Heterologous Biosynthesis of Health-Promoting Baicalein in Lycopersicon esculentum
Abstract
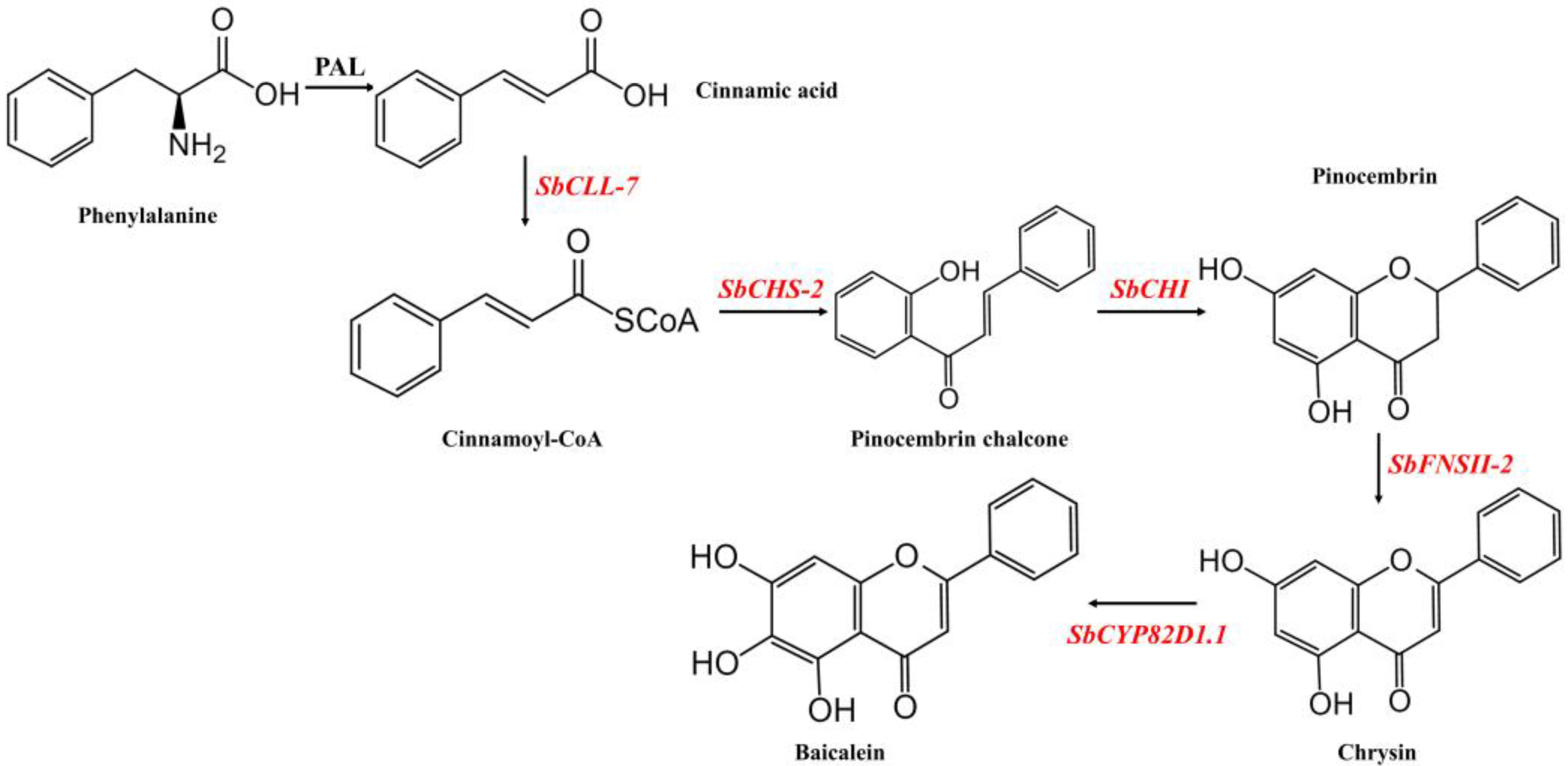
:1. Introduction
2. Results
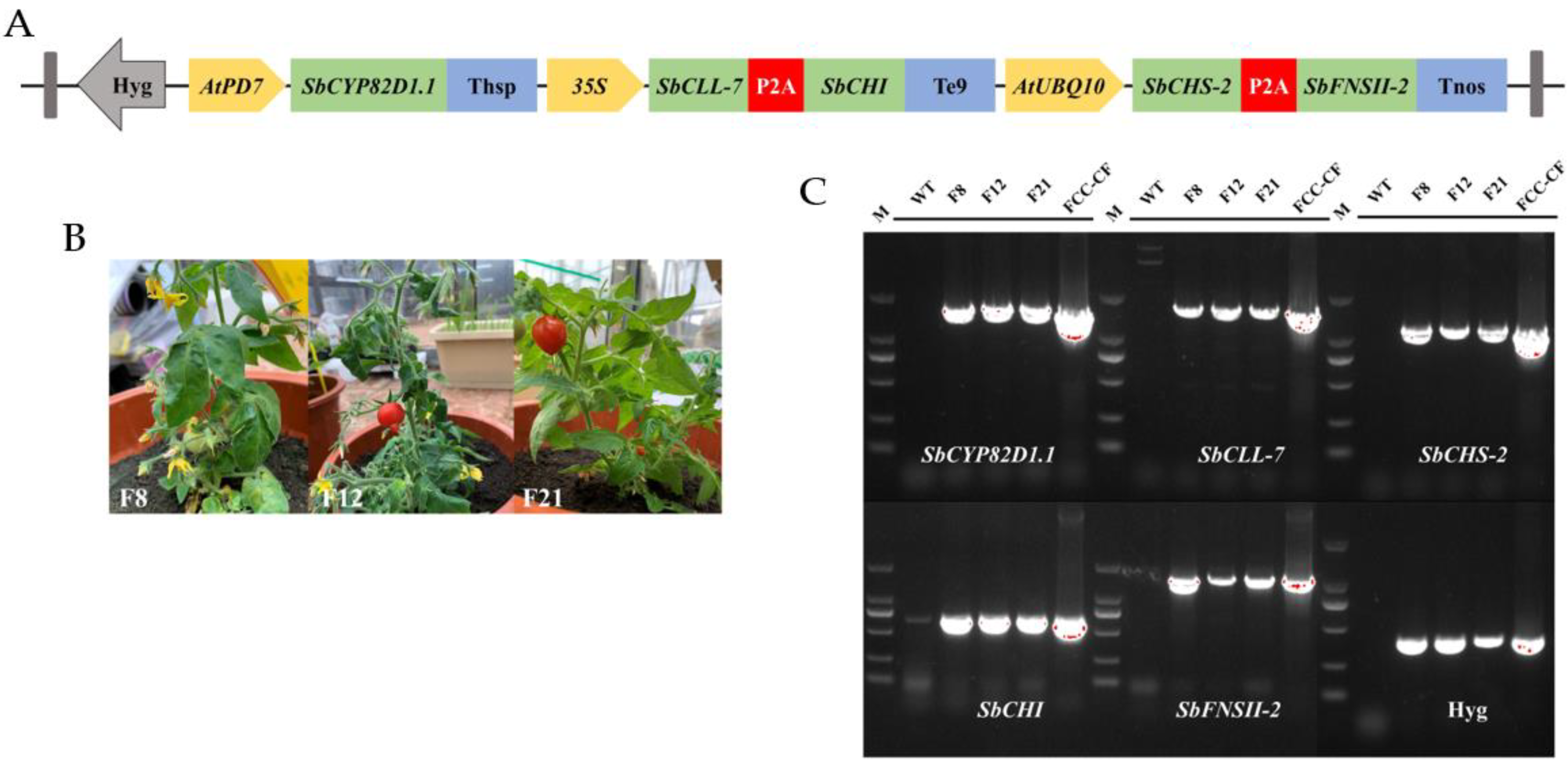
2.1. Design of Multigene Expression Vector
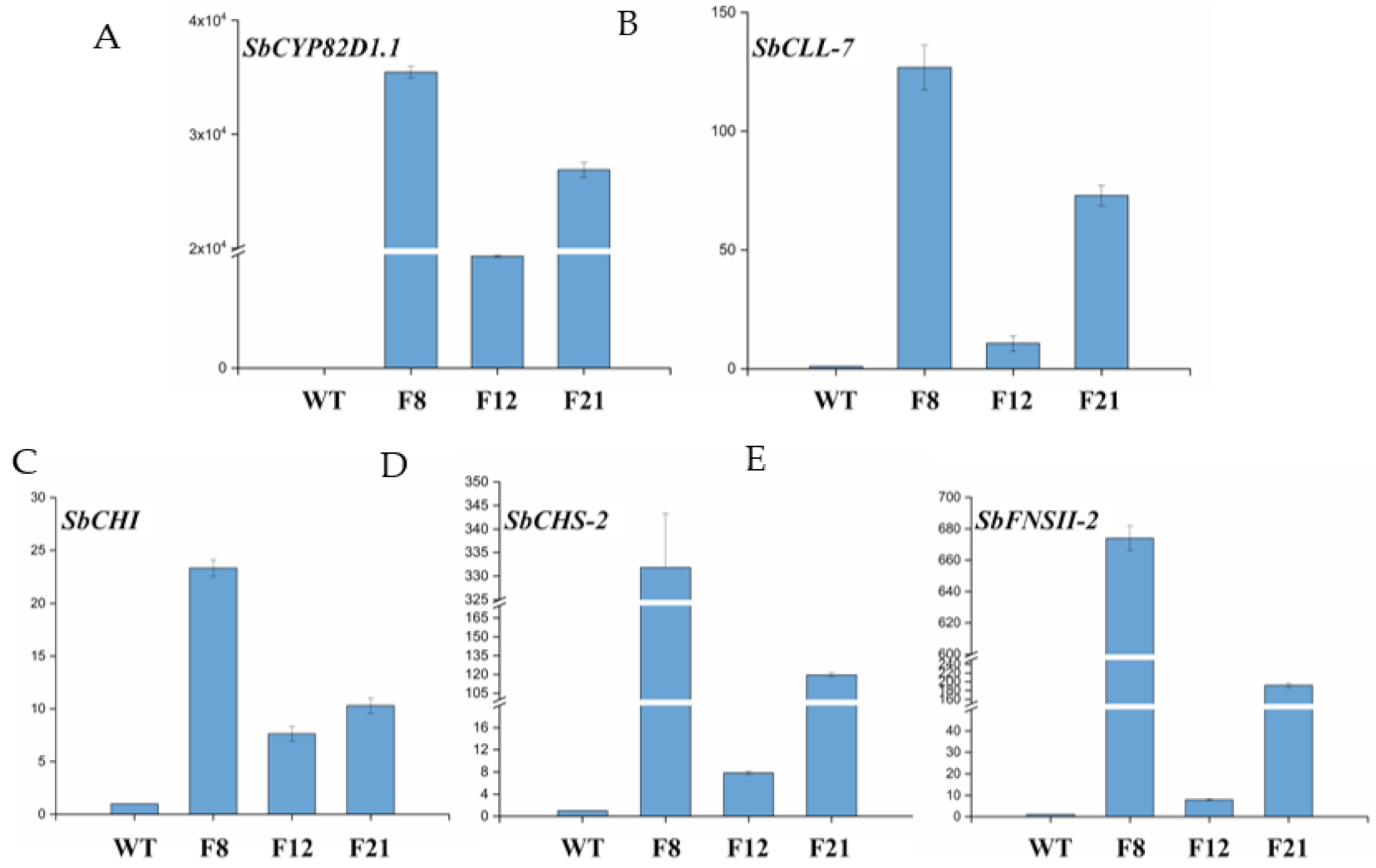
2.2. Genetic Transformation of Tomato Plants and Molecular Analysis
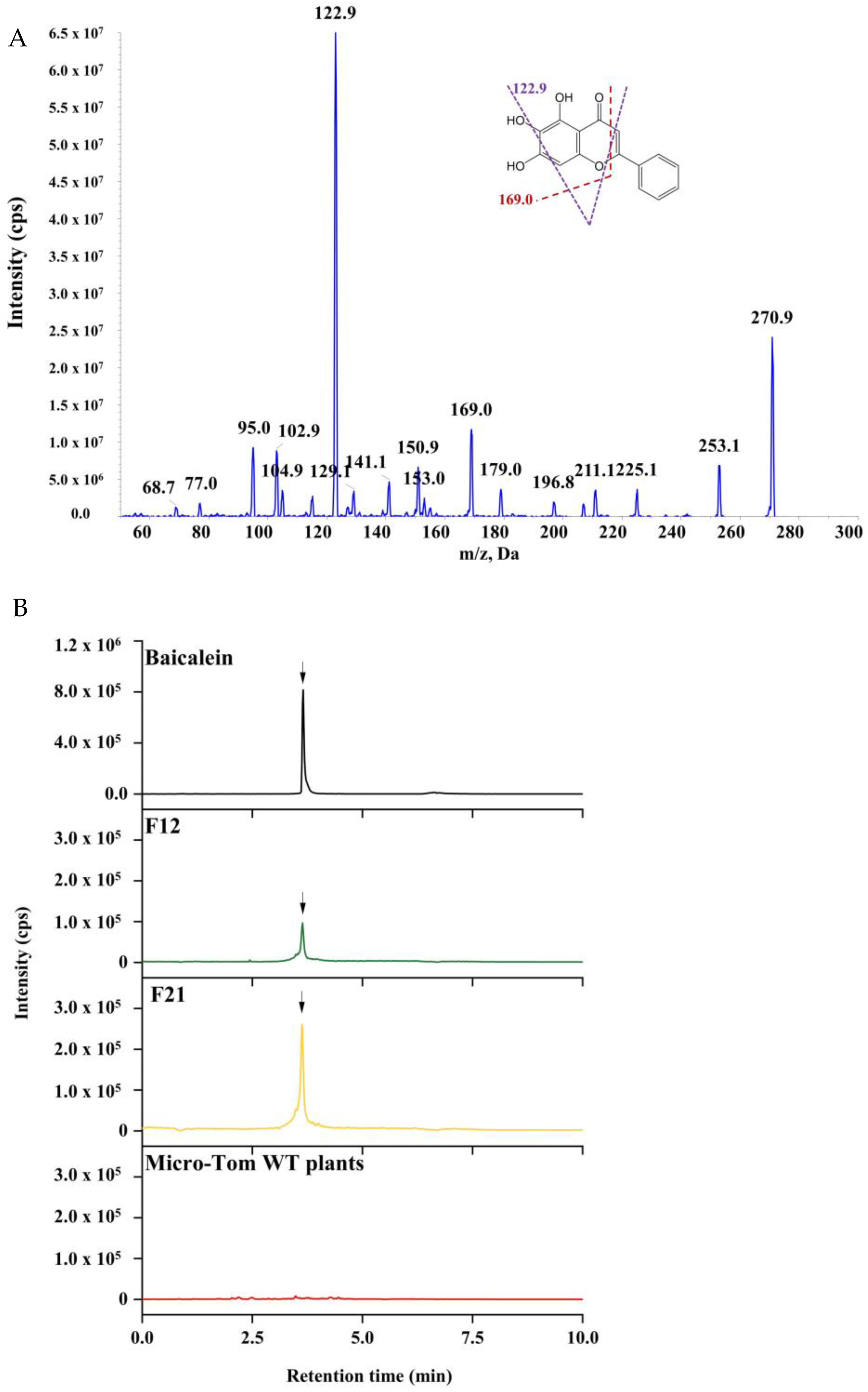
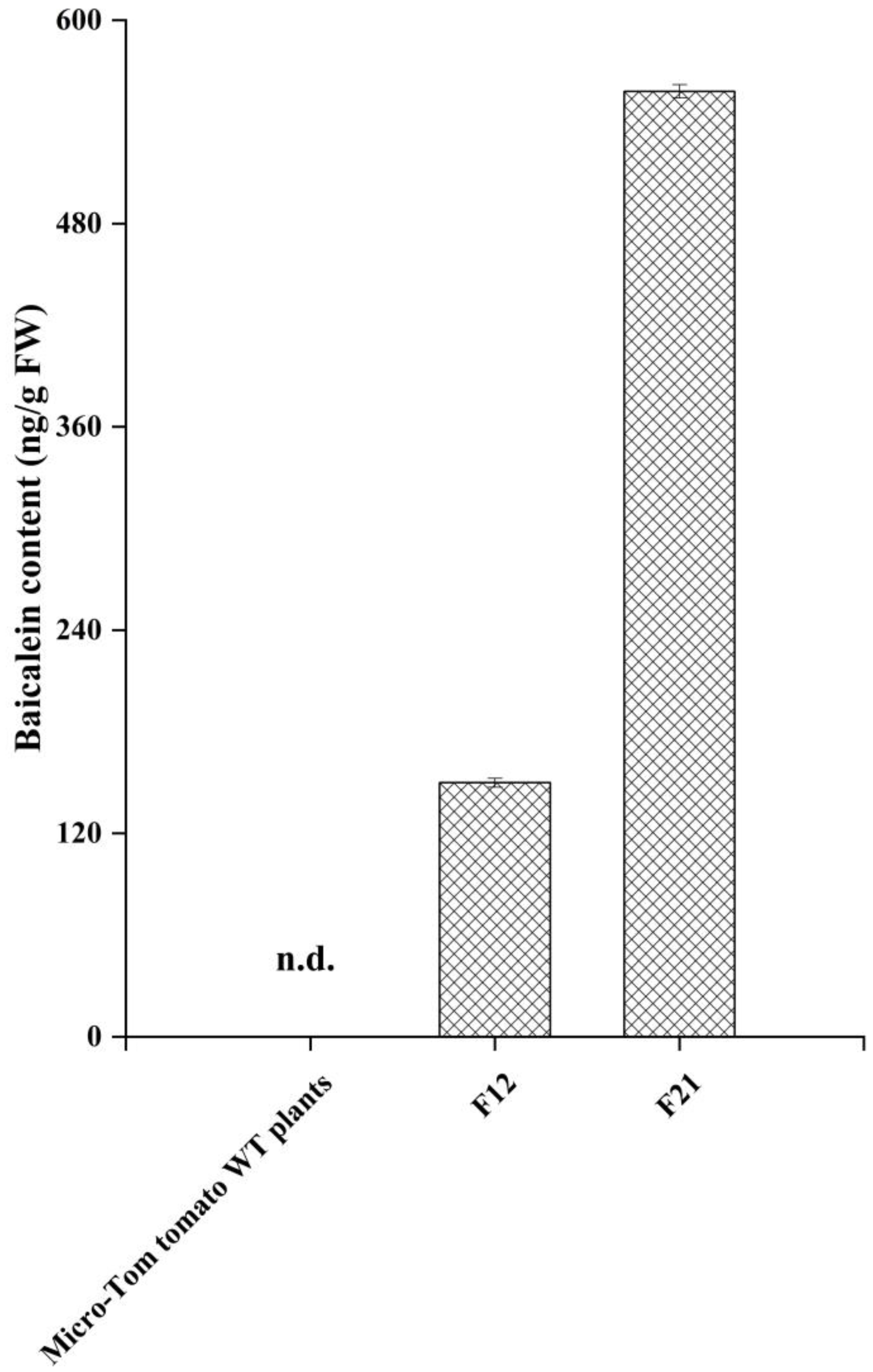
2.3. HPLC-MS/MS Analysis for Baicalein in Transgenic Tomato Lines
3. Discussion
4. Materials and Methods
4.1. Plant Material, Chemicals and Strains
4.2. Multigene Vector Construction
4.3. Agrobacterium-Transformation of Micro-Tom Tomato Plants
4.4. PCR Detection of Transgenic Tomato Plants
4.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.6. Analysis of Baicalein by HPLC-MS/MS
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Gao, Z.; Huang, K.; Yang, X.; Xu, H. Free Radical Scavenging and Antioxidant Activities of Flavonoids Extracted from the Radix of Scutellaria baicalensis Georgi. Biochim. Biophys. Acta Gen. Subj. 1999, 1472, 643–650. [Google Scholar] [CrossRef]
- Patwardhan, R.S.; Sharma, D.; Thoh, M.; Checker, R.; Sandur, S.K. Baicalein Exhibits Anti-Inflammatory Effects via Inhibition of NF-ΚB Transactivation. Biochem. Pharmacol. 2016, 108, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Li, Q.; Wang, Y.; Li, P. A Potent Protective Effect of Baicalein on Liver Injury by Regulating Mitochondria-Related Apoptosis. Apoptosis 2020, 25, 412–425. [Google Scholar] [CrossRef]
- Oo, A.; Rausalu, K.; Merits, A.; Higgs, S.; Vanlandingham, D.; Bakar, S.A.; Zandi, K. Deciphering the Potential of Baicalin as an Antiviral Agent for Chikungunya Virus Infection. Antivir. Res. 2018, 150, 101–111. [Google Scholar] [CrossRef]
- Gao, Y.; Snyder, S.A.; Smith, J.N.; Chen, Y.C. Anticancer Properties of Baicalein: A Review. Med. Chem. Res. 2016, 25, 1515–1523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shagufta; Ahmad, I. An Update on Pharmacological Relevance and Chemical Synthesis of Natural Products and Derivatives with Anti SARS-CoV-2 Activity. ChemistrySelect 2021, 6, 11502–11527. [Google Scholar] [CrossRef]
- Li, J.; Tian, C.; Xia, Y.; Mutanda, I.; Wang, K.; Wang, Y. Production of Plant-Specific Flavones Baicalein and Scutellarein in an Engineered E. Coli from Available Phenylalanine and Tyrosine. Metab. Eng. 2019, 52, 124–133. [Google Scholar] [CrossRef]
- Zhao, Q.; Cui, M.Y.; Levsh, O.; Yang, D.; Liu, J.; Li, J.; Hill, L.; Yang, L.; Hu, Y.; Weng, J.K.; et al. Two CYP82D Enzymes Function as Flavone Hydroxylases in the Biosynthesis of Root-Specific 4′-Deoxyflavones in Scutellaria baicalensis. Mol. Plant 2018, 11, 135–148. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Gao, R.; Pu, X.; Xu, R.; Wang, J.; Zheng, S.; Zeng, Y.; Chen, J.; He, C.; Song, J. Comparative Genome Analysis of Scutellaria baicalensis and Scutellaria barbata Reveals the Evolution of Active Flavonoid Biosynthesis. Genom. Proteom. Bioinform. 2020, 18, 230–240. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, J.; Zhu, X.; Zhang, G.; Yang, S.; Guo, X.; Jiang, H.; Ma, Y. De Novo Biosynthesis of Multiple Pinocembrin Derivatives in Saccharomyces cerevisiae. ACS Synth. Biol. 2020, 9, 3042–3051. [Google Scholar] [CrossRef]
- Ji, D.; Li, J.; Xu, F.; Ren, Y.; Wang, Y. Improve the Biosynthesis of Baicalein and Scutellarein via Manufacturing Self-Assembly Enzyme Reactor In Vivo. ACS Synth. Biol. 2021, 10, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Wang, B.; Tan, J.; Liu, T.; Li, L.; Liu, Y.G. Plant Synthetic Metabolic Engineering for Enhancing Crop Nutritional Quality. Plant Commun. 2020, 1, 100017. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Liu, X.; Liu, T.; Wang, Y.; Ahmed, N.; Li, Z.; Jiang, H. Synthetic Biology of Plant Natural Products: From Pathway Elucidation to Engineered Biosynthesis in Plant Cells. Plant Commun. 2021, 2, 100229. [Google Scholar] [CrossRef] [PubMed]
- Paine, J.A.; Shipton, C.A.; Chaggar, S.; Howells, R.M.; Kennedy, M.J.; Vernon, G.; Wright, S.Y.; Hinchliffe, E.; Adams, J.L.; Silverstone, A.L.; et al. Improving the Nutritional Value of Golden Rice through Increased Pro-Vitamin A Content. Nat. Biotechnol. 2005, 23, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Naqvi, S.; Breitenbach, J.; Sandmann, G.; Christou, P.; Capell, T. Combinatorial Genetic Transformation Generates a Library of Metabolic Phenotypes for the Carotenoid Pathway in Maize. Proc. Natl. Acad. Sci. USA 2008, 105, 18232–18237. [Google Scholar] [CrossRef] [Green Version]
- Shewmaker, C.K.; Sheehy, J.A.; Daley, M.; Colburn, S.; Ke, D.Y. Seed-Specific Overexpression of Phytoene Synthase: Increase in Carotenoids and Other Metabolic Effects. Plant J. 1999, 20, 401–412. [Google Scholar] [CrossRef]
- Diretto, G.; Al-Babili, S.; Tavazza, R.; Papacchioli, V.; Beyer, P.; Giuliano, G. Metabolic Engineering of Potato Carotenoid Content through Tuber-Specific Overexpression of a Bacterial Mini-Pathway. PLoS ONE 2007, 2, e350. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Kim, J.K.; Kim, H.J.; Pak, J.H.; Lee, J.H.; Kim, D.H.; Choi, H.K.; Jung, H.W.; Lee, J.D.; Chung, Y.S.; et al. Genetic Modification of the Soybean to Enhance the β-Carotene Content through Seed-Specific Expression. PLoS ONE 2012, 7, e48287. [Google Scholar] [CrossRef] [Green Version]
- Paul, J.Y.; Khanna, H.; Kleidon, J.; Hoang, P.; Geijskes, J.; Daniells, J.; Zaplin, E.; Rosenberg, Y.; James, A.; Mlalazi, B.; et al. Golden Bananas in the Field: Elevated Fruit pro-Vitamin A from the Expression of a Single Banana Transgene. Plant Biotechnol. J. 2017, 15, 520–532. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; Yu, S.; Zeng, D.; Liu, H.; Wang, H.; Yang, Z.; Xie, X.; Shen, R.; Tan, J.; Li, H.; et al. Development of “Purple Endosperm Rice” by Engineering Anthocyanin Biosynthesis in the Endosperm with a High-Efficiency Transgene Stacking System. Mol. Plant 2017, 10, 918–929. [Google Scholar] [CrossRef] [Green Version]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of Tomato Fruit with Health-Promoting Anthocyanins by Expression of Select Transcription Factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Liu, X.; Yang, W.; Mu, B.; Li, S.; Li, Y.; Zhou, X.; Zhang, C.; Fan, Y.; Chen, R. Engineering of ‘Purple Embryo Maize’ with a Multigene Expression System Derived from a Bidirectional Promoter and Self-Cleaving 2A Peptides. Plant Biotechnol. J. 2018, 16, 1107–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Q.; Zeng, D.; Yu, S.; Cui, C.; Li, J.; Li, H.; Chen, J.; Zhang, R.; Zhao, X.; Chen, L.; et al. From Golden Rice to ASTARice: Bioengineering Astaxanthin Biosynthesis in Rice Endosperm. Mol. Plant 2018, 11, 1440–1448. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.C.; Zhong, Y.J.; Liu, J.; Sandmann, G.; Chen, F. Metabolic Engineering of Tomato for High-Yield Production of Astaxanthin. Metab. Eng. 2013, 17, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y.; Baek, S.H.; Jo, H.J.; Yun, D.W.; Choi, Y.E. Genetically Modified Rice Produces Ginsenoside Aglycone (Protopanaxadiol). Planta 2019, 250, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Farhi, M.; Marhevka, E.; Ben-Ari, J.; Algamas-Dimantov, A.; Liang, Z.; Zeevi, V.; Edelbaum, O.; Spitzer-Rimon, B.; Abeliovich, H.; Schwartz, B.; et al. Generation of the Potent Anti-Malarial Drug Artemisinin in Tobacco. Nat. Biotechnol. 2011, 29, 1072–1074. [Google Scholar] [CrossRef]
- Khairul Ikram, N.K.B.; Beyraghdar Kashkooli, A.; Peramuna, A.V.; van der Krol, A.R.; Bouwmeester, H.; Simonsen, H.T. Stable Production of the Antimalarial Drug Artemisinin in the Moss Physcomitrella patens. Front. Bioeng. Biotechnol. 2017, 5, 47. [Google Scholar] [CrossRef] [Green Version]
- Moco, S.; Capanoglu, E.; Tikunov, Y.; Bino, R.J.; Boyacioglu, D.; Hall, R.D.; Vervoort, J.; De Vos, R.C.H. Tissue Specialization at the Metabolite Level Is Perceived during the Development of Tomato Fruit. J. Exp. Bot. 2007, 58, 4131–4146. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, H.; Zhang, Y.; Martin, C. Can the World’s Favorite Fruit, Tomato, Provide an Effective Biosynthetic Chassis for High-Value Metabolites? Plant Cell Rep. 2018, 37, 1443–1450. [Google Scholar] [CrossRef] [Green Version]
- Martí, E.; Gisbert, C.; Bishop, G.J.; Dixon, M.S.; García-Martínez, J.L. Genetic and Physiological Characterization of Tomato Cv. Micro-Tom. J. Exp. Bot. 2006, 57, 2037–2047. [Google Scholar] [CrossRef] [Green Version]
- Grützner, R.; Schubert, R.; Horn, C.; Yang, C.; Vogt, T.; Marillonnet, S. Engineering Betalain Biosynthesis in Tomato for High Level Betanin Production in Fruits. Front. Plant Sci. 2021, 12, 682443. [Google Scholar] [CrossRef] [PubMed]
- Breitel, D.; Brett, P.; Alseekh, S.; Fernie, A.R.; Butelli, E.; Martin, C. Metabolic Engineering of Tomato Fruit Enriched in L-DOPA. Metab. Eng. 2021, 65, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Khojasteh, A.; Mirjalili, M.H.; Hidalgo, D.; Corchete, P.; Palazon, J. New Trends in Biotechnological Production of Rosmarinic Acid. Biotechnol. Lett. 2014, 36, 2393–2406. [Google Scholar] [CrossRef] [PubMed]
- Farré, G.; Blancquaert, D.; Capell, T.; Van Der Straeten, D.; Christou, P.; Zhu, C. Engineering Complex Metabolic Pathways in Plants. Annu. Rev. Plant Biol. 2014, 65, 187–223. [Google Scholar] [CrossRef] [PubMed]
- Halpin, C. Gene Stacking in Transgenic Plants—The Challenge for 21st Century Plant Biotechnology. Plant Biotechnol. J. 2005, 3, 141–155. [Google Scholar] [CrossRef]
- Zhao, Z.; Xie, X.; Liu, W.; Huang, J.; Tan, J.; Yu, H.; Zong, W.; Tang, J.; Zhao, Y.; Xue, Y.; et al. STI PCR: An Efficient Method for Amplification and de Novo Synthesis of Long DNA Sequences. Mol. Plant 2022, 15, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Benders, G.A.; Axelrod, K.C.; Zaveri, J.; Algire, M.A.; Moodie, M.; Montague, M.G.; Venter, J.C.; Smith, H.O.; Hutchison, C.A. One-Step Assembly in Yeast of 25 Overlapping DNA Fragments to Form a Complete Synthetic Mycoplasma genitalium Genome. Proc. Natl. Acad. Sci. USA 2008, 105, 20404–20409. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.J.; Zhou, H.M.; Chen, J.; Wang, X.C. A Gateway-Based Platform for Multigene Plant Transformation. Plant Mol. Biol. 2006, 62, 927–936. [Google Scholar] [CrossRef]
- Sleight, S.C.; Bartley, B.A.; Lieviant, J.A.; Sauro, H.M. In-Fusion Biobrick Assembly and Re-Engineering. Nucleic Acids Res. 2010, 38, 2624–2636. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Zhang, L.; Xu, Y.; Yang, D.; Zhang, L.; Yang, S.; Zhang, W.; Wang, J.; Tian, S.; Yang, S.; et al. The Comprehensive Study on the Therapeutic Effects of Baicalein for the Treatment of COVID-19 In Vivo and In Vitro. Biochem. Pharmacol. 2021, 183, 114302. [Google Scholar] [CrossRef]
- Moghaddam, E.; Teoh, B.T.; Sam, S.S.; Lani, R.; Hassandarvish, P.; Chik, Z.; Yueh, A.; Abubakar, S.; Zandi, K. Baicalin, a Metabolite of Baicalein with Antiviral Activity against Dengue Virus. Sci. Rep. 2014, 4, 5452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nayak, M.K.; Agrawal, A.S.; Bose, S.; Naskar, S.; Bhowmick, R.; Chakrabarti, S.; Sarkar, S.; Chawla-Sarkar, M. Antiviral Activity of Baicalin against Influenza Virus H1N1-Pdm09 Is Due to Modulation of NS1-Mediated Cellular Innate Immune Responses. J. Antimicrob. Chemother. 2014, 69, 1298–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Z.; Kuang, X.P.; Zhou, Q.Q.; Yan, C.Y.; Li, W.; Gong, H.B.; Kurihara, H.; Li, W.X.; Li, Y.F.; He, R.R. Inhibitory Effects of Baicalein against Herpes Simplex Virus Type 1. Acta Pharm. Sin. B 2020, 10, 2323–2338. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, L.; Liu, X.; Xin, L.; Wu, S.; Chen, X. Development of Potent Promoters That Drive the Efficient Expression of Genes in Apple Protoplasts. Hortic. Res. 2021, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Zhang, K.; Ding, Z.; He, Q.; Li, W.; Zhu, S.; Cheng, W.; Zhang, K.; Li, K. Characterization of a Strong and Constitutive Promoter from the Arabidopsis Serine Carboxypeptidase-like Gene AtSCPL30 as a Potential Tool for Crop Transgenic Breeding. BMC Biotechnol. 2018, 18, 59. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, J.; Xie, L.; Liu, T.; Mo, C.; Cui, S.; Jia, X.; Huang, X.; Luo, Z.; Ma, X. Heterologous Biosynthesis of Health-Promoting Baicalein in Lycopersicon esculentum. Molecules 2022, 27, 3086. https://doi.org/10.3390/molecules27103086
Liao J, Xie L, Liu T, Mo C, Cui S, Jia X, Huang X, Luo Z, Ma X. Heterologous Biosynthesis of Health-Promoting Baicalein in Lycopersicon esculentum. Molecules. 2022; 27(10):3086. https://doi.org/10.3390/molecules27103086
Chicago/Turabian StyleLiao, Jingjing, Lei Xie, Tingyao Liu, Changming Mo, Shengrong Cui, Xunli Jia, Xiyang Huang, Zuliang Luo, and Xiaojun Ma. 2022. "Heterologous Biosynthesis of Health-Promoting Baicalein in Lycopersicon esculentum" Molecules 27, no. 10: 3086. https://doi.org/10.3390/molecules27103086
APA StyleLiao, J., Xie, L., Liu, T., Mo, C., Cui, S., Jia, X., Huang, X., Luo, Z., & Ma, X. (2022). Heterologous Biosynthesis of Health-Promoting Baicalein in Lycopersicon esculentum. Molecules, 27(10), 3086. https://doi.org/10.3390/molecules27103086





