Photoprotective Bioactivity Present in a Unique Marine Bacteria Collection from Portuguese Deep Sea Hydrothermal Vents
Abstract
:1. Introduction

2. Results and Discussion
2.1. New Marine Prokaryotes Isolates from MAR Vents
| Type of sample | ||||||
|---|---|---|---|---|---|---|
| Water | Animals | Sediments | Chimneys | Total | ||
| Phenptypic operational group | I | 18 | 87 | 24 | 11 | 140 |
| II | 11 | 56 | 44 | 14 | 125 | |
| III | 0 | 3 | 0 | 21 | 24 | |
| Total | 29 | 146 | 68 | 46 | 289 | |
| (a) | ||||||
| Hydrothermal vent | ||||||
| Menez Gwen | Lucky Strike | Mount Saldanha | Rainbow | Total | ||
| Phenptypic operational group | I | 57 | 32 | 3 | 48 | 140 |
| II | 82 | 15 | 0 | 28 | 125 | |
| III | 1 | 11 | 0 | 12 | 24 | |
| Total | 140 | 58 | 3 | 88 | 289 | |
2.2. MAR Vents Prokaryotic Biodiversity

2.3. A MAR Vents Bacteria Collection Suitable for Industry
| Type of sample | |||||
|---|---|---|---|---|---|
| Group | Cluster | Water | Animals | Sediments | Chimneys |
| I | A | 1 | 11 | 1 | 3 |
| B | 1 | 16 | 2 | ||
| C | 9 | 1 | 2 | ||
| D | 21 | 11 | |||
| E | 8 | 2 | |||
| F | 4 | ||||
| G | 2 | 2 | |||
| H | 2 | ||||
| I | 1 | 1 | |||
| J | 1 | ||||
| II | L | 5 | 28 | 3 | |
| M | 2 | ||||
| N | 12 | ||||
| O | 4 | 3 | 23 | 4 | |
| P | 3 | 18 | 1 | ||
| Q | 5 | 7 | |||
| R | 4 | ||||
| III | S | 1 | |||
| T | 3 | ||||
| U | 3 | ||||
| V | 2 | ||||
| W | 3 | 9 | |||
| X | 1 | ||||
| (a) | |||||
| Hydrothermal vent | |||||
| Group | Cluster | Menez Gwen | Lucky Strike | Mount Saldanha | Rainbow |
| I | A | 3 | 2 | 11 | |
| B | 6 | 4 | 9 | ||
| C | 2 | 9 | 1 | ||
| D | 24 | 5 | 3 | ||
| E | 2 | 2 | 6 | ||
| F | 4 | ||||
| G | 2 | 2 | |||
| H | 2 | ||||
| I | 1 | 1 | |||
| J | 1 | ||||
| II | L | 10 | 8 | 18 | |
| M | 2 | ||||
| N | 12 | ||||
| O | 31 | 1 | 2 | ||
| P | 18 | 4 | |||
| Q | 9 | 3 | |||
| R | 4 | ||||
| III | S | 1 | |||
| T | 3 | ||||
| U | 3 | ||||
| V | 2 | ||||
| W | 1 | 8 | 3 | ||
| X | 1 | ||||
| Marine Strain | Vent | Type of sample | Depth (m) | T (°C) |
|---|---|---|---|---|
| MG BA 001 to 006 | Menez Gwen | Bathymordiolus azoricus | 825 | 8.4 |
| MG MS 007 to 012 | Microcaris sp. | 825 | 8.2 | |
| MG GA 013 to 016 | Gastropode | 825 | 8.4 | |
| MG SA 017 and 018 | Sediment A | 825 | 8.7 | |
| MG CR 019 to 023 | Crab | 825 | 8.2 | |
| MG SC 024 to 036 | Sediment C | 869 | 8.3 | |
| RB RS 037 to 043 | Rainbow | Rimicaris sp. | 2296 | 3.7 |
| RB PS 045 to 050 | Pachicara sp. | 2297 | 3.7 | |
| RB BA 051 to 062 | Bathymordiolus azoricus | 2290 | 3.8 | |
| LS BA 063 to 069 | Lucky Strike | Bathymordiolus azoricus | 1693 | 4.4 |
| LS WA 070 to 081 | Water | 1728 | 4.3 | |
| MG SD 082 | Menez Gwen | Sediment D | 808 | 9.1 |
| MG SA 083 | Sediment A | 825 | 8.7 | |
| MG MS 098O2 | Microcaris sp. | 825 | 8.2 | |
| RB BA 124O2, 125O2, 127O2, 128O2 and 131O2 | Rainbow | Bathymordiolus azoricus | 2290 | 3.8 |
| LS BA 136O2, 137O2, 138O2, 139O2 and 146O2 | Lucky Strike | Bathymordiolus azoricus | 1693 | 4.4 |
| MG MS 168O2 and 184O2 | Menez Gwen | Microcaris sp. | 825 | 8.2 |
| MG CR 184O2, 185O2 and 186O2 | Crab | 825 | 8.2 | |
| MG CC 194O2 | Chimney C | 808 | 266.2 | |
| MG CB 195O2 and 196O2 | Chimney B | 865 | 266.2 | |
| RB WC 199O2, 200O2, 201O2 and 202O2 | Rainbow | Water | 2301 | 3.7 |
| MG CR 203O2 and 209O2 | Menez Gwen | Crab | 825 | 8.2 |
| MG CB 210O2 and 211O2 | Chimney B | 865 | 266.2 | |
| MG CC 212O2 and 213O2 | Chimney C | 808 | 266.2 | |
| LS CA 214O2 | Lucky Strike | Chimney A | 1728 | 231.3 |
| RB BA 215O2, 216O2 , 217O2, 218O2 and 219O2 | Rainbow | Bathymordiolus azoricus | 2290 | 3.8 |
| RB WC 220O2 and 221O2 | Water | 2301 | 3.7 | |
| LS BA 225O2 and 226O2 | Lucky Strike | Bathymordiolus azoricus | 1693 | 4.4 |
| LS CA 227O2 and 228O2 | Chimney A | 1728 | 231.3 | |
| LS SD 230O2, 231O2 and 232O2 | Sediment D | 1691 | 4.3 | |
| MG CC 235O2 | Menez Gwen | Chimney C | 808 | 266.2 |
| MG MS 240O2 and 241O2 | Microcaris sp. | 825 | 8.2 | |
| RB RS 247O2, 248O2, 249O2, 250O2,251O2, 252O2 and 253O2 | Rainbow | Rimicaris sp. | 2296 | 3.7 |
| MG SD 258O2, 259O2 and 260O2 | Mount Saldanha | Sediment D | 2200 | 3.8 |
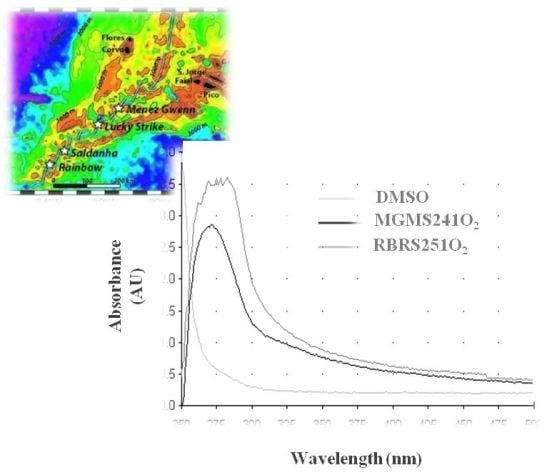
2.4. MAR Vents Bacteria Extracts Possess Photoprotection Capacity


3. Experimental Section
3.1. Marine Bacteria Isolation and Culturing
3.2. PCR Fingerprinting and Whole-Cell Protein Profiling and Phylogenetic Analysis
3.3. Bacteria Collection Selection and Extracts Preparation and Analysis
3.4. Anti-UV Yeast-Based Assay Development and Screening
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef]
- Pettit, G.R.; Fujii, Y.; Hasler, J.A.; Schmidt, J.M. Isolation and characterization of palystatins A–D. J. Nat. Prod. 1982, 45, 272–276. [Google Scholar] [CrossRef]
- Sudek, S.; Lopanik, N.B.; Waggoner, L.E.; Hildebrand, M.; Anderson, C.; Liu, H.; Patel, A.; Sherman, D.H.; Haygood, M.G. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 2007, 70, 67–74. [Google Scholar] [CrossRef]
- Arrieta, J.M.; Arnaud-Haond, S.; Duarte, C.M. What lies underneath: Conserving the oceans’ genetic resources. Proc. Natl. Acad. Sci. USA 2010, 107, 18318–18324. [Google Scholar]
- Takai, K.; Nakamura, K. Archaeal diversity and community development in deep-sea hydrothermal vents. Curr. Opin. Microbiol. 2011, 14, 282–291. [Google Scholar] [CrossRef]
- Rona, P.A.; Klinkhammer, G.; Nelsen, T.A.; Trefry, J.H.; Elderfield, H. Black smokers, massive sulfides and vent biota at the mid-atlantic ridge. Nature 1986, 321, 33–37. [Google Scholar]
- Thornburg, C.C.; Zabriskie, T.M.; McPhail, K.L. Deep-sea hydrothermal vents: Potential hot spots for natural products discovery? J. Nat. Prod. 2010, 73, 489–499. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar]
- Jeanthon, C. Molecular ecology of hydrothermal vent microbial communities. Antonie Van Leeuwenhoek 2000, 77, 117–133. [Google Scholar] [CrossRef]
- Vandamme, P.; Pot, B.; Gillis, M.; de Vos, P.; Kersters, K.; Swings, J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996, 60, 407–438. [Google Scholar]
- Pot, B.; Peter, V.; Kersters, K. Analysis of Electrophoretic Whole-Organism Protein Fingerprints. In Chemical Methods in Prokaryotic Systematics; Goodfellow, M., O’Donnell, A.G., Eds.; Jon Wiley and Sons: London, UK, 1994; pp. 493–517. [Google Scholar]
- Davidson, J.N. The effect of ultraviolet light on living yeast cells. Biochem. J. 1940, 34, 1537–1539. [Google Scholar]
- Holick, M.F. Vitamin D: A millenium perspective. J. Cell. Biochem. 2003, 88, 296–307. [Google Scholar]
- Torma, H.; Berne, B.; Vahlquist, A. UV irradiation and topical vitamin a modulate retinol esterification in hairless mouse epidermis. Acta Derm. Venereol. 1988, 68, 291–299. [Google Scholar]
- Kollias, N.; Sayre, R.M.; Zeise, L.; Chedekel, M.R. Photoprotection by melanin. J. Photochem. Photobiol. B Biol. 1991, 9, 135–160. [Google Scholar] [CrossRef]
- Plonka, P.M.; Grabacka, M. Melanin synthesis in microorganisms—biotechnological and medical aspects. Acta Biochim. Pol. 2006, 53, 429–443. [Google Scholar]
- Gao, Q.; Garcia-Pichel, F. Microbial ultraviolet sunscreens. Nat. Rev. Microbiol. 2011, 9, 791–802. [Google Scholar] [CrossRef]
- Pallela, R.; Na-Young, Y.; Kim, S.K. Anti-photoaging and photoprotective compounds derived from marine organisms. Mar. Drugs 2010, 8, 1189–1202. [Google Scholar] [CrossRef]
- Gadanho, M.; Sampaio, J.P. Occurrence and diversity of yeasts in the mid-atlantic ridge hydrothermal fields near the azores archipelago. Microb. Ecol. 2005, 50, 408–417. [Google Scholar] [CrossRef]
- Gevers, D.; Cohan, F.M.; Lawrence, J.G.; Spratt, B.G.; Coenye, T.; Feil, E.J.; Stackebrandt, E.; van de Peer, Y.; Vandamme, P.; Thompson, F.L.; et al. Opinion: Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005, 3, 733–739. [Google Scholar] [CrossRef]
- Turick, C.E.; Tisa, L.S.; Caccavo, F., Jr. Melanin production and use as a soluble electron shuttle for Fe(III) oxide reduction and as a terminal electron acceptor by Shewanella algae BrY. Appl. Environ. Microbiol. 2002, 68, 2436–2444. [Google Scholar] [CrossRef]
- Valeru, S.P.; Rompikuntal, P.K.; Ishikawa, T.; Vaitkevicius, K.; Sjoling, A.; Dolganov, N.; Zhu, J.; Schoolnik, G.; Wai, S.N. Role of melanin pigment in expression of Vibrio cholerae virulence factors. Infect. Immun. 2009, 77, 935–942. [Google Scholar] [CrossRef]
- Tudor, L.; Togoe, I.; Enache, L.; Georgescu, B. UV influence over Vibrio bacterial species. Roum. Biotechnol. Lett. 2002, 3, 767–772. [Google Scholar]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Chambel, L.; Sol, M.; Fernandes, I.; Barbosa, M.; Zilhao, I.; Barata, B.; Jordan, S.; Perni, S.; Shama, G.; Adriao, A.; et al. Occurrence and persistence of Listeria spp. In the environment of ewe and cow’s milk cheese dairies in portugal unveiled by an integrated analysis of identification, typing and spatial-temporal mapping along production cycle. Int. J. Food Microbiol. 2007, 116, 52–63. [Google Scholar] [CrossRef]
- Ludwig, J.A.; Reynolds, J.F. Diversity Indices. In Statistical Ecology: A Primer on Methods and Computing; John Wiley & Sons: New York, NY, USA, 1988; Volume 8, pp. 85–101. [Google Scholar]
- Kremer, A.; Petit, R.; Pons, O. Measures of Polymorphism within and among Populations. In Molecular Tools for Screening Biodiversity; Karp, A., Isaac, P., Ingram, D., Eds.; Chapman & Hall: London, UK, 1998. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Larget, B.; Simon, D.L. Markov chain monte carlo algorithms for the bayesian analysis of phylogenetic trees. Mol. Biol. Evol. 1999, 16, 750–759. [Google Scholar] [CrossRef]
- Sarker, S.D.; Latif, Z.; Gray, A.I. Natural Products Isolation, 2nd ed; Humana Press: New York, NY, USA, 2006. [Google Scholar]
- Mayer, A.M.; Rodriguez, A.D.; Berlinck, R.G.; Fusetani, N. Marine pharmacology in 2007–8: Marine compounds with antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial, antiprotozoal, antituberculosis, and antiviral activities; Affecting the immune and nervous system, and other miscellaneous mechanisms of action. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 191–222. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2011, 28, 196–268. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Labes, A.; Wiese, J. Bio-mining the microbial treasures of the ocean: New natural products. Biotechnol. Adv. 2011, 29, 468–482. [Google Scholar] [CrossRef]
- Glass Market Research Reports. Available online: http://www.giiresearch.com/topics/MR08_en.shtml (accessed on 21 February 2012).
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Martins, A.; Tenreiro, T.; Andrade, G.; Gadanho, M.; Chaves, S.; Abrantes, M.; Calado, P.; Tenreiro, R.; Vieira, H. Photoprotective Bioactivity Present in a Unique Marine Bacteria Collection from Portuguese Deep Sea Hydrothermal Vents. Mar. Drugs 2013, 11, 1506-1523. https://doi.org/10.3390/md11051506
Martins A, Tenreiro T, Andrade G, Gadanho M, Chaves S, Abrantes M, Calado P, Tenreiro R, Vieira H. Photoprotective Bioactivity Present in a Unique Marine Bacteria Collection from Portuguese Deep Sea Hydrothermal Vents. Marine Drugs. 2013; 11(5):1506-1523. https://doi.org/10.3390/md11051506
Chicago/Turabian StyleMartins, Ana, Tania Tenreiro, Gonçalo Andrade, Mário Gadanho, Sandra Chaves, Marta Abrantes, Patrícia Calado, Rogério Tenreiro, and Helena Vieira. 2013. "Photoprotective Bioactivity Present in a Unique Marine Bacteria Collection from Portuguese Deep Sea Hydrothermal Vents" Marine Drugs 11, no. 5: 1506-1523. https://doi.org/10.3390/md11051506
APA StyleMartins, A., Tenreiro, T., Andrade, G., Gadanho, M., Chaves, S., Abrantes, M., Calado, P., Tenreiro, R., & Vieira, H. (2013). Photoprotective Bioactivity Present in a Unique Marine Bacteria Collection from Portuguese Deep Sea Hydrothermal Vents. Marine Drugs, 11(5), 1506-1523. https://doi.org/10.3390/md11051506