Abstract
Two new antimycin A analogues, antimycin B1 and B2 (1–2), were isolated from a spent broth of a marine-derived bacterium, Streptomyces lusitanus. The structures of 1 and 2 were established on the basis of spectroscopic analyses and chemical methods. The isolated compounds were tested for their anti-bacterial potency. Compound 1 was found to be inactive against the bacteria Bacillus subtilis, Staphyloccocus aureus, and Loktanella hongkongensis. Compound 2 showed antibacterial activities against S. aureus and L. hongkongensis with MIC values of 32.0 and 8.0 μg/mL, respectively.
1. Introduction
Marine actinomycetes are chemically rich sources of structurally diverse secondary metabolites. The vast majority of these secondary metabolites is mainly derived from members of the genus Streptomyces [1,2]. Approximately 289 secondary metabolites from the marine-derived genus of Streptomyces are reported in the Marinlit database, covering a wide variety of chemical structures, including peptides, macrolides, lactones, indoles, terpenes, and quinones. These compounds show an extensive range of activities, such as cytotoxic, antibacterial, antifungal and antimalarial [3]. The bioactive secondary metabolites from marine-derived Streptomyces have thus attracted increasing interest during the last decades [4,5,6].
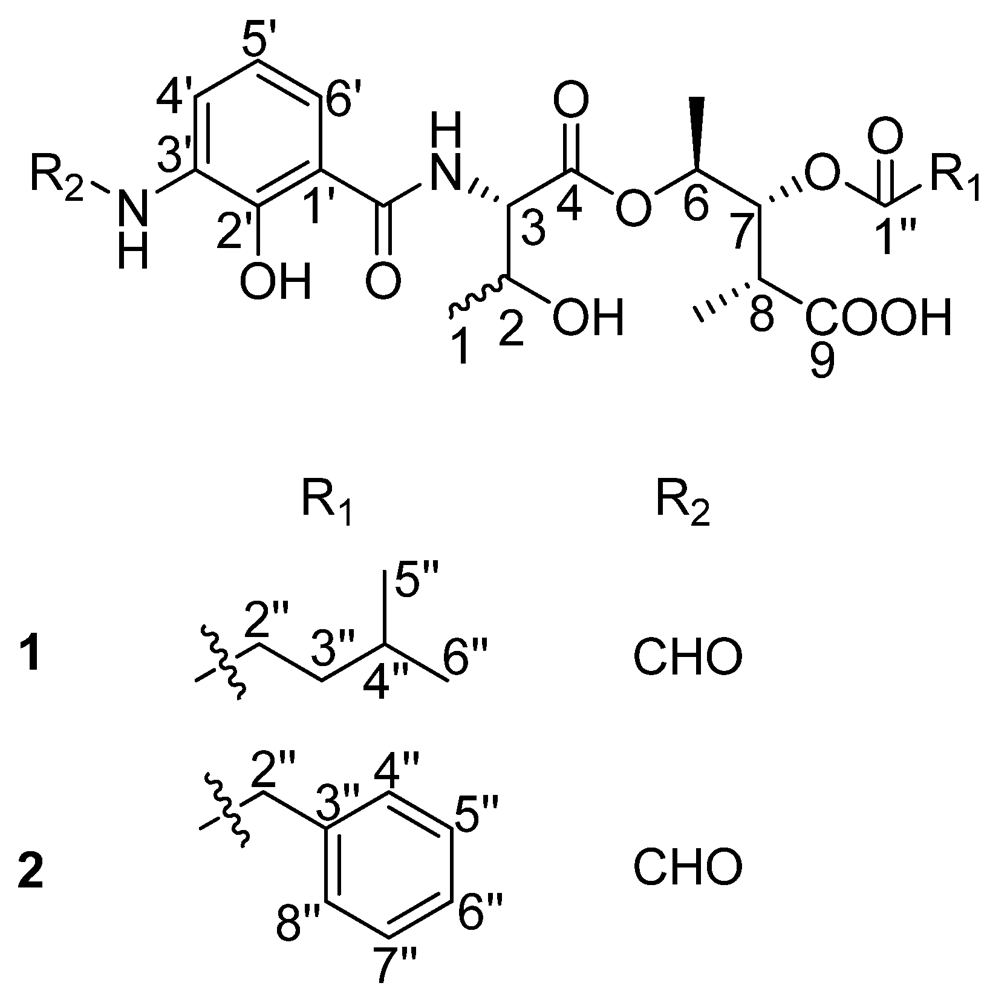
In our endeavor to search for novel antibacterial secondary metabolites from marine Streptomyces using bioassay and LC-UV/Vis-MS guided methods, we isolated two new antimycin A analogues. Antimycin A is characterized by the presence of a nine-membered dilactone ring, a carboxyl phenol amido unit and two alkyl side chains of varying lengths, with the only exception being antimycin A9, which has an aromatic 8-acyl residue [7]. The first group of antimycin A compounds was isolated as fungicides from Streptomyces sp. in 1949 [8]. Molecular studies showed that these compounds could inhibit the mitochondrial electron transport chain between cytochromes b and c [9]. To date, 20 antimycin A compounds have been reported [10]. A recent report analyzing the biosynthetic gene clusters of antimycin A from Streptomyces sp. S4 revealed that these compounds were synthesized by a hybrid NRPS/PKS [11]. Here, we report on the discovery of the first naturally occurring ring-opened antimycin A analogues which were named as antimycin B1 and B2 (Figure 1) as well as their antibacterial activity.
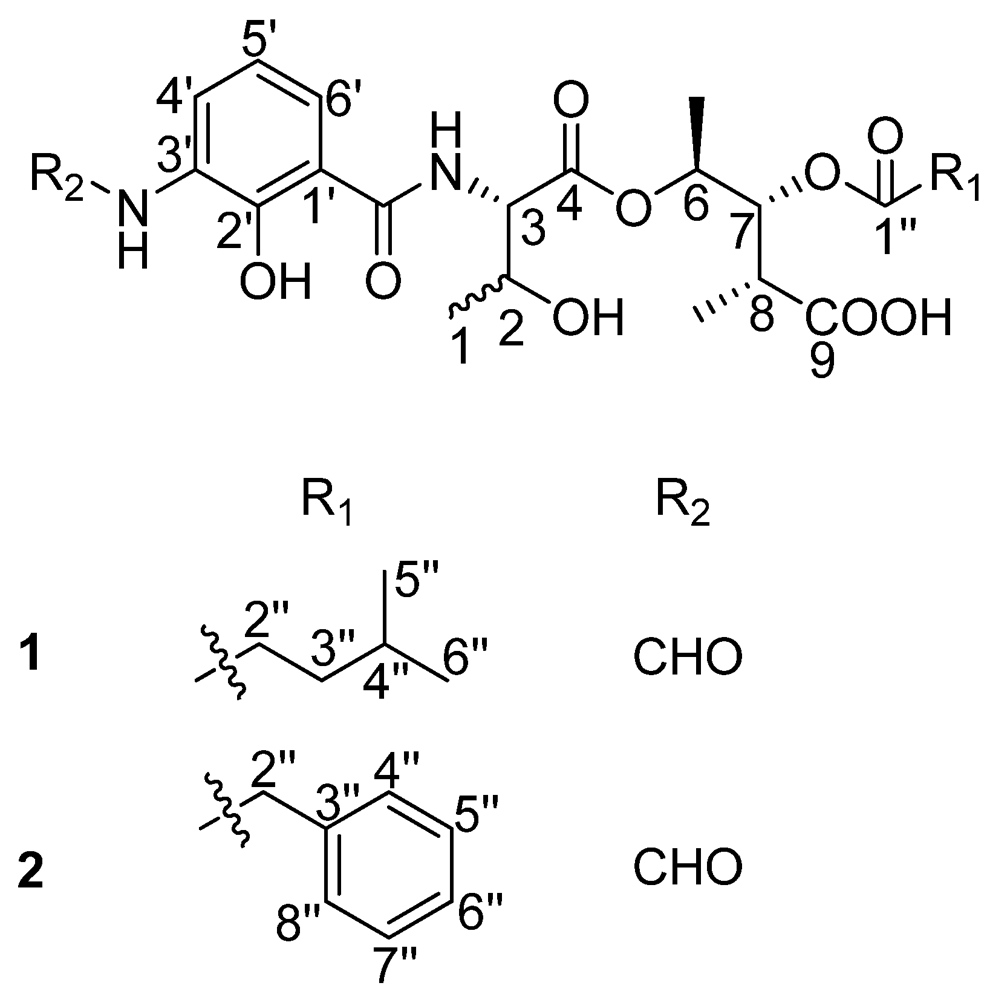
Figure 1.
The structures of compounds 1 and 2.
Figure 1.
The structures of compounds 1 and 2.
2. Results and Discussion
Compound 1 was obtained as a light-brown amorphous solid with a molecular formula of C24H34N2O10, which was deduced from NMR and HRESITOFMS data (Obsd [M + H]+ at m/z 511.2290), which suggested nine degrees of unsaturation. The UV spectrum indicated absorption at λmax (ε) 220 (22,900) and 317 (5800) nm. The 1H NMR data for 1 (Table 1) indicated the presence of five exchangeable protons at δH 13.00 (s), 12.60 (s), 9.83 (brs), 8.81 (d, J = 8.0 Hz), and 5.09 (s), a formyl proton at δH 8.32 (d, J = 1.5 Hz), three aromatic protons at δH 8.27 (d, J = 8.0 Hz),7.80 (d, J = 8.0 Hz), and 6.92 (t, J = 8.0 Hz), four methine protons directly connected to heteroatoms δH 5.23 (dd, J = 10.0, 5.5 Hz), 4.98 (dq, J = 6.5, 5.5 Hz), 4.45 (dd, J = 8.0, 4.5 Hz), and 4.20 (m), one methine signal at δH 1.50 (m), and two methylene signals at δH 2.26 (m) and 1.37 (m) together with five methyl groups at δH 1.19 (d, J = 6.5 Hz), 1.16 (d, J = 6.5 Hz), 1.00 (d, J = 7.0 Hz), 0.84 (d, J = 7.0 Hz), and 0.82 (d, J = 6.5 Hz). These data suggested that 1 is an antimycin A analogue [8].

Table 1.
1H and 13C NMR data of compounds 1 and 2 (DMSO-d6, δ in ppm, J in Hz).
| Position | Compound 1 | Compound 2 | |||
|---|---|---|---|---|---|
| δC a | δH (J in Hz) b | δC a | δH (J in Hz) b | ||
| 1 | 20.04 | 1.16 d (6.5) | 20.02 | 1.14 d (6.5) | |
| 2 | 65.7 | 4.20 m | 65.7 | 4.16 m | |
| 2-OH | 5.09 s | 5.05 s | |||
| 3 | 58.6 | 4.45 dd (8.0, 4.5) | 58.7 | 4.45 dd (7.8, 4.3) | |
| 4 | 169.3 | 169.3 | |||
| 6 | 69.3 | 4.98 dq (6.5, 5.5) | 69.3 | 4.96 dq (6.5, 6.5) | |
| 7 | 73.0 | 5.23 dd (10.0, 5.5) | 73.4 | 5.22 dd (7.0, 6.5) | |
| 8 | 38.8 | 2.69 m | 38.8 | 2.67 m | |
| 9 | 174.5 | 12.60 s | 174.6 | 12.60 s | |
| 6-Me | 14.88 | 1.19 d (6.5) | 14.59 | 1.14 d (6.5) | |
| 8-Me | 11.22 | 1.00 d (7.0) | 11.25 | 0.96 d (7.0) | |
| 1′ | 113.9 | 113.6 | |||
| 2′ | 150.7 | 150.5 | |||
| 3′ | 127.0 | 126.6 | |||
| 4′ | 124.2 | 8.27 d (8.0) | 124.3 | 8.26 d (8.0) | |
| 5′ | 117.8 | 6.92 t (8.0) | 117.8 | 6.91 t (8.0) | |
| 6′ | 122.9 | 7.80 d (8.0) | 122.1 | 7.77 d (8.0) | |
| 1′-CONH | 170.0 | 8.81 d (8.0) | 169.6 | 8.80 brs | |
| 2′-OH | 13.00 s | 13.00 s | |||
| 3′-NHCHO | 9.83 brs | 9.83 brs | |||
| 3′-NHCHO | 160.5 | 8.32 d (1.5) | 160.3 | 8.32 d (2.0) | |
| 1′′ | 172.2 | 170.5 | |||
| 2′′ | 31.2 | 2.26 m | 39.9 | 3.66 s | |
| 3′′ | 32.8 | 1.37 m | 133.8 | ||
| 4′′ | 26.6 | 1.50 m | 129.1 | 7.23 d (8.0) | |
| 5′′ | 21.74 | 0.84 d (7.0) | 128.0 | 7.31 t (8.0) | |
| 6′′ | 21.74 | 0.82 d (6.5) | 126.6 | 7.25 t (8.0) | |
| 7′′ | 128.0 | 7.31 t (8.0) | |||
| 8′′ | 129.1 | 7.23 d (8.0) | |||
a Recorded at 500 MHz; b Recorded at 125 MHz.
Two substructures I and II were assigned by analyzing the 1D and 2D NMR spectra (Figure 2). Substructure I was assembled starting from the amino acid threonine moiety that was inferred from the 1H–1H COSY correlations of δH 4.20 (m, 2-H) with δH 4.45 (dd, J = 8.0, 4.5 Hz, 3-H) and δH 1.16 (d, J = 6.5, 1-H), of δH 4.45 (3-H) with δH 8.81 (d, J = 8.0 Hz, 1′-CONH) and HMBC correlations of 3-H with δC 169.3 (C-4). The 1H–1H COSY correlations from δH 6.92 (t, J = 8.0 Hz, 5-H′) to δH 8.27 (d, J = 8.0, 4-H′) and 7.80 (d, J = 8.0 Hz, 6-H′) indicated the presence of an 1,2,3-trisubstituted benzene. The HMBC correlations from phenolic hydroxyl proton observed at δH 13.00 (s, 2′-OH) to δC 113.9 (C-1′), 150.7 (C-2′) and 127.0 (C-3′), from the formamide proton observed at δH 9.83 (brs, 3′-NHCHO) to C-3′, from H-6′ to δC 170.0 1′-CONH, suggested that 2′-OH, 3′-NHCHO, and 1′-CONH groups were attached on C-2′, C-3′ and C-1′, respectively. HMBC correlations from H-6′, 3-NH and H-3 to the carbonyl carbon δC 170.0 (1′-CONH), allowed the trisubstituted aromatic ring moiety and the threonine moiety to be connected.
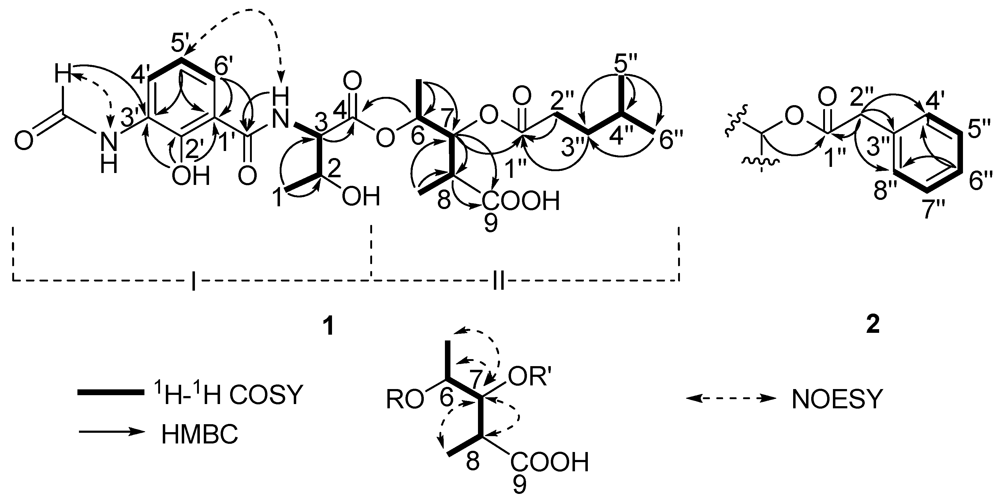
Figure 2.
The key 2D correlations of compounds 1 and 2.
Figure 2.
The key 2D correlations of compounds 1 and 2.
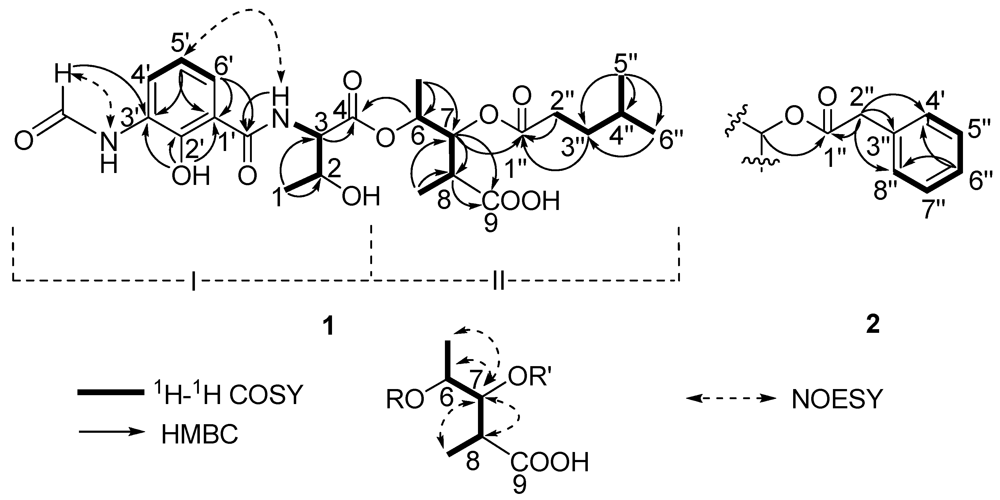
Substructure II was assembled from the following 1H–1H COSY and HMBC spectral data. 1H–1H COSY correlations from δH 4.98 (dq, J = 6.5, 5.5 Hz, 6-H) to δH 1.19 (d, J = 6.5 Hz, 6-Me) and δH 5.23 (dd, J = 10.0, 5.5 Hz, 7-H), from δH 2.69 (m, 8-H) to 7-H and δH 1.00 (d, J = 7.0, 8-Me) suggested the presence of a 2,3,4-trisubstituted pentanoic acid group. HMBC correlations from 7-H, 8-H and 8-Me to δC 174.5 (C-9) indicated a carbonyl group attached on C-8. 1H-1H COSY correlations from δH 1.50 (m, 4′′-H) to 0.84 (d, J = 7.0 Hz, 5′′-H), δH 0.82 (d, J = 6.5, 6′′-H) and δH 1.37 (m, 3′′-H), and from 3′′-H to δH 2.26 (m, 2′′-H), together with HMBC correlations from 3′′-H and 2′′-H to δC 172.2 (C-1′′) indicated the presence of a 4-methylvaleryl group. The long-range coupling between 7-H and C-1′′ revealed that the 4-methylvaleryl residue was connected to C-7 via an ester bond. The HMBC correlation of 6-H with C-4 allowed the substructures I and II to be connected.
There are five stereocenters in compound 1. To assign the absolute configurations, two reactions were considered. The strategy was to establish the absolute stereochemistry of the threonine group using Marfey’s reagent, followed by lactonization to form the nine-membered dilactone ring found in cyclized analogs. If lactonization was successful, then NOEs could be obtained via NOESY or ROESY NMR experiments to complete the assignments. The threonine moiety of 1 was defined by acid hydrolysis and Marfey’s method [12,13], using standard amino acids, L-Threonine, L-allo-Threonine, D-Threonine and D-allo-Threonine. The derivatized threonine residue from compound 1 gave the same retention time as that prepared from standard L-Threonine and L-allo-Threonine (RT = 29.3 min). Thus, the absolute configuration of the C-3 was determined as 3S, but C-2 remains unresolved. The relative configurations at C-6, C-7 and C-8 were assigned as 6S*, 7R*, 8R* based on comparison of 1H and 13C chemical shifts and 1H-1H coupling constant data with literature values [14]. To determine the absolute configurations of C-2, C-6, C-7 and C-8, lactonization was attempted using a modification of Shiina’s method [15]. The mixture products were checked using LC-MS. However, no expected dehydrated molecule mass data was observed. Due to the limited amount of the compound, it was not possible to perform additional experiments to determine the absolute configurations of C-2, C-6, C-7 and C-8. Thus, the structure of 1 was determined as depicted in Figure 1; it is named antimycin B1.
Compound 2 had a molecular formula of C26H31N2O10 deduced from its HRESITOFMS data (Obsd [M + H]+ at m/z 531.1985) and NMR spectra. Comparison of the 1H and 13C NMR spectral data (Table 1) revealed close similarities between 2 and 1. The difference between them was the absence of the 4-methylvaleryl residue in Substructure II of 1 and the appearance of a phenyl-acetyl group in 2, which is verified by HMBC correlations from δH 3.66 (s, 2′′-H) to carbonyl carbon δC 170.5 (C-1′′) and carbons on the second aromatic ring δC 133.8 (C-3′′), 129.1 (C-4′′, 8′′). The correlation between δH 5.22 (dd, J = 7.0, 6.5 Hz, 7-H) and C-1′′ revealed that the phenyl-acetyl residue was connected to C-7 by an ester bond. The absolute configuration of C-3 in 2 was also determined as 3S by applying Marfey’s method. The relative configurations at C-6, C-7 and C-8 were assigned as 6S*, 7R*, 8R* as in compound 1. Thus, the structure of compound 2, named antimycin B2, was determined and is also shown in Figure 1.
Compounds 1 and 2 were evaluated for their antibacterial activities against Staphylococcus aureus, Bacillus subtilis and Loktanella hongkongensis. Compound 1 did not show any activity against any of the three strains. Compound 2 showed selective activity against two strains, S. aureus and L. hongkongensis. See Table 2.

Table 2.
Antibacterial activities of compounds 1 and 2.
| Compound | Antibacterial (MIC, μg/mL) | ||
|---|---|---|---|
| Staphylococcus aureus | Loktanella hongkongensis | Bacillus subtillis | |
| 1 | NA | NA | NA |
| 2 | 32.00 | 8.00 | NA |
| Penicillin G | 0.25 | 2.00 | 0.10 |
| Streptomycin | 8.00 | 16.00 | 8.00 |
NA means there is no bioactivity with a MIC > 32.00 μg/mL.
3. Experimental Section
3.1. General Experimental Procedures
The 1D and 2D NMR spectral data were obtained on Varian Inova 500 MHz NMR spectrometers. UV spectra were recorded on a Varian Cary 50 Conc UV Visible spectrophotometer with a path length of 1 cm. Optical rotations were measured using a Rudolph Research Autopol III Automatic polarimeter with a 10 cm cell. OD measurements of the antibacterial experiments were recorded at 600 nm on a Biorad Model 680 microplate reader. High-resolution mass spectra were acquired from UPLC-TOF-MS. The UPLC system was a Waters ACQUITY UPLC system (Waters, Manchester, UK) coupled to a Bruker microTOF-q II mass spectrometer (Brucker Daltonics GmbH, Bremen, German).
3.2. Isolation of Strain XM52, Identification, Cultivation, and Extraction
Streptomyces strain XM52 was isolated from the rhizosphere of the mangrove plant Avicennia mariana from Fujian Province, China. Strain XM52 was classified as a Streptomyces lusitanus based on nearly complete 16S rDNA analysis, which showed 99% sequence identity with Streptomyces lusitanus strain NBRC13464 (the sequence data has been deposited in GenBank under accession number NR 041143.1). The strain was cultured in 90 × 1.0 L volumes of GPY media (10.0 g glucose, 5.0 g peptone, 3.0 g yeast extract, 10.0 g sea salts, 1.0 L double distilled water, pH 7.2) while shaking at 160 rpm for 14 days at 23 °C. The fermented culture (90.0 L) was filtered through cheese cloth (8 layers) to separate the mycelia. The filtrate was extracted with ethyl acetate three times, while the mycelia were extracted by 80% acetone under ultrasonication. Evaporation of the acetone in vacuo left a wet residue that was partitioned with EtOAc. The combined two EtOAc extracts were evaporated in vacuo at 40 °C to yield 7.9 g of brown oily residue. The EtOAc extract (7.9 g) was subjected to reversed-phase C18 flash chromatography successively eluted with solvent mixtures of H2O-MeOH (9:1), H2O-MeOH (7:3), H2O-MeOH (5:5), H2O-MeOH (3:7), H2O-MeOH (1:9), and 100% MeOH. The H2O-MeOH (3:7) eluting fraction (labeled Fr. 4) showed potent antibacterial activity against S. aureus, L. hongkongensis, The Fr. 4 fraction was then subjected to Sephadex LH-20 eluted with MeOH to yield 15 sub-fractions (labeled Fr.4-1 to Fr.4-15), Fr. 4-12 were purified by C-18 semi-preparative HPLC (Phenomenex Luna C18 (2) 250 × 10 mm column) with MeCN-H2O (38:62–42:58) to obtain pure compounds 1 (3.5 mg) and 2 (2.4 mg).
Antimycin B1 (1): Light-brown amorphous solid, 3.5 mg, [α]25D = 0 (c 0.1, CH3OH); UV (MeCN) λmax (ε): 220 (22,900) and 317 (5800) nm. HRESIMS: Obsd m/z 511.2290 [M + H]+ (C24H35N2O10 requires 511.2292). Table 1 presents the 1H and 13C NMR data.
Antimycin B2 (2): Light-brown amorphous solid, 2.4 mg, [α]25D = 0 (c 0.1, CH3OH); UV (MeCN) λmax (ε): 220 (23,300) and 317 (5900) nm. HRESIMS: Obsd m/z 531.1985 [M + H]+ (C26H31N2O10 requires 531.1979). Table 1 presents the 1H and 13C NMR data.
3.3. Marfey’s Analysis of the Threonine Moiety
A 0.2 mg-portion of compound 1 was placed in a sealed glass tube, dissolved in 6 N HCl (1.0 mL) and heated to 110 °C for 20 h. Hydrolysates were evaporated to dryness and then resuspended in water (40.0 μL). A solution of (1-fluoro-2,4-dinitrophenyl)-5-L-alanine amide (FDAA) (4.2 μmol) in acetone (150.0 μL) and then 1 N NaHCO3 (20.0 μL) was added to each reaction vessel and the mixtures were stirred at 40 °C for 2 h. A 2 N HCl solution (10.0 μL) was added to each reaction vessel to stop the reaction and the solution was evaporated in vacuo. The residues were then resuspended in 200.0 μL of MeOH and subjected to HPLC (Phenomenex Luna C18 (2) 250 × 4.5 mm column) using 50 mM TEAP/acetonitrile linear gradient elution (pH 3.0, from 10% to 40% acetonitrile during 45 min, flow rate 1.0 mL/min, at 340 nm), four standard Threonine-FDLA derivatives that had been prepared using the same method were compared. HPLC analysis of Marfey’s derivatives from the direct hydrolysis of 1 established the following retention times of the derivatized amino acids (reference derivatives tR): L-Thr 29.3 min (L-Thr 29.3 min, L-allo-Thr 29.3 min, D-Thr 31.6 min, D-allo-Thr 31.6 min).
3.4. Lactonization
A solution of compound 1 (2.0 mg) in dry toluene (1.0 mL) was slowly added over 7 h through a syringe pump to a toluene solution (0.5 mL) of 2-methyl-6-nitrobenzoic anhydride (MNBA) (1.3 mg, 3.75 μmol), 4-(dimethylamino) pyridine (DMAP) (1.4 mg, 15.0 μmol), stirred at room temperature under an atmosphere of N2. After the completion of the addition, the resulting mixture was stirred for another 13 h. The reaction mixture was then centrifuged. The suspension was diluted with EtOAc(50 mL), washed with aqueous saturated NaHCO3, water, and brine, and dried over anhydrous Na2SO4. The dried sample was then subjected to LC-MS to check the products.
3.5. Evaluation of Antibacterial Activity
The antibacterial activities of compounds 1 and 2 were evaluated by MIC assays against Staphylococcus aureus, Bacillus subtilis, and Loktanella hongkongensis. Briefly, the bacterial strains were inoculated in YP Broth (0.2% yeast extract, 0.1% peptone, 1.7% sea salts) and were incubated at 28 °C for 12 h. A stock solution of the sample was prepared at 50 mg/mL in DMSO and further diluted to varying concentrations in 96 well plates that contained the incubated microbial strains. The plates were incubated at 28 °C overnight. Cell growth was checked by measuring the optical density at 600 nm; growth inhibition was compared to that caused by varying concentrations of Penicillin G and Streptomycin (serving as the positive controls).
4. Conclusions
Two new antimycin A analogues, antimycin B1 (1) and antimycin B2 (2), were isolated from a spent broth of marine-derived actinomycete Streptomyces lusitanus. The key structural features of compounds 1 and 2 were characterized by spectroscopic analyses and chemical methods. The compounds were suspected to be artificial products originated during the treatment process. A previous degradation study of antimycin A compounds revealed that mild alkaline hydrolysis of antimycin A compounds yielded blastmycic acid and antimycin lactone, while blastmycic acid would be further degraded into antimycic acid under more severe conditions [16]. However, due to intramolecular reaction [17], no ring-opened dilactone products were observed. Thus, the antimycin B compounds described here are the first naturally occurring antimycin A dilactone opened products to be identified. Antibacterial activities were evaluated using MIC assays. Compound 2 showed moderate antibacterial activities against S. aureus, L. hongkongensis and B. subtilis, with the activity against L. hongkongensis being stronger than that of streptomycin. Compound 1 did not show any activities against these three bacterial strains.
Acknowledgments
The authors thank Rui Feng, Division of Life Science, Hong Kong University of Science and Technology, for the NMR spectra. This work was financially supported by a grant (KZCXZ-YW-T001) from the CAS/SAFEA International Partnership program for Creative Research Teams, a grant from the Research Grants Council of the Hong Kong Special Administrative Region (N_HKUST602/09), and a grant from China Ocean Mineral Resource Research and Development Association to P.Y. Qian.
References
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar]
- Pathom-aree, W.; Stach, J.E.M.; Ward, A.C.; Horikoshi, K.; Bull, A.T.; Goodfellow, M. Diversity of actinomycetes isolated from challenger deep sediment (10,898 m) from the Mariana Trench. Extremophiles 2006, 10, 181–189. [Google Scholar]
- Marinlit Database. Available online: http://www.chem.canterbury.ac.nz/marinlit/marinlit.shtml (accessed on 29 August 2011). University of Canterbury, Christchurch, New Zealand..
- Lam, K.S. Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 2006, 9, 245–251. [Google Scholar]
- Olano, C.; Méndez, C.; Salas, J. Antitumor compounds from marine Actinomycetes. Mar. Drugs 2009, 7, 210–248. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2011, 28, 196–268. [Google Scholar]
- Shiomi, K.; Hatae, K.; Hatano, H.; Matsumoto, A.; Takahashi, Y.; Jiang, C.L.; Tomoda, H.; Kobayashi, S.; Tanaka, H.; Omura, S. A new antibiotic, antimycin A9, produced by Streptomyces sp. K01-0031. J. Antibiot. 2005, 58, 74–78. [Google Scholar] [CrossRef]
- Dunshee, B.R.; Leben, C.; Keitt, G.W.; Strong, F.M. The isolation and properties of antimycin A. J. Am. Chem. Soc. 1949, 71, 2436–2437. [Google Scholar]
- Keilin, D.; Hartree, E.F. Relationship between certain components of the cytochrome system. Nature 1955, 176, 200–206. [Google Scholar] [CrossRef]
- Xu, L.Y.; Quan, X.S.; Wang, C.; Sheng, H.F.; Zhou, G.X.; Lin, B.R.; Jiang, R.W.; Yao, X.S. Antimycins A(19) and A(20), two new antimycins produced by marine actinomycete Streptomyces antibioticus H74-18. J. Antibiot. 2011, 64, 661–665. [Google Scholar] [CrossRef]
- Seipke, R.F.; Barke, J.; Brearley, C.; Hill, L.; Yu, D.W.; Goss, R.G.; Hutchings, M.I. A single Streptomyces mutualist makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus. PLoS One 2011, 6, e22028. [Google Scholar]
- Marfey, P.; Ottesen, M. Determination of D-amino acids. I. Hydrolysis of DNP-L-amino acid methyl esters with carboxypeptidase-Y. Carlsberg Res.Commun. 1984, 49, 585–590. [Google Scholar] [CrossRef]
- Marfey, P. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591–596. [Google Scholar] [CrossRef]
- Chakraborty, T.K.; Chattopadhyay, A.K.; Ghosh, S. Total synthesis of (+)-blastmycinone and formal synthesis of (+)-antimycin A3b. Tetrahedron Lett. 2007, 48, 1139–1142. [Google Scholar]
- Wu, Y.; Yang, Y.Q. An expeditious enantioselective synthesis of antimycin A3b. J. Org. Chem. 2006, 71, 4296–4301. [Google Scholar]
- Liu, W.C.; van Tamelen, E.E.; Strong, F.M. The chemistry of antimycin A. VIII. Degradation of antimycin A1. J. Am. Chem. Soc. 1960, 82, 1652–1654. [Google Scholar]
- van Tamelen, E.E.; Dickie, J.P.; Loomans, M.E.; Dewey, R.S.; Strong, F.M. The chemistry of antimycin A. X. Structure of the antimycins. J. Am. Chem. Soc. 1961, 83, 1639–1646. [Google Scholar]
- Samples Availability: Available from the authors.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
