PD
2.3. Molecular Dynamics Simulation Study
2.4. Frontier Molecular Orbitals (FMOs) and Density Functional Theory (DFT) Analyses
3. Materials and Methods
3.1. Physicochemical and Drug-Likeness Properties of Karanjin
3.2. ADMET Properties of Karanjin
3.3. In Silico Study of Karanjin against AD and PD
3.4. Molecular Dynamics Simulation Study
3.5. FMOs and DFT Analyses
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali, M.; Muhammad, S.; Shah, M.R.; Khan, A.; Rashid, U.; Farooq, U.; Ullah, F.; Sadiq, A.; Ayaz, M.; Ali, M. Neurologically potent molecules from Crataegus oxyacantha; isolation, anticholinesterase inhibition, and molecular docking. Front. Pharmacol. 2017, 8, 327. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, M.; Sadiq, A.; Junaid, M.; Ullah, F.; Subhan, F.; Ahmed, J. Neuroprotective and anti-aging potentials of essential oils from aromatic and medicinal plants. Front. Aging Neurosci. 2017, 9, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, C. World alzheimer report 2018. In The State of the Art of Dementia Research: New Frontiers; Alzheimer’s Disease International Edition: London, UK, 2018; p. 148. [Google Scholar]
- WHO. Dementia. Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 15 February 2021).
- ADFM. Alzheimer’s Disease. Available online: https://adfm.org.my/ (accessed on 15 February 2021).
- Ayaz, M.; Junaid, M.; Ullah, F.; Subhan, F.; Sadiq, A.; Ali, G.; Ovais, M.; Shahid, M.; Ahmad, A.; Wadood, A. Anti-Alzheimer’s studies on β-sitosterol isolated from Polygonum hydropiper L. Front. Pharmacol. 2017, 8, 697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalil, A.T.; Ayaz, M.; Ovais, M.; Wadood, A.; Ali, M.; Shinwari, Z.K.; Maaza, M. In vitro cholinesterase enzymes inhibitory potential and in silico molecular docking studies of biogenic metal oxides nanoparticles. Inorg. Nano-Met. Chem. 2018, 48, 441–448. [Google Scholar] [CrossRef]
- Ovais, M.; Ayaz, M.; Khalil, A.T.; Shah, S.A.; Jan, M.S.; Raza, A.; Shahid, M.; Shinwari, Z.K. HPLC-DAD finger printing, antioxidant, cholinesterase, and α-glucosidase inhibitory potentials of a novel plant Olax nana. BMC Complement. Altern. Med. 2018, 18, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayaz, M.; Ullah, F.; Sadiq, A.; Kim, M.O.; Ali, T. Natural products-based drugs: Potential therapeutics against Alzheimer’s disease and other neurological disorders. Front. Pharmacol. 2019, 10, 1417. [Google Scholar] [CrossRef] [Green Version]
- Ovais, M.; Zia, N.; Ahmad, I.; Khalil, A.T.; Raza, A.; Ayaz, M.; Sadiq, A.; Ullah, F.; Shinwari, Z.K. Phyto-therapeutic and nanomedicinal approaches to cure alzheimer’s disease: Present status and future opportunities. Front. Aging Neurosci. 2018, 10, 284. [Google Scholar] [CrossRef] [Green Version]
- Rocca, W.A. The burden of Parkinson’s disease: A worldwide perspective. Lancet Neurol. 2018, 17, 928–929. [Google Scholar] [CrossRef] [Green Version]
- Klemann, C.J.; Martens, G.J.; Sharma, M.; Martens, M.; Isacson, O.; Gasser, T.; Visser, J.; Poelmans, G. Integrated molecular landscape of Parkinson’s disease. NPJ Parkinsons Dis. 2017, 3, 14. [Google Scholar] [CrossRef]
- Twelves, D.; Perkins, K.S.; Counsell, C. Systematic review of incidence studies of Parkinson’s disease. Mov. Disord. 2003, 18, 19–31. [Google Scholar] [CrossRef]
- Pringsheim, T.; Jette, N.; Frolkis, A.; Steeves, T.D. The prevalence of Parkinson’s disease: A systematic review and meta-analysis. Mov. Disord. 2014, 29, 1583–1590. [Google Scholar] [CrossRef] [PubMed]
- Dorsey, E.R.; Constantinescu, R.; Thompson, J.; Biglan, K.; Holloway, R.; Kieburtz, K.; Marshall, F.; Ravina, B.; Schifitto, G.; Siderowf, A. Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology 2007, 68, 384–386. [Google Scholar] [CrossRef] [PubMed]
- Calabresi, P.; Di Filippo, M.; Ghiglieri, V.; Tambasco, N.; Picconi, B. Levodopa-induced dyskinesias in patients with Parkinson’s disease: Filling the bench-to-bedside gap. Lancet Neurol. 2010, 9, 1106–1117. [Google Scholar] [CrossRef]
- Diaz, N.L.; Waters, C.H. Current strategies in the treatment of Parkinson’s disease and a personalized approach to management. Expert Rev. Neurother. 2009, 9, 1781–1789. [Google Scholar] [CrossRef]
- Muller, T.; Hefter, H.; Hueber, R.; Jost, W.H.; Leenders, K.L.; Odin, P.; Schwarz, J. Is levodopa toxic? J. Neurol. 2004, 251, 44–46. [Google Scholar] [CrossRef]
- Tamminga, C.A. Partial dopamine agonists in the treatment of psychosis. J. Neural Transm. 2002, 109, 411–420. [Google Scholar] [CrossRef]
- Cummings, J.; Lee, G.; Ritter, A.; Sabbagh, M.; Zhong, K. Alzheimer’s disease drug development pipeline: 2020. Alzheimer’s Dement. 2020, 6, e12050. [Google Scholar] [CrossRef]
- McFarthing, K.; Buff, S.; Rafaloff, G.; Dominey, T.; Wyse, R.K.; Stott, S.R. Parkinson’s disease drug therapies in the clinical trial pipeline: 2020. J. Parkinson’s Dis. 2020, 10, 757–774. [Google Scholar] [CrossRef]
- Bui, T.T.; Nguyen, T.H. Natural product for the treatment of Alzheimer’s disease. J. Basic Clin. Physiol. Pharmacol. 2017, 28, 413–423. [Google Scholar] [CrossRef]
- Rahman, M.; Bajgai, J.; Fadriquela, A.; Sharma, S.; Trinh, T.T.; Akter, R.; Jeong, Y.J.; Goh, S.H.; Kim, C.-S.; Lee, K.-J. Therapeutic potential of natural products in treating neurodegenerative disorders and their future prospects and challenges. Molecules 2021, 26, 5327. [Google Scholar] [CrossRef]
- Maher, P. The potential of flavonoids for the treatment of neurodegenerative diseases. Int. J. Mol. Sci. 2019, 20, 3056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noor, A.A.M.; Othman, S.N.N.; Lum, P.T.; Mani, S.; Shaikh, M.F.; Sekar, M. Molecules of interest–Karanjin–A review. Pharmacogn. J. 2020, 12. [Google Scholar]
- Saini, P.; Lakshmayya, L.; Bisht, V.S. Anti-Alzheimer activity of isolated karanjin from Pongamia pinnata (L.) pierre and embelin from Embelia ribes Burm.f. Ayu 2017, 38, 76. [Google Scholar] [PubMed]
- Kim, G.H.; Kim, J.E.; Rhie, S.J.; Yoon, S. The role of oxidative stress in neurodegenerative diseases. Exp. Neurobiol. 2015, 24, 325. [Google Scholar] [CrossRef] [PubMed]
- Schyman, P.; Liu, R.; Desai, V.; Wallqvist, A. vNN web server for ADMET predictions. Front. Pharmacol. 2017, 8, 889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martell, R.E.; Brooks, D.G.; Wang, Y.; Wilcoxen, K. Discovery of novel drugs for promising targets. Clin. Ther. 2013, 35, 1271–1281. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef] [Green Version]
- Swain, S.S.; Paidesetty, S.K.; Dehury, B.; Das, M.; Vedithi, S.C.; Padhy, R.N. Computer-aided synthesis of dapsone-phytochemical conjugates against dapsone-resistant Mycobacterium leprae. Sci. Rep. 2020, 10, 6839. [Google Scholar] [CrossRef] [Green Version]
- Heifetz, A.; Southey, M.; Morao, I.; Townsend-Nicholson, A.; Bodkin, M.J. Computational methods used in hit-to-lead and lead optimization stages of structure-based drug discovery. In Computational Methods for GPCR Drug Discovery; Springer: Cham, Switzerland, 2018; pp. 375–394. [Google Scholar]
- Cruz-Vicente, P.; Passarinha, L.A.; Silvestre, S.; Gallardo, E. Recent developments in new therapeutic agents against alzheimer and parkinson diseases: In silico approaches. Molecules 2021, 26, 2193. [Google Scholar] [CrossRef]
- Amin, M.L. P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights 2013, 7, 27–34. [Google Scholar] [CrossRef]
- Chang, R.; Yee, K.-L.; Sumbria, R.K. Tumor necrosis factor α inhibition for Alzheimer’s disease. J. Cent. Nerv. Syst. Dis. 2017, 9, 1179573517709278. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Yang, L.; He, P.; Li, R.; Shen, Y. Differential activation of tumor necrosis factor receptors distinguishes between brains from Alzheimer’s disease and non-demented patients. J. Alzheimer’s Dis. 2010, 19, 621–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syaifie, P.H.; Hemasita, A.W.; Nugroho, D.W.; Mardliyati, E.; Anshori, I. In silico investigation of propolis compounds as potential neuroprotective agent. Biointerface Res. Appl. Chem. 2022, 12, 8285–8306. [Google Scholar] [CrossRef]
- Kölsch, H.; Jessen, F.; Freymann, N.; Kreis, M.; Hentschel, F.; Maier, W.; Heun, R. ACE I/D polymorphism is a risk factor of Alzheimer’s disease but not of vascular dementia. Neurosci. Lett. 2005, 377, 37–39. [Google Scholar] [CrossRef] [PubMed]
- Monastero, R.; Caldarella, R.; Mannino, M.; Cefalu, A.B.; Lopez, G.; Noto, D.; Camarda, C.; Camarda, L.K.; Notarbartolo, A.; Averna, M.R. Lack of association between angiotensin converting enzyme polymorphism and sporadic Alzheimer’s disease. Neurosci. Lett. 2002, 335, 147–149. [Google Scholar] [CrossRef]
- Goh, N.-Y.; Razif, M.F.M.; Yap, Y.H.-Y.; Ng, C.L.; Fung, S.-Y. In silico analysis and characterization of medicinal mushroom cystathionine-synthase as an angiotensin converting enzyme (ACE) inhibitory protein. Comput. Biol. Chem. 2021, 107620. [Google Scholar] [CrossRef]
- Citron, M. Emerging Alzheimer’s disease therapies: Inhibition of β-secretase. Neurobiol. Aging 2002, 23, 1017–1022. [Google Scholar] [CrossRef]
- Hooper, C.; Killick, R.; Lovestone, S. The GSK3 hypothesis of Alzheimer’s disease. J. Neurochem. 2008, 104, 1433–1439. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.-S.; Tran, T.-D.; Mai, T.-T.; Nguyen, N.-L.; Thai, K.-M.; Le, M.-T. Synthesis, In silico and In vitro Evaluation of Some Flavone Derivatives for Acetylcholinesterase and BACE-1 Inhibitory Activity. Molecules 2020, 25, 4064. [Google Scholar] [CrossRef]
- Subramanian, G.; Poda, G. In silico ligand-based modeling of hBACE-1 inhibitors. Chem. Biol. Drug Des. 2018, 91, 817–827. [Google Scholar] [CrossRef]
- Coimbra, J.R.; Baptista, S.J.; Dinis, T.C.; Silva, M.; Moreira, P.I.; Santos, A.E.; Salvador, J.A. Combining virtual screening protocol and in vitro evaluation towards the discovery of BACE1 inhibitors. Biomolecules 2020, 10, 535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, R.; Munjal, N.S.; Singh, T.R. Identification of novel small molecules against GSK3β for Alzheimer’s disease using chemoinformatics approach. J. Mol. Graph. Modell. 2019, 91, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.; Gupta, M.; Vishwakarma, R.A.; Kumar, A.; Bharate, S.B. (Z)-2-(3-Chlorobenzylidene)-3,4-dihydro-N-(2-methoxyethyl)-3-oxo-2H-benzo[b][1,4]oxazine-6-carboxamide as GSK-3β inhibitor: Identification by virtual screening and its validation in enzyme-and cell-based assay. Chem. Biol. Drug Des. 2017, 89, 964–971. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Han, C.; Li, G.; Guo, H.; Wang, Y.; Hu, Y.; Lin, Z.; Wang, Y. In silico design novel (5-imidazol-2-yl-4-phenylpyrimidin-2-yl)[2-(2-pyridylamino)ethyl]amine derivatives as inhibitors for glycogen synthase kinase 3 based on 3D-QSAR, molecular docking and molecular dynamics simulation. Comput. Biol. Chem. 2020, 88, 107328. [Google Scholar] [CrossRef]
- Ranjan, A.; Chauhan, A.; Jindal, T. In silico and in vitro evaluation of human acetylcholinesterase inhibition by organophosphates. Environ. Toxicol. Pharmacol. 2018, 57, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Castro-Silva, E.; Bello, M.; Hernández-Rodríguez, M.; Correa-Basurto, J.; Murillo-Alvarez, J.; Rosales-Hernandez, M.; Muñoz-Ochoa, M. In vitro and in silico evaluation of fucosterol from Sargassum horridum as potential human acetylcholinesterase inhibitor. J. Biomol. Struct. Dyn. 2018, 37, 3259–3268. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Kumar, V.; Prashar, V.; Saini, S.; Dwivedi, A.R.; Bajaj, B.; Mehta, D.; Parkash, J.; Kumar, V. Dipropargyl substituted diphenylpyrimidines as dual inhibitors of monoamine oxidase and acetylcholinesterase. Eur. J. Med. Chem. 2019, 177, 221–234. [Google Scholar] [CrossRef]
- Morelli, M.; Simola, N.; Wardas, J. The Adenosinergic System: A Non-Dopaminergic Target in Parkinson’s Disease; Springer: Cham, Swizterland, 2015; Volume 10. [Google Scholar]
- Jenner, P. A2A antagonists as novel non-dopaminergic therapy for motor dysfunction in PD. Neurology 2003, 61, S32–S38. [Google Scholar] [CrossRef]
- Chartier-Harlin, M.-C.; Kachergus, J.; Roumier, C.; Mouroux, V.; Douay, X.; Lincoln, S.; Levecque, C.; Larvor, L.; Andrieux, J.; Hulihan, M. α-synuclein locus duplication as a cause of familial Parkinson’s disease. The Lancet 2004, 364, 1167–1169. [Google Scholar] [CrossRef]
- Ibanez, P.; Bonnet, A.; Debarges, B.; Lohmann, E.; Tison, F.; Agid, Y.; Dürr, A.; Brice, A.; Pollak, P.; Group, F.P.s.D.G.S. Causal relation between α-synuclein locus duplication as a cause of familial Parkinson’s disease. The Lancet 2004, 364, 1169–1171. [Google Scholar] [CrossRef]
- Elmer, L.W.; Bertoni, J.M. The increasing role of monoamine oxidase type B inhibitors in Parkinson’s disease therapy. Expert Opin. Pharmacother. 2008, 9, 2759–2772. [Google Scholar] [CrossRef] [PubMed]
- Bonifácio, M.J.; Palma, P.N.; Almeida, L.; Soares-da-Silva, P. Catechol-O-methyltransferase and its inhibitors in Parkinso’s disease. CNS Drug Rev. 2007, 13, 352–379. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zhang, S.; Zheng, L.; Hu, Z.; Cheng, L.; Chen, L.; Li, J.; Shi, Z. In silico identification of A1 agonists and A2a inhibitors in pain based on molecular docking strategies and dynamics simulations. Purinergic Signal. 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rohilla, S.; Bansal, R.; Chauhan, P.; Kachler, S.; Klotz, K.-N. A New Series of 1, 3-Dimethylxanthine Based Adenosine A2A Receptor Antagonists as a Non-Dopaminergic Treatment of Parkinson’s Disease. Curr. Drug Discov. Technol. 2021, 18, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Benito, M.; Granado, N.; García-Sanz, P.; Michel, A.; Dumoulin, M.; Moratalla, R. Modeling Parkinson’s disease with the alpha-synuclein protein. Front. Pharmacol. 2020, 11, 356. [Google Scholar] [CrossRef]
- Kumar, R.; Bavi, R.; Jo, M.G.; Arulalapperumal, V.; Baek, A.; Rampogu, S.; Kim, M.O.; Lee, K.W. New compounds identified through in silico approaches reduce the α-synuclein expression by inhibiting prolyl oligopeptidase in vitro. Sci. Rep. 2017, 7, 10827. [Google Scholar] [CrossRef] [Green Version]
- Kozioł, E.; Luca, S.V.; Ağalar, H.G.; Sağlık, B.N.; Demirci, F.; Marcourt, L.; Wolfender, J.-L.; Jóźwiak, K.; Skalicka-Woźniak, K. Rutamarin: Efficient Liquid–Liquid Chromatographic Isolation from Ruta graveolens L. and Evaluation of Its In vitro and in silico MAO-B Inhibitory Activity. Molecules 2020, 25, 2678. [Google Scholar] [CrossRef]
- Nam, M.-H.; Park, M.; Park, H.; Kim, Y.; Yoon, S.; Sawant, V.S.; Choi, J.W.; Park, J.-H.; Park, K.D.; Min, S.-J. Indole-substituted benzothiazoles and benzoxazoles as selective and reversible MAO-B inhibitors for treatment of Parkinson’s disease. ACS Chem. Neurosci. 2017, 8, 1519–1529. [Google Scholar] [CrossRef]
- Załuski, M.; Schabikowski, J.; Schlenk, M.; Olejarz-Maciej, A.; Kubas, B.; Karcz, T.; Kuder, K.; Latacz, G.; Zygmunt, M.; Synak, D. Novel multi-target directed ligands based on annelated xanthine scaffold with aromatic substituents acting on adenosine receptor and monoamine oxidase B. Synthesis, in vitro and in silico studies. Bioorg. Med. Chem. 2019, 27, 1195–1210. [Google Scholar] [CrossRef]
- Cuyàs, E.; Verdura, S.; Lozano-Sánchez, J.; Viciano, I.; Llorach-Parés, L.; Nonell-Canals, A.; Bosch-Barrera, J.; Brunet, J.; Segura-Carretero, A.; Sanchez-Martinez, M. The extra virgin olive oil phenolic oleacein is a dual substrate-inhibitor of catechol-O-methyltransferase. Food Chem. Toxicol. 2019, 128, 35–45. [Google Scholar] [CrossRef]
- Govindasamy, H.; Magudeeswaran, S.; Poomani, K. Identification of novel flavonoid inhibitor of Catechol-O-Methyltransferase enzyme by molecular screening, quantum mechanics/molecular mechanics and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2020, 38, 5307–5319. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.N.; Georrge, J.J.; Modi, K.M.; Narechania, M.B.; Patel, D.P.; Gonzalez, F.J.; Pandya, H.A. Pharmacophore-based virtual screening of catechol-o-methyltransferase (COMT) inhibitors to combat Alzheimer’s disease. J. Biomol. Struct. Dyn. 2018, 36, 3938–3957. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 100633, Karanjin. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Karanjin (accessed on 17 February 2021).
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Zhang, M.-Q.; Wilkinson, B. Drug discovery beyond the ‘rule-of-five’. Curr. Opin. Biotechnol. 2007, 18, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swain, S.S.; Paidesetty, S.K.; Dehury, B.; Sahoo, J.; Vedithi, S.C.; Mahapatra, N.; Hussain, T.; Padhy, R.N. Molecular docking and simulation study for synthesis of alternative dapsone derivative as a newer antileprosy drug in multidrug therapy. J. Cell. Biochem. 2018, 119, 9838–9852. [Google Scholar] [CrossRef]
- Swain, S.S.; Paidesetty, S.K.; Padhy, R.N. Synthesis of novel thymol derivatives against MRSA and ESBL producing pathogenic bacteria. Nat. Prod. Res. 2019, 33, 3181–3189. [Google Scholar] [CrossRef]
- Li, X.; Buxbaum, J.N. Transthyretin and the brain re-visited: Is neuronal synthesis of transthyretin protective in Alzheimer’s disease? Mol. Neurodegener. 2011, 6, 79. [Google Scholar] [CrossRef] [Green Version]
- Fridman, C.; Gregório, S.P.; Dias Neto, E.; Ojopi, É.P.B. Alterações genéticas na doença de Alzheimer. Arch. Clin. Psychiatry (São Paulo) 2004, 31, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Vassar, R. BACE1 inhibitor drugs in clinical trials for Alzheimer’s disease. Alzheimer’s Res. Ther. 2014, 6, 89. [Google Scholar] [CrossRef] [Green Version]
- Koelsch, G. BACE1 function and inhibition: Implications of intervention in the amyloid pathway of Alzheimer’s disease pathology. Molecules 2017, 22, 1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridler, C. BACE1 inhibitors block new Aβ plaque formation. Nat. Rev. Neurol. 2018, 14, 126. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Qin, M.; Mahaman, Y.A.R.; Zhang, B.; Huang, F.; Zeng, K.; Xia, Y.; Ke, D.; Wang, Q.; Liu, R. BACE1 SUMOylation increases its stability and escalates the protease activity in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2018, 115, 3954–3959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eldar-Finkelman, H.; Martinez, A. GSK-3 inhibitors: Preclinical and clinical focus on CNS. Front. Mol. Neurosci. 2011, 4, 32. [Google Scholar] [CrossRef] [Green Version]
- Bhat, R.; Xue, Y.; Berg, S.; Hellberg, S.; Ormo, M.; Nilsson, Y.; Radesäter, A.-C.; Jerning, E.; Markgren, P.-O.; Borgegård, T. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418. J. Biol. Chem. 2003, 278, 45937–45945. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Li, M.; Wang, Y.; Li, Q.; Deng, G.; Wan, J.; Yang, Q.; Chen, Q.; Wang, J. GSK-3β inhibitor TWS119 attenuates rtPA-induced hemorrhagic transformation and activates the Wnt/β-catenin signaling pathway after acute ischemic stroke in rats. Mol. Neurobiol. 2016, 53, 7028–7036. [Google Scholar] [CrossRef]
- Kremer, A.; Louis, J.V.; Jaworski, T.; Van Leuven, F. GSK3 and Alzheimer’s disease: Facts and fiction. Front. Mol. Neurosci. 2011, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Dickson, D.W. The pathogenesis of senile plaques. J. Neuropathol. Exp. Neurol. 1997, 56, 321–339. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Bickler, P. Interaction of isoflurane, tumor necrosis factor-α and β-amyloid on long-term potentiation in rat hippocampal slices. Anesth. Analg. 2017, 124, 582–587. [Google Scholar] [CrossRef]
- Rees, T.M.; Brimijoin, S. The role of acetylcholinesterase in the pathogenesis of Alzheimer’s disease. Drugs Today 2003, 39, 75–83. [Google Scholar] [CrossRef]
- Wilson, C.N.; Mustafa, S.J. Adenosine Receptors in Health and Disease; Springer: Cham, Switzerland, 2009. [Google Scholar]
- Olanow, C.W.; Brundin, P. Parkinson’s disease and alpha synuclein: Is Parkinson’s disease a prion-like disorder? Mov. Disord. 2013, 28, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Jauand, M.; Sitges, C.; Rodriguez, V.; Picornell, A.; Ramon, M.; Buskila, D.; Montoya, P. Pain sensitivity in fibromyalgia is associated with catechol-O-methyltransferase (COMT) gene. Eur. J. Pain 2013, 17, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Teo, K.C.; Ho, S.-L. Monoamine oxidase-B (MAO-B) inhibitors: Implications for disease-modification in Parkinson’s disease. Transl. Neurodegener. 2013, 2, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.W.; Jang, B.K.; Cho, N.-C.; Park, J.-H.; Yeon, S.K.; Ju, E.J.; Lee, Y.S.; Han, G.; Pae, A.N.; Kim, D.J. Synthesis of a series of unsaturated ketone derivatives as selective and reversible monoamine oxidase inhibitors. Bioorg. Med. Chem. 2015, 23, 6486–6496. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.S.; Singh, S.R.; Sahoo, A.; Panda, P.K.; Hussain, T.; Pati, S. Integrated bioinformatics-cheminformatics approach toward locating pseudo-potential antiviral marine alkaloids against SARS-CoV-2-Mpro. Proteins 2022. [Google Scholar] [CrossRef]
- Al-Wahaibi1, L.H.; Joubert, J.; Blacque, O.; Al-Shaalan, N.H.; El-Emam, A.A. Crystal structure, Hirshfeld surface analysis and DFT studies of 5-(adamantan-1-yl)-3-[(4-chlorobenzyl)sulfanyl]-4-methyl-4H-1,2,4-triazole, a potential 11βHSD1 inhibitor. Sci. Rep. 2019, 9, 19745. [Google Scholar] [CrossRef]












| Property | Result (vNN-ADMET, swissADME and admetSAR Tools) |
|---|---|
| Molecular formula | C18H12O4 |
| Molecular weight | 292.30 |
| Hydrogen bond donors | 0 |
| Hydrogen bond acceptors | 4 |
| Rotatable bonds | 2 |
| Log P (Partition coefficient, Predicted value) | 2.54 or 3.43 |
| Melting point | 157–159 °C (in the crystallized form) |
| Molar refractivity | 81.027 cm3 or 84.18 cm3 |
| Molar volume | 214.875 cm3 |
| Topological polar surface area | 48.7 Å2 or 52.58 Å2 |
| Lipinski’s rule of five | Passed |
| Ghose filter | Passed |
| Veber’s rule | Passed |
| BBB likeness rule | Passed |
| Unweighted QED | Passed |
| Weighted QED | Passed |
| GI absorption | High |
| BBB Permeant | Yes |
| CYP1A2, CYP2C19, CYP2C9, CYP2D6 and CYP3A4 inhibitors | Yes |
| Bioavailability score | 0.55 |
| Karanjin/Standards | Selected Targets Associated with AD | |||||||||
| ACE (PBD ID: 1O86) | BACE1 (PBD ID: 4DJU) | GSK-3 (PDB ID: 1Q5K) | TACE (PDB ID: 2OI0) | AChE (PDB ID: 6ZWE) | ||||||
| AutoDock | Molegro Virtual Docker | AutoDock | Molegro Virtual Docker | AutoDock | Molegro Virtual Docker | AutoDock | Molegro Virtual Docker | AutoDock | Molegro Virtual Docker | |
| Karanjin | −7.54 | −85.48 | −8.79 | −77.11 | −8.23 | −69.63 | −9.16 | −1289.34 | −9.40 | −107.87 |
| Donepezil * | −8.88 | −120.35 | −9.21 | −68.34 | −7.69 | −106.51 | −11.00 | −1642.78 | −11.00 | −80.81 |
| Galantamine * | −7.42 | −100.24 | −7.06 | −96.06 | −6.68 | −79.57 | −8.48 | −1096.31 | −8.20 | −108.12 |
| Rivastigmine * | −6.47 | −84.66 | −6.66 | −86.59 | −5.31 | −65.71 | −7.57 | −1191.48 | −7.30 | −98.58 |
| Selected Targets Associated with PD | ||||||||||
| A2AAR (PDB ID: 3EML) | ASN (PDB ID: 1XQ8) | COMT (PDB ID: 1H1D) | MAO_B (PDB ID: 2C65) | -- | ||||||
| AutoDock | Molegro Virtual Docker | AutoDock | Molegro Virtual Docker | AutoDock | Molegro Virtual Docker | AutoDock | Molegro Virtual Docker | -- | -- | |
| Karanjin | −8.39 | −88.37 | −4.75 | −83.35 | −8.95 | −90.88 | −9.22 | −145.14 | -- | -- |
| Dopamine * | −5.69 | −59.07 | −5.16 | −67.09 | −7.36 | −87.40 | −6.59 | −82.62 | -- | -- |
| Rasagiline * | −6.89 | −72.66 | −5.51 | −66.95 | −8.44 | −104.91 | −7.57 | −97.88 | -- | -- |
| Selegiline * | −5.53 | −62.34 | −4.23 | −70.15 | −7.56 | −106.52 | −6.98 | −95.33 | -- | -- |
| Protein Code | Van der Waal Energy kJ/mol | Electrostatic Energy kJ/mol | Polar Solvation Energy kJ/mol | Binding Energy kJ/mol |
|---|---|---|---|---|
| 2C65 | −205.968 | −4.908 | 66.203 | −161.262 |
| 6ZWE | −197.955 | −2.742 | 49.001 | −168.652 |
| Disease | Targets | Reason for Selected Targets | References |
|---|---|---|---|
| AD | ACE | It has been shown to block memory consolidation in some investigations. | Li and Buxbaum [74] Kölsch et al. [38] Monastero et al. [39] Fridman et al. [75] |
| BACE1 | BACE1, a β-secretase involved in the formation of β-amyloid peptide, which is a dominant component in AD. | Vassar [76] Koelsch [77] Ridler [78] Bao et al. [79] | |
| GSK3 | GSK3 phosphorylates the Tau protein, whose expression is associated to AD. | Eldar-Finkelman and Martinez [80] Bhat et al. [81] Wang et al. [82] Kremer et al. [83] | |
| TACE | TNF-α is normally kept at relatively low levels, but, as AD progresses, the levels rise. | Chang et al. [35] Dickson [84] Cheng et al. [36] Zhou and Bickler [85] | |
| AChE | AChE inhibition may affect amyloid precursor protein processing and protect neurons against a variety of insults. | Rees and Brimijoin [86] | |
| PD | A2AAR | The basal ganglia have a more selective and extensive distribution of A2A. This selective receptor distribution may help to ensure fewer side effects, making nondopaminergic antagonists against PD. | Wilson and Mustafa [87] |
| ASN | PD is caused by a doubling or tripling of the α-synuclein. | Olanow and Brundin [88] Chartier-Harlin et al. [54] Ibanez et al. [55] | |
| COMT | The COMT gene codes for an enzyme which degrades catecholamines, and this process is slowed in people with PD. | Martínez-Jauand et al. [89] | |
| MAO-B | MAO-B expression has been found in human brains, specifically in the substantia nigra of patients with PD. | Teo and Ho [90] Choi et al. [91] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gnanaraj, C.; Sekar, M.; Fuloria, S.; Swain, S.S.; Gan, S.H.; Chidambaram, K.; Rani, N.N.I.M.; Balan, T.; Stephenie, S.; Lum, P.T.; et al. In Silico Molecular Docking Analysis of Karanjin against Alzheimer’s and Parkinson’s Diseases as a Potential Natural Lead Molecule for New Drug Design, Development and Therapy. Molecules 2022, 27, 2834. https://doi.org/10.3390/molecules27092834
Gnanaraj C, Sekar M, Fuloria S, Swain SS, Gan SH, Chidambaram K, Rani NNIM, Balan T, Stephenie S, Lum PT, et al. In Silico Molecular Docking Analysis of Karanjin against Alzheimer’s and Parkinson’s Diseases as a Potential Natural Lead Molecule for New Drug Design, Development and Therapy. Molecules. 2022; 27(9):2834. https://doi.org/10.3390/molecules27092834
Chicago/Turabian StyleGnanaraj, Charles, Mahendran Sekar, Shivkanya Fuloria, Shasank S. Swain, Siew Hua Gan, Kumarappan Chidambaram, Nur Najihah Izzati Mat Rani, Tavamani Balan, Sarah Stephenie, Pei Teng Lum, and et al. 2022. "In Silico Molecular Docking Analysis of Karanjin against Alzheimer’s and Parkinson’s Diseases as a Potential Natural Lead Molecule for New Drug Design, Development and Therapy" Molecules 27, no. 9: 2834. https://doi.org/10.3390/molecules27092834
APA StyleGnanaraj, C., Sekar, M., Fuloria, S., Swain, S. S., Gan, S. H., Chidambaram, K., Rani, N. N. I. M., Balan, T., Stephenie, S., Lum, P. T., Jeyabalan, S., Begum, M. Y., Chandramohan, V., Thangavelu, L., Subramaniyan, V., & Fuloria, N. K. (2022). In Silico Molecular Docking Analysis of Karanjin against Alzheimer’s and Parkinson’s Diseases as a Potential Natural Lead Molecule for New Drug Design, Development and Therapy. Molecules, 27(9), 2834. https://doi.org/10.3390/molecules27092834