Design, Synthesis, Antibacterial Evaluations and In Silico Studies of Novel Thiosemicarbazides and 1,3,4-Thiadiazoles
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Antibacterial Evaluation
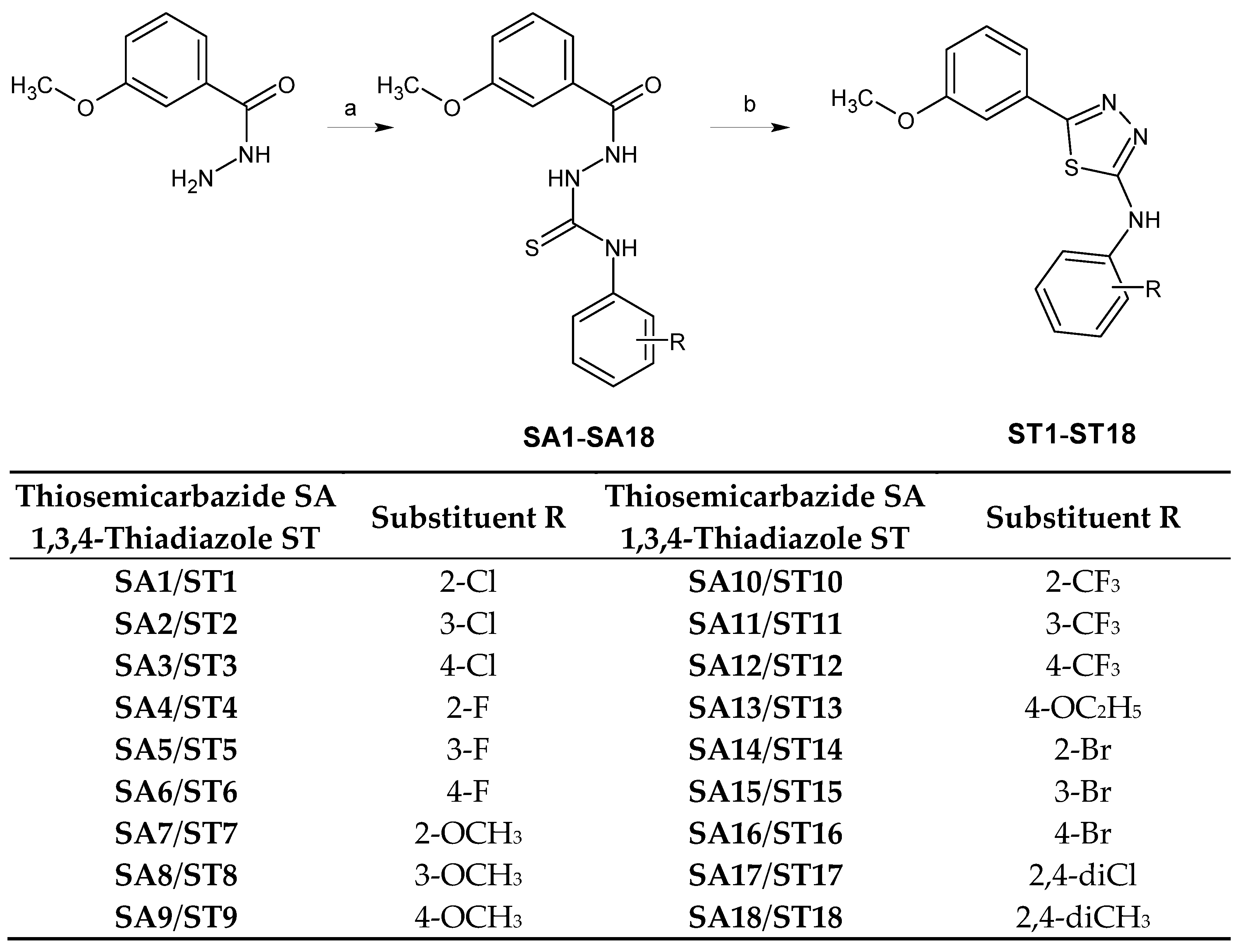
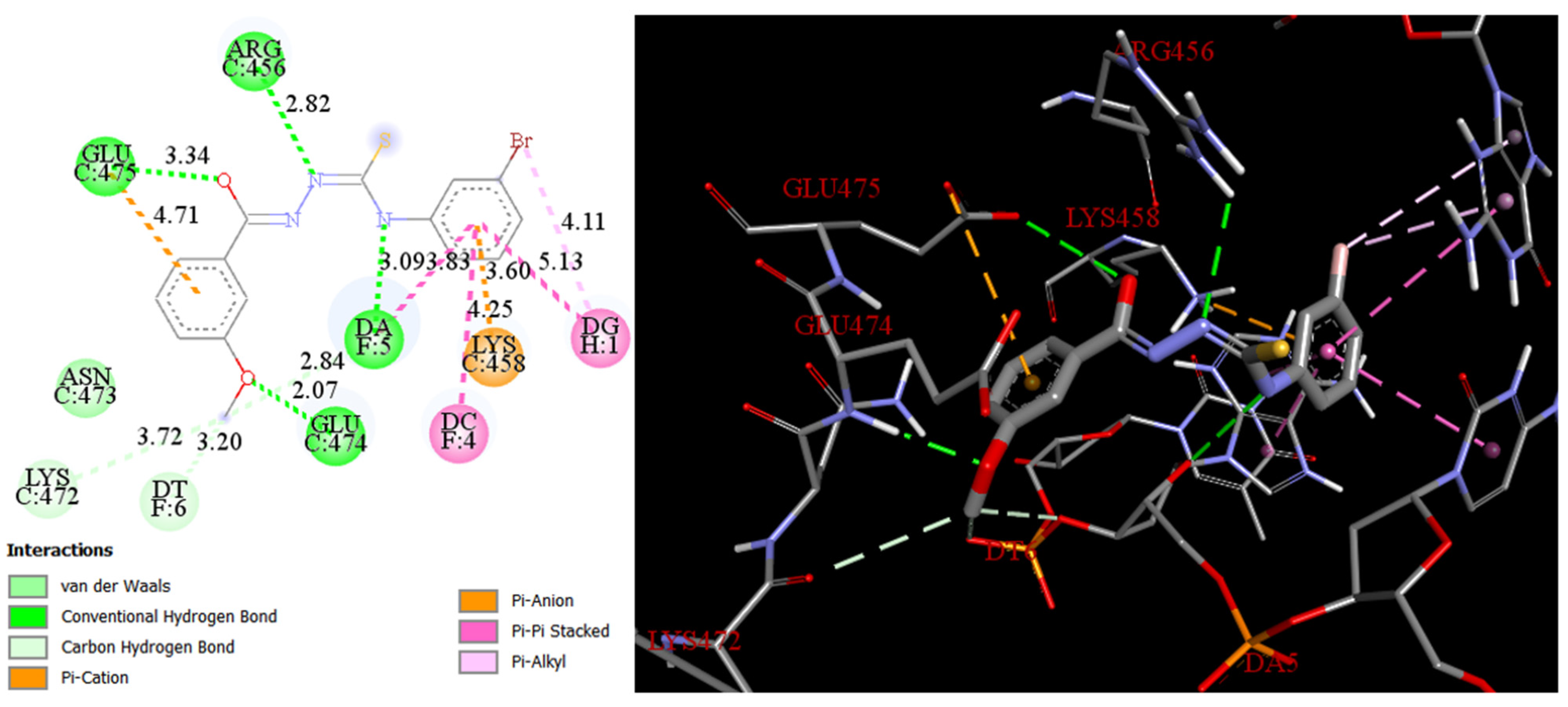
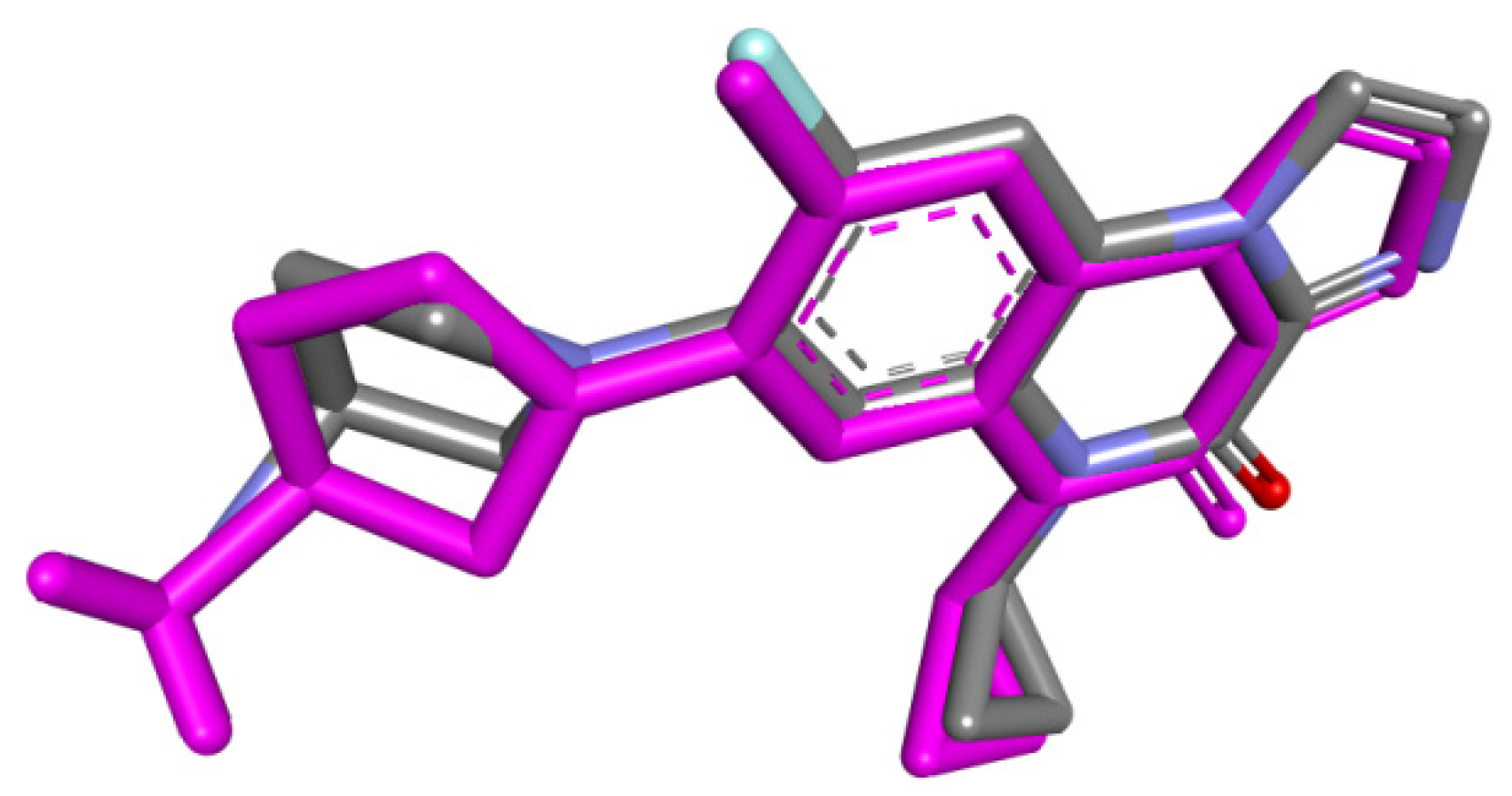
2.3. Docking
3. Experimental
3.1. Chemistry
3.1.1. General Comments
3.1.2. Synthesis of Thiosemicarbazide Derivatives SA13-SA18
3.1.3. Synthesis of 5-(3-Methoxyphenyl)-2-Substituted-1,3,4-Thiadiazole ST13-ST18
3.2. Microbiology
3.3. Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Othman, A.A.; Kihel, M.; Amara, S. 1,3,4-Oxadiazole, 1,3,4-thiadiazole and 1, 2, 4-triazole derivatives as potential antibacterial agents. Arab. J. Chem. 2019, 12, 1660–1675. [Google Scholar] [CrossRef] [Green Version]
- Wayne, P.A.; Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Inform. Suppl. 2011, 31, 100–121. [Google Scholar]
- Valliammai, A.; Sethupathy, S.; Priya, A.; Selvaraj, A.; Bhaskar, J.P.; Krishnan, V.; Pandian, S.K. 5-Dodecanolide interferes with biofilm formation and reduces the virulence of Methicillin-resistant Staphylococcus aureus (MRSA) through up regulation of agr system. Sci. Rep. 2019, 9, 13744. [Google Scholar] [CrossRef] [PubMed]
- El Sayed, H.; Awad, L.F.; Badawy, M.E.; Rabea, E.I.; Ibrahim, N.A.; Abd Al Moaty, M.N. Synthesis, antibacterial, antioxidant, and molecular docking studies of 6-methylpyrimidin-4(3H)-one and oxo-1,2,4-triazolo[4,3-a]pyrimidine derivatives. J. Mol. Struct. 2022, 1249, 131551. [Google Scholar]
- Khan, S.A.; Asiri, A.M.; Al-Ghamdi, N.S.M.; Asad, M.; Zayed, M.E.; Elroby, S.A.; Sharma, K. Microwave assisted synthesis of chalcone and its polycyclic heterocyclic analogues as promising antibacterial agents: In vitro, in silico and DFT studies. J. Mol. Struct. 2019, 1190, 77–85. [Google Scholar] [CrossRef]
- Amin, N.H.; El-Saadi, M.T.; Ibrahim, A.A.; Abdel-Rahman, H.M. Design, synthesis and mechanistic study of new 1,2,4-triazole derivatives as antimicrobial agents. Bioorg. Chem. 2021, 111, 104841. [Google Scholar] [CrossRef]
- Fang, Z.; Zheng, S.; Chan, K.F.; Yuan, W.; Guo, Q.; Wu, W.; Sun, N. Design, synthesis and antibacterial evaluation of 2,4-disubstituted-6-thiophenyl-pyrimidines. Eur. J. Med. Chem. 2019, 161, 141–153. [Google Scholar] [CrossRef]
- Dawoud, N.T.; El-Fakharany, E.M.; Abdallah, A.E.; El-Gendi, H.; Lotfy, D.R. Synthesis, and docking studies of novel heterocycles incorporating the indazolylthiazole moiety as antimicrobial and anticancer agents. Sci. Rep. 2022, 12, 3424. [Google Scholar] [CrossRef]
- Rodrigues, M.; Sharath, B.S.; Bennehalli, B.; Vagdevi, H.M. Synthesis, biological evaluation and molecular docking studies of heterocycle encompassed benzoxazole derivatives as antimicrobial agents. Mater. Today Proc. 2022, 49, 849–853. [Google Scholar] [CrossRef]
- Salgın-Gökşen, U.; Gökhan-Kelekçi, N.; Göktaş, Ö.; Köysal, Y.; Kılıç, E.; Işık, Ş.; Özalp, M. 1-Acylthiosemicarbazides, 1,2,4-triazole-5(4H)-thiones, 1,3,4-thiadiazoles and hydrazones containing 5-methyl-2-benzoxazolinones: Synthesis, analgesic, anti-inflammatory and antimicrobial activities. Bioorg. Med. Chem. Lett. 2007, 15, 5738–5751. [Google Scholar] [CrossRef]
- Tehranchian, S.; Akbarzadeh, T.; Fazeli, M.R.; Jamalifar, H.; Shafiee, A. Synthesis and antibacterial activity of 1-[1,2,4-triazol-3-yl] and 1-[1,3,4-thiadiazol-2-yl]-3-methylthio-6,7-dihydrobenzo[c]thiophen-4(5H)ones. Bioorg. Med. Chem. Lett. 2005, 15, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Caleb, A.A.; Ballo, D.; Rachid, B.; Amina, H.; Mostapha, B.; Abdelfettah, Z.; Mokhtar, E.E. Synthesis and antibacterial activity of new spiro[thiadiazoline-quinoxaline]derivatives. Arkivoc 2011, 2, 217–226. [Google Scholar]
- Narendhar, B.; Alagarsamy, V.; Krishnan, C. Design and Synthesis of novel 1-substituted-3-(3-(3-nitrophenyl)-4-oxo-3,4-dihydrobenzopyrimidin-2-ylamino)isothioureas for their anti-HIV, antibacterial activities, graph theoretical analysis, in silico modeling, prediction of toxicity and metabolic studies. Drug Res. 2020, 70, 348–355. [Google Scholar]
- Toan, V.N.; Tri, N.M.; Thanh, N.D. Substituted 4-formyl-2H-chromen-2-ones: Their reaction with N-(2,3,4,6-tetra-O-acetyl-β-D-galactopyranosyl)thiosemicarbazide, antibacterial and antifungal activity of their thiosemicarbazone products. Curr. Org. Chem. 2020, 24, 2272–2282. [Google Scholar] [CrossRef]
- Molnar, M.; Tomić, M.; Pavić, V. Coumarinyl thiosemicarbazides as antimicrobial agents. Pharm. Chem. J. 2018, 51, 1078–1081. [Google Scholar] [CrossRef]
- Muğlu, H.; Akın, M.; Çavuş, M.S.; Yakan, H.; Şaki, N.; Güzel, E. Exploring of antioxidant and antibacterial properties of novel 1,3,4-thiadiazole derivatives: Facile synthesis, structural elucidation and DFT approach to antioxidant characteristics. Comput. Biol. Chem. 2022, 96, 107618. [Google Scholar] [CrossRef]
- Ernazarova, B.; Zhusubaliev, T.; Bakirova, A.; Zhusupbaeva, G.; Akparalieva, O.; Abdullaeva, Z.; Orozmatova, A. Study of Biological Activity and Toxicity of Thiosemicarbazides Carbohydrate Derivatives by in Silico, In Vitro and In Vivo Methods. J. Agric. Chem. Environ. 2022, 11, 15–23. [Google Scholar]
- Bhakiaraj, D.; Elavarasan, T.; Mathavan, M.; Megala, S.; Enbaraj, E.; Gopalakrishnan, M. Synthesis, spectral analysis, antibacterial activity and molecular docking studies of some novel derivatives of combined tetrazole and thiosemicarbazide moieties. J. Adv. Sci. Res. 2021, 12, 210–218. [Google Scholar] [CrossRef]
- Han, M.İ.; İnce, U.; Gündüz, M.G.; Küçükgüzel, Ş.G. Synthesis, Antimicrobial Evaluation, and Molecular Modeling Studies of New Thiosemicarbazide-Triazole Hybrid Derivatives of (S)-Naproxen. Chem. Biodivers. 2022, 19, e202100900. [Google Scholar] [CrossRef]
- Sriram, D.; Yogeeswari, P.; Thirumurugan, R.; Pavana, R.K. Discovery of new antitubercular oxazolyl thiosemicarbazones. J. Med. Chem. 2006, 49, 3448–3450. [Google Scholar] [CrossRef]
- García, C.C.; Brousse, B.N.; Carlucci, M.J.; Moglioni, A.G.; Alho, M.M.; Moltrasio, G.Y.; Damonte, E.B. Inhibitory effect of thiosemicarbazone derivatives on Junin virus replication in vitro. Antivir. Chem. Chemother. 2003, 14, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Plech, T.; Wujec, M.; Siwek, A.; Kosikowska, U.; Malm, A. Synthesis and antimicrobial activity of thiosemicarbazides, s-triazoles and their Mannich bases bearing 3-chlorophenyl moiety. Eur. J. Med. Chem. 2011, 46, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Kalhor, M.; Shabani, M.; Nikokar, I.; Banisaeed, S.R. Synthesis, characterization and antibacterial activity of some novel thiosemicarbazides, 1,2,4-triazol-3-thiols and their S-substituted derivatives. Iran J. Pharm. Res. 2015, 14, 67. [Google Scholar] [PubMed]
- Bhat, M.A.; Khan, A.A.; Ghabbour, H.A.; Quah, C.K.; Fun, H.K. Synthesis, characterization, x-ray structure and antimicrobial activity of N-(4-chlorophenyl)-2-(pyridin-4-ylcarbonyl) hydrazinecarbothioamide. Trop. J. Pharm. Res. 2016, 15, 1751–1757. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Geng, J.; Liu, Y.; Yu, S.; Zhao, G. Thiadiazole—A promising structure in medicinal chemistry. ChemMedChem 2013, 8, 27–41. [Google Scholar] [CrossRef]
- Siwek, A.; Stączek, P.; Wujec, M.; Stefańska, J.; Kosikowska, U.; Malm, A.; Paneth, P. Biological and docking studies of topoisomerase IV inhibition by thiosemicarbazides. J. Mol. Model. 2011, 17, 2297–2303. [Google Scholar] [CrossRef]
- Kowalczyk, A.; Paneth, A.; Trojanowski, D.; Paneth, P.; Zakrzewska-Czerwińska, J.; Stączek, P. Thiosemicarbazide Derivatives Decrease the ATPase Activity of Staphylococcus aureus Topoisomerase IV, Inhibit Mycobacterial Growth, and Affect Replication in Mycobacterium smegmatis. Int. J. Mol. Sci. 2021, 22, 3881. [Google Scholar] [CrossRef]
- Plech, T.; Paneth, A.; Kaproń, B.; Kosikowska, U.; Malm, A.; Strzelczyk, A.; Stączek, P. Structure–activity Relationship studies of microbiologically active thiosemicarbazides derived from hydroxybenzoic acid hydrazides. Chem. Biol. Drug. Des. 2015, 85, 315–325. [Google Scholar] [CrossRef]
- Paneth, A.; Stączek, P.; Plech, T.; Strzelczyk, A.; Dzitko, K.; Wujec, M.; Paneth, P. Biological evaluation and molecular modelling study of thiosemicarbazide derivatives as bacterial type IIA topoisomerases inhibitors. J. Enzyme Inhib. Med. Chem. 2016, 31, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Janowska, S.; Khylyuk, D.; Bielawska, A.; Szymanowska, A.; Gornowicz, A.; Bielawski, K.; Noworól, J.; Mandziuk, S.; Wujec, M. New 1,3,4-Thiadiazole Derivatives with Anticancer Activity. Molecules 2022, 27, 1814. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Naghavi, M. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Germe, T.; Vörös, J.; Jeannot, F.; Taillier, T.; Stavenger, R.A.; Bacqué, E.; Bax, B.D. A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance. Nucleic Acids Res. 2018, 46, 4114–4128. [Google Scholar] [CrossRef] [PubMed]
- Laponogov, I.; Pan, X.S.; Veselkov, D.A.; McAuley, K.E.; Fisher, L.M.; Sanderson, M.R. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS ONE 2010, 5, 11338. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI Supplement M100; Clinical Lab Standards Institute: Malvern, PA, USA, 2022; Volume 42, pp. 34–78. [Google Scholar]
- Hashem, H.E.; Amr, A.E.G.E.; Nossier, E.S.; Elsayed, E.A.; Azmy, E.M. Synthesis, antimicrobial activity and molecular docking of novel thiourea derivatives tagged with thiadiazole, imidazole and triazine moieties as potential DNA gyrase and topoisomerase iv inhibitors. Molecules 2020, 25, 2766. [Google Scholar] [CrossRef] [PubMed]
- El-Hachem, N.; Haibe-Kains, B.; Khalil, A.; Kobeissy, F.H.; Nemer, G. AutoDock and AutoDockTools for protein-ligand docking: Beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) as a case study. In Neuroproteomics; Humana Press: New York, NY, USA, 2017; pp. 391–403. [Google Scholar]
- Available online: https://www.3ds.com/products-services/biovia/products/scientific-informatics/biovia-draw/ (accessed on 20 February 2022).
- Deeb, O.; Rosales-Hernández, M.C.; Gómez-Castro, C.; Garduño-Juárez, R.; Correa-Basurto, J. Exploration of human serum albumin binding sites by docking and molecular dynamics flexible ligand–protein interactions. Biopolymers 2010, 93, 161–170. [Google Scholar] [CrossRef]
- Gasteiger, J.; Marsili, M. A new model for calculating atomic charges in molecules. Tetrahedron Lett. 1978, 19, 3181–3184. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef] [Green Version]
- Yusuf, D.; Davis, A.M.; Kleywegt, G.J.; Schmitt, S. An alternative method for the evaluation of docking performance: RSR vs RMSD. J. Chem. Inf. Model. 2008, 48, 1411–1422. [Google Scholar] [CrossRef]

| Compounds | Staphylococcus aureus ATCC 25923 | Staphylococcus aureus ATCC 43300 | Staphylococcus epidermidis ATCC 12228 | Micrococcus luteus ATCC 10240 | ||||
|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| SA1 | 62.5 | >1000 | 62.5 | >1000 | 62.5 | >1000 | 62.5 | >1000 |
| SA2 | 1000 | nd | >1000 | nd | 500 | nd | 15.63 | >1000 |
| SA3 | 1000 | nd | 500 | nd | 125 | >1000 | 250 | >1000 |
| SA4 | 1000 | nd | >1000 | nd | 1000 | nd | >1000 | nd |
| SA5 | 125 | >1000 | 1000 | nd | >1000 | nd | >1000 | nd |
| SA6 | 500 | nd | 1000 | nd | 250 | >1000 | 250 | >1000 |
| SA7 | 250 | >1000 | 1000 | nd | 62.5 | 1000 | 500 | nd |
| SA10 | 250 | >1000 | 250 | >1000 | 62.5 | >1000 | 15.63 | 15.63 |
| SA11 | 250 | >1000 | 250 | >1000 | 31.25 | >1000 | 3.9 | 3.9 |
| SA12 | 500 | nd | >1000 | nd | 125 | >1000 | 3.9 | 3.9 |
| SA13 | 1000 | nd | 500 | nd | 125 | >1000 | 250 | 1000 |
| SA14 | 125 | >1000 | 250 | >1000 | 125 | >1000 | 250 | 250 |
| SA15 | 500 | nd | 1000 | nd | 250 | >1000 | 15.63 | 15.63 |
| SA16 | 125 | >1000 | >1000 | nd | 500 | nd | 15.63 | 15.63 |
| SA17 | 62.5 | >1000 | >1000 | nd | >1000 | nd | 15.63 | 500 |
| ST2 | 1000 | nd | 1000 | nd | 62.5 | 1000 | 500 | nd |
| ST3 | 1000 | nd | 500 | nd | 500 | nd | 250 | >1000 |
| ST4 | 500 | nd | >1000 | nd | 62.5 | >1000 | 31.25 | 125 |
| ST6 | >1000 | nd | 500 | nd | 1000 | nd | 500 | nd |
| ST7 | >1000 | nd | >1000 | nd | 31.25 | >1000 | 31.25 | 31.25 |
| ST8 | >1000 | nd | 1000 | nd | 31.25 | 1000 | 500 | nd |
| ST12 | >1000 | nd | 1000 | nd | 250 | >1000 | 1000 | nd |
| ST14 | >1000 | nd | 1000 | nd | 500 | nd | >1000 | nd |
| ST15 | >1000 | nd | 500 | nd | 250 | >1000 | >1000 | nd |
| ST16 | 250 | >1000 | 500 | nd | 31.25 | 1000 | 15.63 | 15.63 |
| ST17 | 250 | >1000 | 1000 | nd | 500 | >1000 | 125 | 125 |
| Compounds | Topoisomerase IV (3LTN) | DNA Gyrase (6FQM) | ||
|---|---|---|---|---|
| Binding Energy Kcal/mol | Inhibition Constant, Ki µM | Binding Energy Kcal/mol | Inhibition Constant, Ki µM | |
| SA1 | −8.38 | 0.72 | −7.76 | 2.06 |
| SA2 | −8.22 | 0.94 | −8.40 | 0.7 |
| SA3 | −8.27 | 0.86 | −7.97 | 1.44 |
| SA4 | −8.34 | 0.77 | −7.66 | 2.42 |
| SA5 | −7.81 | 1.89 | −7.92 | 1.57 |
| SA6 | −7.59 | 2.73 | −7.64 | 2.53 |
| SA7 | −7.75 | 2.09 | −6.90 | 8.79 |
| SA10 | −7.90 | 1.63 | −7.53 | 3.05 |
| SA11 | −8.18 | 1.01 | −6.93 | 8.36 |
| SA12 | −8.28 | 0.85 | −8.06 | 1.23 |
| SA 13 | −8.24 | 0.91 | −7.62 | 2.61 |
| SA14 | −7.86 | 1.74 | −8.93 | 0.28 |
| SA15 | −9.11 | 0.21 | −8.84 | 0.33 |
| SA16 | −7.72 | 2.21 | −7.85 | 1.77 |
| SA17 | −8.74 | 0.39 | −8.00 | 1.36 |
| ST2 | −8.58 | 0.51 | −8.67 | 0.44 |
| ST3 | −8.58 | 0.51 | −8.67 | 0.44 |
| ST4 | −8.12 | 1.12 | −8.13 | 1.11 |
| ST6 | −7.44 | 3.52 | −7.68 | 2.33 |
| ST7 | −8.22 | 0.93 | −8.16 | 1.04 |
| ST8 | −8.56 | 0.53 | −8.12 | 1.13 |
| ST12 | −7.25 | 4.88 | −7.60 | 2.71 |
| ST14 | −8.31 | 0.81 | −8.96 | 0.27 |
| ST15 | −8.64 | 0.46 | −8.16 | 1.04 |
| ST16 | −7.72 | 2.21 | −7.85 | 1.77 |
| ST17 | −7.85 | 1.76 | −7.67 | 2.41 |
| PD 0305970 | −8.38 | 0.72 | - | - |
| E32 | - | - | −9.29 | 0.155 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janowska, S.; Khylyuk, D.; Andrzejczuk, S.; Wujec, M. Design, Synthesis, Antibacterial Evaluations and In Silico Studies of Novel Thiosemicarbazides and 1,3,4-Thiadiazoles. Molecules 2022, 27, 3161. https://doi.org/10.3390/molecules27103161
Janowska S, Khylyuk D, Andrzejczuk S, Wujec M. Design, Synthesis, Antibacterial Evaluations and In Silico Studies of Novel Thiosemicarbazides and 1,3,4-Thiadiazoles. Molecules. 2022; 27(10):3161. https://doi.org/10.3390/molecules27103161
Chicago/Turabian StyleJanowska, Sara, Dmytro Khylyuk, Sylwia Andrzejczuk, and Monika Wujec. 2022. "Design, Synthesis, Antibacterial Evaluations and In Silico Studies of Novel Thiosemicarbazides and 1,3,4-Thiadiazoles" Molecules 27, no. 10: 3161. https://doi.org/10.3390/molecules27103161






