Conotoxins Targeting Nicotinic Acetylcholine Receptors: An Overview
Abstract
:1. Cone Snails, the New Gold Mines?
2. Alpha-Conotoxins, the Largest Characterized Group of Conotoxins

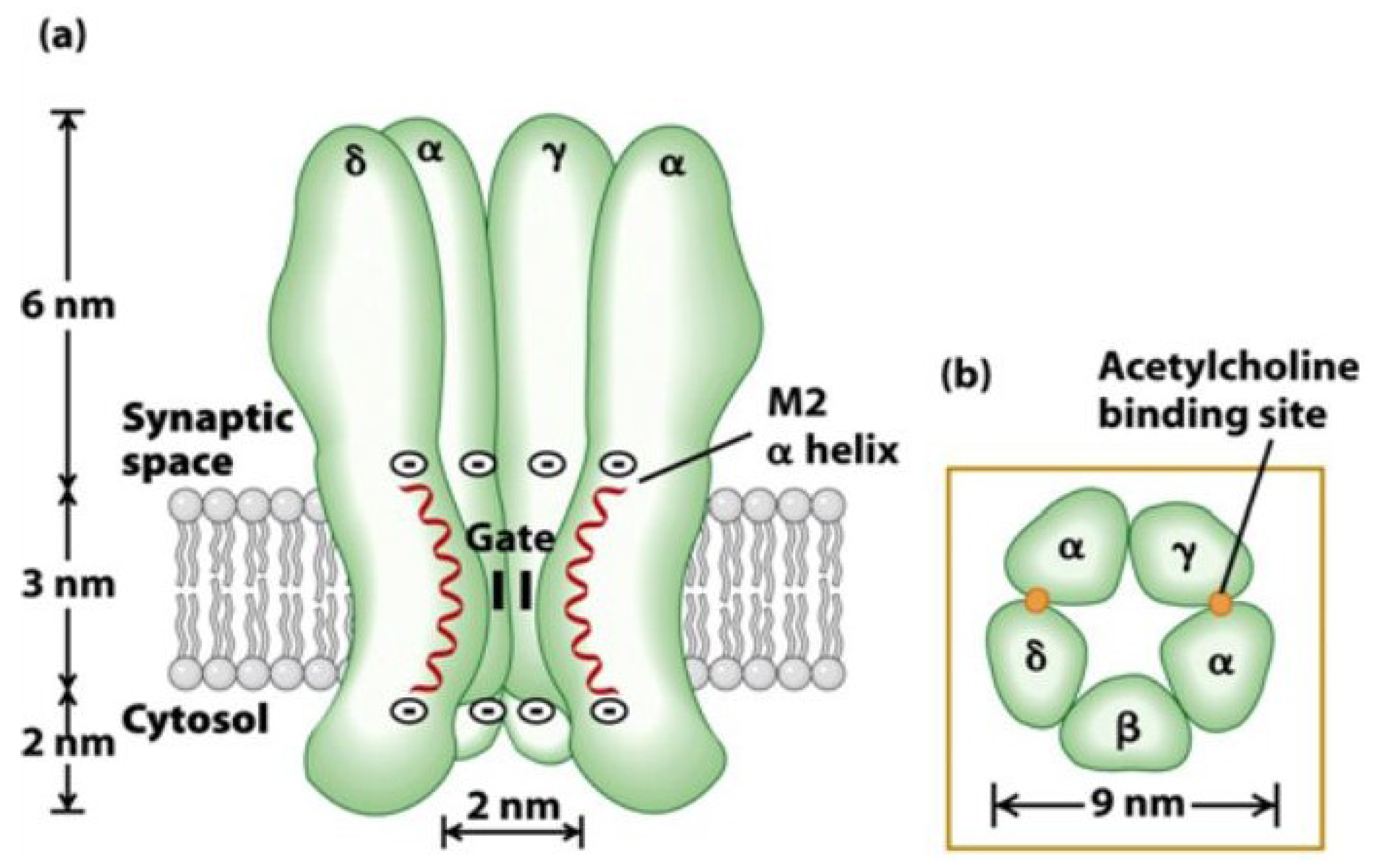
3. Nicotinic Acetylcholine Receptors (nAChRs)

4. α-Conotoxins and Their Mode of Action—State of the Art


5. Alpha-Conotoxins and Their Mode of Action
5.1. Neuronal Subtype nAChRs
5.1.1. α7 nAChRs Selective α-Conotoxins



5.1.2. α3β2 nAChR Selective α-Conotoxins

5.1.3. α3β4 nAChR Selective α-Conotoxins

5.1.4. α4β2 nAChR Selective α-Conotoxins

5.1.5. α6* nAChR Selective α-Conotoxins

5.1.6. α9α10 nAChR Selective α-Conotoxins

5.2. Muscle Subtype nAChRs: α1β1δε (adult) and α1β1γδ (fetal) nAChRs


6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef]
- Milne, T.J.; Abbenante, G.; Tyndall, J.D.; Halliday, J.; Lewis, R.J. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J. Biol. Chem. 2003, 278, 31105–31110. [Google Scholar]
- Davis, J.; Jones, A.; Lewis, R.J. Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides 2009, 30, 1222–1227. [Google Scholar] [CrossRef]
- Olivera, B.M.; Rivier, J.; Clark, C.; Ramilo, C.A.; Corpuz, G.P.; Abogadie, F.C.; Mena, E.E.; Woodward, S.R.; Hillyard, D.R.; Cruz, L.J. Diversity of Conus neuropeptides. Science 1990, 249, 257–263. [Google Scholar]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus venom peptide pharmacology. Pharmacol. Rev. 2012, 64, 259–298. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Santos, A.D.; Olivera, B.M. Conus peptides targeted to specific nicotinic acetylcholine receptor subtypes. Annu. Rev. Biochem. 1999, 68, 59–88. [Google Scholar] [CrossRef]
- Dutertre, S.; Ulens, C.; Buttner, R.; Fish, A.; van Elk, R.; Kendel, Y.; Hopping, G.; Alewood, P.F.; Schroeder, C.; Nicke, A.; et al. AChBP-targeted alpha-conotoxin correlates distinct binding orientations with nAChR subtype selectivity. EMBO J. 2007, 26, 3858–3867. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Yoshikami, D.; Mahe, E.; Nielsen, D.B.; Rivier, J.E.; Gray, W.R.; Olivera, B.M. A nicotinic acetylcholine receptor ligand of unique specificity, alpha-conotoxin ImI. J. Biol. Chem. 1994, 269, 16733–16739. [Google Scholar]
- Terlau, H.; Olivera, B.M. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- Janes, R.W. alpha-Conotoxins as selective probes for nicotinic acetylcholine receptor subclasses. Curr. Opin. Pharmacol. 2005, 5, 280–292. [Google Scholar] [CrossRef]
- Livett, B.G.; Sandall, D.W.; Keays, D.; Down, J.; Gayler, K.R.; Satkunanathan, N.; Khalil, Z. Therapeutic applications of conotoxins that target the neuronal nicotinic acetylcholine receptor. Toxicon 2006, 48, 810–829. [Google Scholar] [CrossRef]
- Vincler, M.; Wittenauer, S.; Parker, R.; Ellison, M.; Olivera, B.M.; McIntosh, J.M. Molecular mechanism for analgesia involving specific antagonism of alpha9alpha10 nicotinic acetylcholine receptors. Proc. Natl. Acad. Sci. USA 2006, 103, 17880–17884. [Google Scholar] [CrossRef]
- Sandall, D.W.; Satkunanathan, N.; Keays, D.A.; Polidano, M.A.; Liping, X.; Pham, V.; Down, J.G.; Khalil, Z.; Livett, B.G.; Gayler, K.R. A novel alpha-conotoxin identified by gene sequencing is active in suppressing the vascular response to selective stimulation of sensory nerves in vivo. Biochemistry 2003, 42, 6904–6911. [Google Scholar] [CrossRef]
- Satkunanathan, N.; Livett, B.; Gayler, K.; Sandall, D.; Down, J.; Khalil, Z. Alpha-conotoxin Vc1.1 alleviates neuropathic pain and accelerates functional recovery of injured neurones. Brain Res. 2005, 1059, 149–158. [Google Scholar] [CrossRef]
- PyMOL version 1.3. Available online: http://pymol.sourceforge.net/ (accessed on 10 April 2013).
- Yuan, D.D.; Han, Y.H.; Wang, C.G.; Chi, C.W. From the identification of gene organization of alpha conotoxins to the cloning of novel toxins. Toxicon 2007, 49, 1135–1149. [Google Scholar] [CrossRef]
- Olivera, B.M.E.E. Just Lecture, 1996. Conus venom peptides, receptor and ion channel targets, and drug design: 50 million years of neuropharmacology. Mol. Biol. Cell 1997, 8, 2101–2109. [Google Scholar] [CrossRef]
- Dutertre, S.; Nicke, A.; Lewis, R.J. Beta2 subunit contribution to 4/7 alpha-conotoxin binding to the nicotinic acetylcholine receptor. J. Biol. Chem. 2005, 280, 30460–30468. [Google Scholar] [CrossRef]
- Miller, P.S.; Smart, T.G. Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol. Sci. 2010, 31, 161–174. [Google Scholar] [CrossRef]
- Dellisanti, C.D.; Ghosh, B.; Hanson, S.M.; Raspanti, J.M.; Grant, V.A.; Diarra, G.M.; Schuh, A.M.; Satyshur, K.; Klug, C.S.; Czajkowski, C. Site-directed spin labeling reveals pentameric ligand-gated ion channel gating motions. PLoS Biol. 2013, 11, e1001714. [Google Scholar] [CrossRef]
- Gotti, C.; Clementi, F. Neuronal nicotinic receptors: From structure to pathology. Prog. Neurobiol. 2004, 74, 363–396. [Google Scholar] [CrossRef]
- Klee, E.W.; Ebbert, J.O.; Schneider, H.; Hurt, R.D.; Ekker, S.C. Zebrafish for the study of the biological effects of nicotine. Nicotine Tob. Res. 2011, 13, 301–312. [Google Scholar] [CrossRef]
- Tapper, A.R.; McKinney, S.L.; Nashmi, R.; Schwarz, J.; Deshpande, P.; Labarca, C.; Whiteaker, P.; Marks, M.J.; Collins, A.C.; Lester, H.A. Nicotine activation of alpha4* receptors: Sufficient for reward, tolerance, and sensitization. Science 2004, 306, 1029–1032. [Google Scholar] [CrossRef]
- Improgo, M.R.; Scofield, M.D.; Tapper, A.R.; Gardner, P.D. The nicotinic acetylcholine receptor CHRNA5/A3/B4 gene cluster: Dual role in nicotine addiction and lung cancer. Prog. Neurobiol. 2010, 92, 212–226. [Google Scholar] [CrossRef]
- Hurst, R.; Rollema, H.; Bertrand, D. Nicotinic acetylcholine receptors: From basic science to therapeutics. Pharmacol. Ther. 2013, 137, 22–54. [Google Scholar] [CrossRef]
- Albuquerque, E.X.; Pereira, E.F.; Alkondon, M.; Rogers, S.W. Mammalian nicotinic acetylcholine receptors: From structure to function. Physiol. Rev. 2009, 89, 73–120. [Google Scholar] [CrossRef]
- Papke, R.L.; Thinschmidt, J.S.; Moulton, B.A.; Meyer, E.M.; Poirier, A. Activation and inhibition of rat neuronal nicotinic receptors by ABT-418. Br. J. Pharmacol. 1997, 120, 429–438. [Google Scholar] [CrossRef]
- Carson, K.V.; Brinn, M.P.; Robertson, T.A.; To, A.N.R.; Esterman, A.J.; Peters, M.; Smith, B.J. Current and emerging pharmacotherapeutic options for smoking cessation. Subst. Abuse 2013, 7, 85–105. [Google Scholar]
- Rahman, S. Nicotinic receptors as therapeutic targets for drug addictive disorders. CNS Neurol Disord. Drug Targets 2013, 12, 633–640. [Google Scholar] [CrossRef]
- Colovic, M.B.; Krstic, D.Z.; Lazarevic-Pasti, T.D.; Bondzic, A.M.; Vasic, V.M. Acetylcholinesterase inhibitors: Pharmacology and toxicology. Curr. Neuropharmacol. 2013, 11, 315–335. [Google Scholar] [CrossRef]
- Grishin, A.A.; Wang, C.I.; Muttenthaler, M.; Alewood, P.F.; Lewis, R.J.; Adams, D.J. Alpha-conotoxin AuIB isomers exhibit distinct inhibitory mechanisms and differential sensitivity to stoichiometry of alpha3beta4 nicotinic acetylcholine receptors. J. Biol. Chem. 2010, 285, 22254–22263. [Google Scholar]
- Indurthi, D.C.; Pera, E.; Kim, H.L.; Chu, C.; McLeod, M.D.; Michael McIntosh, J.; Absalom, N.L.; Chebib, M. Presence of multiple binding sites on alpha9alpha10 nAChR receptors alludes to stoichiometric dependent action of the alpha-conotoxin, Vc1.1. Biochem. Pharmacol. 2014, 98, 131–140. [Google Scholar]
- Kabbani, N.; Nordman, J.C.; Corgiat, B.A.; Veltri, D.P.; Shehu, A.; Seymour, V.A.; Adams, D.J. Are nicotinic acetylcholine receptors coupled to G proteins? BioEssays 2013, 35, 1025–1034. [Google Scholar] [CrossRef]
- Unwin, N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J. Mol. Biol. 2005, 346, 967–989. [Google Scholar] [CrossRef]
- Unwin, N.; Fujiyoshi, Y. Gating movement of acetylcholine receptor caught by plunge-freezing. J. Mol. Biol. 2012, 422, 617–634. [Google Scholar] [CrossRef]
- Brejc, K.; van Dijk, W.J.; Klaassen, R.V.; Schuurmans, M.; van Der Oost, J.; Smit, A.B.; Sixma, T.K. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 2001, 411, 269–276. [Google Scholar] [CrossRef]
- Armishaw, C.; Jensen, A.A.; Balle, T.; Clark, R.J.; Harpsoe, K.; Skonberg, C.; Liljefors, T.; Stromgaard, K. Rational design of alpha-conotoxin analogues targeting alpha7 nicotinic acetylcholine receptors: Improved antagonistic activity by incorporation of proline derivatives. J. Biol. Chem. 2009, 284, 9498–9512. [Google Scholar] [CrossRef]
- Celie, P.H.; van Rossum-Fikkert, S.E.; van Dijk, W.J.; Brejc, K.; Smit, A.B.; Sixma, T.K. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 2004, 41, 907–914. [Google Scholar] [CrossRef]
- Celie, P.H.; Klaassen, R.V.; van Rossum-Fikkert, S.E.; van Elk, R.; van Nierop, P.; Smit, A.B.; Sixma, T.K. Crystal structure of acetylcholine-binding protein from Bulinus truncatus reveals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors. J. Biol. Chem. 2005, 280, 26457–26466. [Google Scholar] [CrossRef]
- Dellisanti, C.D.; Yao, Y.; Stroud, J.C.; Wang, Z.Z.; Chen, L. Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution. Nat. Neurosci. 2007, 10, 953–962. [Google Scholar] [CrossRef]
- Li, S.X.; Huang, S.; Bren, N.; Noridomi, K.; Dellisanti, C.D.; Sine, S.M.; Chen, L. Ligand-binding domain of an alpha7-nicotinic receptor chimera and its complex with agonist. Nat. Neurosci. 2011, 14, 1253–1259. [Google Scholar] [CrossRef]
- Ulens, C.; Hogg, R.C.; Celie, P.H.; Bertrand, D.; Tsetlin, V.; Smit, A.B.; Sixma, T.K. Structural determinants of selective alpha-conotoxin binding to a nicotinic acetylcholine receptor homolog AChBP. Proc. Natl. Acad. Sci. USA 2006, 103, 3615–3620. [Google Scholar] [CrossRef]
- Celie, P.H.; Kasheverov, I.E.; Mordvintsev, D.Y.; Hogg, R.C.; van Nierop, P.; van Elk, R.; van Rossum-Fikkert, S.E.; Zhmak, M.N.; Bertrand, D.; Tsetlin, V.; et al. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an alpha-conotoxin PnIA variant. Nat. Struct. Mol. Biol. 2005, 12, 582–588. [Google Scholar] [CrossRef]
- Harel, M.; Kasher, R.; Nicolas, A.; Guss, J.M.; Balass, M.; Fridkin, M.; Smit, A.B.; Brejc, K.; Sixma, T.K.; Katchalski-Katzir, E.; et al. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron 2001, 32, 265–275. [Google Scholar] [CrossRef]
- Dutertre, S.; Nicke, A.; Tyndall, J.D.; Lewis, R.J. Determination of alpha-conotoxin binding modes on neuronal nicotinic acetylcholine receptors. J. Mol. Recognit. 2004, 17, 339–347. [Google Scholar] [CrossRef]
- Couturier, S.; Bertrand, D.; Matter, J.M.; Hernandez, M.C.; Bertrand, S.; Millar, N.; Valera, S.; Barkas, T.; Ballivet, M. A neuronal nicotinic acetylcholine receptor subunit (alpha 7) is developmentally regulated and forms a homo-oligomeric channel blocked by alpha-BTX. Neuron 1990, 5, 847–856. [Google Scholar] [CrossRef]
- Rubboli, F.; Court, J.A.; Sala, C.; Morris, C.; Chini, B.; Perry, E.; Clementi, F. Distribution of nicotinic receptors in the human hippocampus and thalamus. Eur. J. Neurosci. 1994, 6, 1596–1604. [Google Scholar] [CrossRef]
- Wevers, A.; Jeske, A.; Lobron, C.; Birtsch, C.; Heinemann, S.; Maelicke, A.; Schroder, R.; Schroder, H. Cellular distribution of nicotinic acetylcholine receptor subunit mRNAs in the human cerebral cortex as revealed by non-isotopic in situ hybridization. Brain Res. Mol. Brain Res. 1994, 25, 122–128. [Google Scholar] [CrossRef]
- Breese, C.R.; Adams, C.; Logel, J.; Drebing, C.; Rollins, Y.; Barnhart, M.; Sullivan, B.; Demasters, B.K.; Freedman, R.; Leonard, S. Comparison of the regional expression of nicotinic acetylcholine receptor alpha7 mRNA and [125I]-alpha-bungarotoxin binding in human postmortem brain. J. Comp. Neurol 1997, 387, 385–398. [Google Scholar] [CrossRef]
- Sacco, K.A.; Bannon, K.L.; George, T.P. Nicotinic receptor mechanisms and cognition in normal states and neuropsychiatric disorders. J. Psychopharmacol. 2004, 18, 457–474. [Google Scholar] [CrossRef]
- Steinlein, O.K.; Bertrand, D. Nicotinic receptor channelopathies and epilepsy. Pflugers Arch. 2010, 460, 495–503. [Google Scholar] [CrossRef]
- Prince, R.J.; Sine, S.M. Molecular dissection of subunit interfaces in the acetylcholine receptor. Identification of residues that determine agonist selectivity. J. Biol. Chem. 1996, 271, 25770–25777. [Google Scholar] [CrossRef]
- Dutertre, S.; Lewis, R.J. Computational approaches to understand alpha-conotoxin interactions at neuronal nicotinic receptors. Eur. J. Biochem. 2004, 271, 2327–2334. [Google Scholar] [CrossRef]
- Taly, A.; Corringer, P.J.; Grutter, T.; Prado de Carvalho, L.; Karplus, M.; Changeux, J.P. Implications of the quaternary twist allosteric model for the physiology and pathology of nicotinic acetylcholine receptors. Proc. Natl. Acad. Sci. USA 2006, 103, 16965–16970. [Google Scholar]
- Fainzilber, M.; Hasson, A.; Oren, R.; Burlingame, A.L.; Gordon, D.; Spira, M.E.; Zlotkin, E. New mollusc-specific alpha-conotoxins block Aplysia neuronal acetylcholine receptors. Biochemistry 1994, 33, 9523–9529. [Google Scholar] [CrossRef]
- Hogg, R.C.; Miranda, L.P.; Craik, D.J.; Lewis, R.J.; Alewood, P.F.; Adams, D.J. Single amino acid substitutions in alpha-conotoxin PnIA shift selectivity for subtypes of the mammalian neuronal nicotinic acetylcholine receptor. J. Biol. Chem. 1999, 274, 36559–36564. [Google Scholar]
- Luo, S.; Nguyen, T.A.; Cartier, G.E.; Olivera, B.M.; Yoshikami, D.; McIntosh, J.M. Single-residue alteration in alpha-conotoxin PnIA switches its nAChR subtype selectivity. Biochemistry 1999, 38, 14542–14548. [Google Scholar] [CrossRef]
- Hogg, R.C.; Hopping, G.; Alewood, P.F.; Adams, D.J.; Bertrand, D. Alpha-conotoxins PnIA and [A10L]PnIA stabilize different states of the alpha7-L247T nicotinic acetylcholine receptor. J. Biol. Chem. 2003, 278, 26908–26914. [Google Scholar]
- Quiram, P.A.; McIntosh, J.M.; Sine, S.M. Pairwise interactions between neuronal alpha(7) acetylcholine receptors and alpha-conotoxin PnIB. J. Biol. Chem. 2000, 275, 4889–4896. [Google Scholar] [CrossRef]
- Quiram, P.A.; Sine, S.M. Structural elements in alpha-conotoxin ImI essential for binding to neuronal alpha7 receptors. J. Biol. Chem. 1998, 273, 11007–11011. [Google Scholar] [CrossRef]
- Quiram, P.A.; Sine, S.M. Identification of residues in the neuronal alpha7 acetylcholine receptor that confer selectivity for conotoxin ImI. J. Biol. Chem. 1998, 273, 11001–11006. [Google Scholar] [CrossRef]
- Quiram, P.A.; Jones, J.J.; Sine, S.M. Pairwise interactions between neuronal alpha7 acetylcholine receptors and alpha-conotoxin ImI. J. Biol. Chem. 1999, 274, 19517–19524. [Google Scholar] [CrossRef]
- Hansen, S.B.; Sulzenbacher, G.; Huxford, T.; Marchot, P.; Taylor, P.; Bourne, Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005, 24, 3635–3646. [Google Scholar] [CrossRef]
- Whiteaker, P.; Christensen, S.; Yoshikami, D.; Dowell, C.; Watkins, M.; Gulyas, J.; Rivier, J.; Olivera, B.M.; McIntosh, J.M. Discovery, synthesis, and structure activity of a highly selective alpha7 nicotinic acetylcholine receptor antagonist. Biochemistry 2007, 46, 6628–6638. [Google Scholar] [CrossRef]
- Whiteaker, P.; Marks, M.J.; Christensen, S.; Dowell, C.; Collins, A.C.; McIntosh, J.M. Synthesis and characterization of 125I-alpha-conotoxin ArIB[V11L;V16A], a selective alpha7 nicotinic acetylcholine receptor antagonist. J. Pharmacol. Exp. Ther. 2008, 325, 910–919. [Google Scholar] [CrossRef]
- Inserra, M.C.; Kompella, S.N.; Vetter, I.; Brust, A.; Daly, N.L.; Cuny, H.; Craik, D.J.; Alewood, P.F.; Adams, D.J.; Lewis, R.J. Isolation and characterization of alpha-conotoxin LsIA with potent activity at nicotinic acetylcholine receptors. Biochem. Pharmacol. 2013, 86, 791–799. [Google Scholar] [CrossRef]
- Marks, M.J.; Smith, K.W.; Collins, A.C. Differential agonist inhibition identifies multiple epibatidine binding sites in mouse brain. J. Pharmacol. Exp. Ther. 1998, 285, 377–386. [Google Scholar]
- Salas, R.; Sturm, R.; Boulter, J.; de Biasi, M. Nicotinic receptors in the habenulo-interpeduncular system are necessary for nicotine withdrawal in mice. J. Neurosci. 2009, 29, 3014–3018. [Google Scholar] [CrossRef]
- Fowler, C.D.; Lu, Q.; Johnson, P.M.; Marks, M.J.; Kenny, P.J. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature 2011, 471, 597–601. [Google Scholar] [CrossRef]
- Paolini, M.; de Biasi, M. Mechanistic insights into nicotine withdrawal. Biochem. Pharmacol. 2011, 82, 996–1007. [Google Scholar] [CrossRef]
- Cartier, G.E.; Yoshikami, D.; Gray, W.R.; Luo, S.; Olivera, B.M.; McIntosh, J.M. A new alpha-conotoxin which targets alpha3beta2 nicotinic acetylcholine receptors. J. Biol. Chem. 1996, 271, 7522–7528. [Google Scholar]
- Harvey, S.C.; McIntosh, J.M.; Cartier, G.E.; Maddox, F.N.; Luetje, C.W. Determinants of specificity for alpha-conotoxin MII on alpha3beta2 neuronal nicotinic receptors. Mol. Pharmacol. 1997, 51, 336–342. [Google Scholar]
- Jin, A.H.; Daly, N.L.; Nevin, S.T.; Wang, C.I.; Dutertre, S.; Lewis, R.J.; Adams, D.J.; Craik, D.J.; Alewood, P.F. Molecular engineering of conotoxins: The importance of loop size to alpha-conotoxin structure and function. J. Med. Chem. 2008, 51, 5575–5584. [Google Scholar] [CrossRef]
- Everhart, D.; Reiller, E.; Mirzoian, A.; McIntosh, J.M.; Malhotra, A.; Luetje, C.W. Identification of residues that confer alpha-conotoxin-PnIA sensitivity on the alpha 3 subunit of neuronal nicotinic acetylcholine receptors. J. Pharmacol. Exp. Ther. 2003, 306, 664–670. [Google Scholar] [CrossRef]
- Luo, S.; Zhangsun, D.; Schroeder, C.I.; Zhu, X.; Hu, Y.; Wu, Y.; Weltzin, M.M.; Eberhard, S.; Kaas, Q.; Craik, D.J.; et al. A novel alpha4/7-conotoxin LvIA from Conus lividus that selectively blocks alpha3beta2 vs. alpha6/alpha3beta2beta3 nicotinic acetylcholine receptors. FASEB J. 2014, 28, 1842–1853. [Google Scholar] [CrossRef]
- Luo, S.; Kulak, J.M.; Cartier, G.E.; Jacobsen, R.B.; Yoshikami, D.; Olivera, B.M.; McIntosh, J.M. alpha-conotoxin AuIB selectively blocks alpha3 beta4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J. Neurosci. 1998, 18, 8571–8579. [Google Scholar]
- Dutton, J.L.; Bansal, P.S.; Hogg, R.C.; Adams, D.J.; Alewood, P.F.; Craik, D.J. A new level of conotoxin diversity, a non-native disulfide bond connectivity in alpha-conotoxin AuIB reduces structural definition but increases biological activity. J. Biol. Chem. 2002, 277, 48849–48857. [Google Scholar]
- Clark, R.J.; Fischer, H.; Nevin, S.T.; Adams, D.J.; Craik, D.J. The synthesis, structural characterization, and receptor specificity of the alpha-conotoxin Vc1.1. J. Biol. Chem. 2006, 281, 23254–23263. [Google Scholar] [CrossRef]
- Innocent, N.; Livingstone, P.D.; Hone, A.; Kimura, A.; Young, T.; Whiteaker, P.; McIntosh, J.M.; Wonnacott, S. Alpha-conotoxin Arenatus IB[V11L,V16D] [corrected] is a potent and selective antagonist at rat and human native alpha7 nicotinic acetylcholine receptors. J. Pharmacol. Exp. Ther. 2008, 327, 529–537. [Google Scholar] [CrossRef]
- Pereira, E.F.; Alkondon, M.; McIntosh, J.M.; Albuquerque, E.X. Alpha-conotoxin-ImI: A competitive antagonist at alpha-bungarotoxin-sensitive neuronal nicotinic receptors in hippocampal neurons. J. Pharmacol. Exp. Ther. 1996, 278, 1472–1483. [Google Scholar]
- Grishin, A.A.; Cuny, H.; Hung, A.; Clark, R.J.; Brust, A.; Akondi, K.; Alewood, P.F.; Craik, D.J.; Adams, D.J. Identifying key amino acid residues that affect alpha-conotoxin AuIB inhibition of alpha3beta4 nicotinic acetylcholine receptors. J. Biol. Chem. 2013, 288, 34428–34442. [Google Scholar] [CrossRef]
- Luo, S.; Zhangsun, D.; Zhu, X.; Wu, Y.; Hu, Y.; Christensen, S.; Harvey, P.J.; Akcan, M.; Craik, D.J.; McIntosh, J.M. Characterization of a Novel alpha-Conotoxin TxID from Conus textile That Potently Blocks Rat alpha3beta4 Nicotinic Acetylcholine Receptors. J. Med. Chem. 2013, 56, 9655–9663. [Google Scholar] [CrossRef]
- Gotti, C.; Zoli, M.; Clementi, F. Brain nicotinic acetylcholine receptors: Native subtypes and their relevance. Trends Pharmacol. Sci. 2006, 27, 482–491. [Google Scholar] [CrossRef]
- Wonnacott, S. Presynaptic nicotinic ACh receptors. Trends Neurosci. 1997, 20, 92–98. [Google Scholar] [CrossRef]
- Grady, S.R.; Salminen, O.; McIntosh, J.M.; Marks, M.J.; Collins, A.C. Mouse striatal dopamine nerve terminals express alpha4alpha5beta2 and two stoichiometric forms of alpha4beta2*-nicotinic acetylcholine receptors. J. Mol. Neurosci. 2010, 40, 91–95. [Google Scholar] [CrossRef]
- Flores, C.M.; Rogers, S.W.; Pabreza, L.A.; Wolfe, B.B.; Kellar, K.J. A subtype of nicotinic cholinergic receptor in rat brain is composed of alpha 4 and beta 2 subunits and is up-regulated by chronic nicotine treatment. Mol. Pharmacol. 1992, 41, 31–37. [Google Scholar]
- Rueter, L.E.; Donnelly-Roberts, D.L.; Curzon, P.; Briggs, C.A.; Anderson, D.J.; Bitner, R.S. A-85380: A pharmacological probe for the preclinical and clinical investigation of the alphabeta neuronal nicotinic acetylcholine receptor. CNS Drug Rev. 2006, 12, 100–112. [Google Scholar] [CrossRef]
- Ebbert, J.O. Emerging drugs for the treatment of tobacco dependence. Expert Opin. Emerg. Drugs 2009, 14, 23–32. [Google Scholar]
- Taly, A.; Corringer, P.J.; Guedin, D.; Lestage, P.; Changeux, J.P. Nicotinic receptors: Allosteric transitions and therapeutic targets in the nervous system. Nat. Rev. Drug Discov. 2009, 8, 733–750. [Google Scholar] [CrossRef]
- Nicke, A.; Loughnan, M.L.; Millard, E.L.; Alewood, P.F.; Adams, D.J.; Daly, N.L.; Craik, D.J.; Lewis, R.J. Isolation, structure, and activity of GID, a novel alpha 4/7-conotoxin with an extended N-terminal sequence. J. Biol. Chem. 2003, 278, 3137–3144. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Dowell, C.; Watkins, M.; Garrett, J.E.; Yoshikami, D.; Olivera, B.M. Alpha-conotoxin GIC from Conus geographus, a novel peptide antagonist of nicotinic acetylcholine receptors. J. Biol. Chem. 2002, 277, 33610–33615. [Google Scholar]
- Loughnan, M.L.; Nicke, A.; Jones, A.; Adams, D.J.; Alewood, P.F.; Lewis, R.J. Chemical and functional identification and characterization of novel sulfated alpha-conotoxins from the cone snail Conus anemone. J. Med. Chem. 2004, 47, 1234–1241. [Google Scholar] [CrossRef]
- Millard, E.L.; Nevin, S.T.; Loughnan, M.L.; Nicke, A.; Clark, R.J.; Alewood, P.F.; Lewis, R.J.; Adams, D.J.; Craik, D.J.; Daly, N.L. Inhibition of neuronal nicotinic acetylcholine receptor subtypes by alpha-Conotoxin GID and analogues. J. Biol. Chem. 2009, 284, 4944–4951. [Google Scholar] [CrossRef]
- Banerjee, J.; Yongye, A.B.; Chang, Y.P.; Gyanda, R.; Medina-Franco, J.L.; Armishaw, C.J. Design and synthesis of alpha-conotoxin GID analogues as selective alpha4beta2 nicotinic acetylcholine receptor antagonists. Biopolymers 2014, 102, 78–87. [Google Scholar] [CrossRef]
- Beissner, M.; Dutertre, S.; Schemm, R.; Danker, T.; Sporning, A.; Grubmuller, H.; Nicke, A. Efficient binding of 4/7 alpha-conotoxins to nicotinic alpha4beta2 receptors is prevented by Arg185 and Pro195 in the alpha4 subunit. Mol. Pharmacol. 2012, 82, 711–718. [Google Scholar] [CrossRef]
- Mackey, E.D.; Engle, S.E.; Kim, M.R.; O’Neill, H.C.; Wageman, C.R.; Patzlaff, N.E.; Wang, Y.; Grady, S.R.; McIntosh, J.M.; Marks, M.J.; et al. alpha6* nicotinic acetylcholine receptor expression and function in a visual salience circuit. J. Neurosci. 2012, 32, 10226–10237. [Google Scholar] [CrossRef]
- Hone, A.J.; Meyer, E.L.; McIntyre, M.; McIntosh, J.M. Nicotinic acetylcholine receptors in dorsal root ganglion neurons include the alpha6beta4* subtype. FASEB J. 2012, 26, 917–926. [Google Scholar] [CrossRef]
- Liu, J.; McGlinn, A.M.; Fernandes, A.; Milam, A.H.; Strang, C.E.; Andison, M.E.; Lindstrom, J.M.; Keyser, K.T.; Stone, R.A. Nicotinic acetylcholine receptor subunits in rhesus monkey retina. Invest. Ophthalmol. Vis. Sci. 2009, 50, 1408–1415. [Google Scholar]
- Klink, R.; de Kerchove d’Exaerde, A.; Zoli, M.; Changeux, J.P. Molecular and physiological diversity of nicotinic acetylcholine receptors in the midbrain dopaminergic nuclei. J. Neurosci. 2001, 21, 1452–1463. [Google Scholar]
- Azam, L.; Winzer-Serhan, U.H.; Chen, Y.; Leslie, F.M. Expression of neuronal nicotinic acetylcholine receptor subunit mRNAs within midbrain dopamine neurons. J. Comp. Neurol 2002, 444, 260–274. [Google Scholar] [CrossRef]
- Champtiaux, N.; Gotti, C.; Cordero-Erausquin, M.; David, D.J.; Przybylski, C.; Lena, C.; Clementi, F.; Moretti, M.; Rossi, F.M.; le Novere, N.; et al. Subunit composition of functional nicotinic receptors in dopaminergic neurons investigated with knock-out mice. J. Neurosci. 2003, 23, 7820–7829. [Google Scholar]
- Pons, S.; Fattore, L.; Cossu, G.; Tolu, S.; Porcu, E.; McIntosh, J.M.; Changeux, J.P.; Maskos, U.; Fratta, W. Crucial role of alpha4 and alpha6 nicotinic acetylcholine receptor subunits from ventral tegmental area in systemic nicotine self-administration. J. Neurosci. 2008, 28, 12318–12327. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.C.; Jin, G.Z.; Wu, J. Mysterious alpha6-containing nAChRs: Function, pharmacology, and pathophysiology. Acta Pharmacol. Sin. 2009, 30, 740–751. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Azam, L.; Staheli, S.; Dowell, C.; Lindstrom, J.M.; Kuryatov, A.; Garrett, J.E.; Marks, M.J.; Whiteaker, P. Analogs of alpha-conotoxin MII are selective for alpha6-containing nicotinic acetylcholine receptors. Mol. Pharmacol. 2004, 65, 944–952. [Google Scholar] [CrossRef]
- Azam, L.; Yoshikami, D.; McIntosh, J.M. Amino acid residues that confer high selectivity of the alpha6 nicotinic acetylcholine receptor subunit to alpha-conotoxin MII[S4A,E11A,L15A]. J. Biol. Chem. 2008, 283, 11625–11632. [Google Scholar] [CrossRef]
- McCallum, S.E.; Parameswaran, N.; Bordia, T.; McIntosh, J.M.; Grady, S.R.; Quik, M. Decrease in alpha3*/alpha6* nicotinic receptors but not nicotine-evoked dopamine release in monkey brain after nigrostriatal damage. Mol. Pharmacol. 2005, 68, 737–746. [Google Scholar]
- Perry, D.C.; Mao, D.; Gold, A.B.; McIntosh, J.M.; Pezzullo, J.C.; Kellar, K.J. Chronic nicotine differentially regulates alpha6- and beta3-containing nicotinic cholinergic receptors in rat brain. J. Pharmacol. Exp. Ther. 2007, 322, 306–315. [Google Scholar] [CrossRef]
- Dowell, C.; Olivera, B.M.; Garrett, J.E.; Staheli, S.T.; Watkins, M.; Kuryatov, A.; Yoshikami, D.; Lindstrom, J.M.; McIntosh, J.M. Alpha-conotoxin PIA is selective for alpha6 subunit-containing nicotinic acetylcholine receptors. J. Neurosci. 2003, 23, 8445–8452. [Google Scholar]
- Hone, A.J.; Scadden, M.; Gajewiak, J.; Christensen, S.; Lindstrom, J.; McIntosh, J.M. alpha-Conotoxin PeIA[S9H,V10A,E14N] potently and selectively blocks alpha6beta2beta3 versus alpha6beta4 nicotinic acetylcholine receptors. Mol. Pharmacol. 2012, 82, 972–982. [Google Scholar] [CrossRef]
- Kim, H.W.; McIntosh, J.M. alpha6 nAChR subunit residues that confer alpha-conotoxin BuIA selectivity. FASEB J. 2012, 26, 4102–4110. [Google Scholar] [CrossRef]
- Luo, S.; Zhangsun, D.; Wu, Y.; Zhu, X.; Hu, Y.; McIntyre, M.; Christensen, S.; Akcan, M.; Craik, D.J.; McIntosh, J.M. Characterization of a novel alpha-conotoxin from conus textile that selectively targets alpha6/alpha3beta2beta3 nicotinic acetylcholine receptors. J. Biol. Chem. 2013, 288, 894–902. [Google Scholar] [CrossRef]
- Plazas, P.V.; Katz, E.; Gomez-Casati, M.E.; Bouzat, C.; Elgoyhen, A.B. Stoichiometry of the alpha 9 alpha 10 nicotinic cholinergic receptor. J. Neurosci. 2005, 25, 10905–10912. [Google Scholar] [CrossRef]
- Elgoyhen, A.B.; Vetter, D.E.; Katz, E.; Rothlin, C.V.; Heinemann, S.F.; Boulter, J. alpha10: A determinant of nicotinic cholinergic receptor function in mammalian vestibular and cochlear mechanosensory hair cells. Proc. Natl. Acad. Sci. USA 2001, 98, 3501–3506. [Google Scholar] [CrossRef]
- Koval, L.; Lykhmus, O.; Zhmak, M.; Khruschov, A.; Tsetlin, V.; Magrini, E.; Viola, A.; Chernyavsky, A.; Qian, J.; Grando, S.; et al. Differential involvement of alpha4beta2, alpha7 and alpha9alpha10 nicotinic acetylcholine receptors in B lymphocyte activation in vitro. Int. J. Biochem. Cell. Biol. 2011, 43, 516–524. [Google Scholar] [CrossRef]
- Peng, H.S.; Ferris, R.L.; Matthews, T.; Hiel, H.; Lopez-Albaitero, A.; Lustig, L.R. Characterization of the human nicotinic acetylcholine receptor subunit alpha (alpha) 9 (CHRNA9) and alpha (alpha) 10 (CHRNAIO) in lymphocytes. Life Sci. 2004, 76, 263–280. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Absalom, N.; Chebib, M.; Elgoyhen, A.B.; Vincler, M. Alpha9 nicotinic acetylcholine receptors and the treatment of pain. Biochem. Pharmacol. 2009, 78, 693–702. [Google Scholar] [CrossRef]
- Wu, C.H.; Lee, C.H.; Ho, Y.S. Nicotinic acetylcholine receptor-based blockade: Applications of molecular targets for cancer therapy. Clin. Cancer Res. 2011, 17, 3533–3541. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Plazas, P.V.; Watkins, M.; Gomez-Casati, M.E.; Olivera, B.M.; Elgoyhen, A.B. A novel alpha-conotoxin, PeIA, cloned from Conus pergrandis, discriminates between rat alpha9alpha10 and alpha7 nicotinic cholinergic receptors. J. Biol. Chem. 2005, 280, 30107–30112. [Google Scholar]
- Ellison, M.; Haberlandt, C.; Gomez-Casati, M.E.; Watkins, M.; Elgoyhen, A.B.; McIntosh, J.M.; Olivera, B.M. Alpha-RgIA: A novel conotoxin that specifically and potently blocks the alpha9alpha10 nAChR. Biochemistry 2006, 45, 1511–1517. [Google Scholar] [CrossRef]
- Callaghan, B.; Adams, D.J. Analgesic alpha-conotoxins Vc1.1 and RgIA inhibit N-type calcium channels in sensory neurons of alpha9 nicotinic receptor knockout mice. Channels 2010, 4, 51–54. [Google Scholar]
- Callaghan, B.; Haythornthwaite, A.; Berecki, G.; Clark, R.J.; Craik, D.J.; Adams, D.J. Analgesic alpha-conotoxins Vc1.1 and Rg1A inhibit N-type calcium channels in rat sensory neurons via GABAB receptor activation. J. Neurosci. 2008, 28, 10943–10951. [Google Scholar] [CrossRef]
- Cuny, H.; de Faoite, A.; Huynh, T.G.; Yasuda, T.; Berecki, G.; Adams, D.J. gamma-Aminobutyric acid type B (GABAB) receptor expression is needed for inhibition of N-type (Cav2.2) calcium channels by analgesic alpha-conotoxins. J. Biol. Chem. 2012, 287, 23948–23957. [Google Scholar]
- Zheng, G.; Zhang, Z.; Dowell, C.; Wala, E.; Dwoskin, L.P.; Holtman, J.R.; McIntosh, J.M.; Crooks, P.A. Discovery of non-peptide, small molecule antagonists of alpha9alpha10 nicotinic acetylcholine receptors as novel analgesics for the treatment of neuropathic and tonic inflammatory pain. Bioorg. Med. Chem. Lett. 2011, 21, 2476–2479. [Google Scholar]
- Napier, I.A.; Klimis, H.; Rycroft, B.K.; Jin, A.H.; Alewood, P.F.; Motin, L.; Adams, D.J.; Christie, M.J. Intrathecal alpha-conotoxins Vc1.1, AuIB and MII acting on distinct nicotinic receptor subtypes reverse signs of neuropathic pain. Neuropharmacology 2012, 62, 2202–2207. [Google Scholar] [CrossRef]
- Adams, D.J.; Berecki, G. Mechanisms of conotoxin inhibition of N-type (Ca(v)2.2) calcium channels. Biochim. Biophys. Acta 2013, 1828, 1619–1628. [Google Scholar] [CrossRef]
- Halai, R.; Clark, R.J.; Nevin, S.T.; Jensen, J.E.; Adams, D.J.; Craik, D.J. Scanning mutagenesis of alpha-conotoxin Vc1.1 reveals residues crucial for activity at the alpha9alpha10 nicotinic acetylcholine receptor. J. Biol. Chem. 2009, 284, 20275–20284. [Google Scholar]
- Yu, R.; Kompella, S.N.; Adams, D.J.; Craik, D.J.; Kaas, Q. Determination of the alpha-conotoxin Vc1.1 binding site on the alpha9alpha10 nicotinic acetylcholine receptor. J. Med. Chem. 2013, 56, 3557–3567. [Google Scholar] [CrossRef]
- Ellison, M.; Feng, Z.P.; Park, A.J.; Zhang, X.; Olivera, B.M.; McIntosh, J.M.; Norton, R.S. Alpha-RgIA, a novel conotoxin that blocks the alpha9alpha10 nAChR: Structure and identification of key receptor-binding residues. J. Mol. Biol. 2008, 377, 1216–1227. [Google Scholar] [CrossRef]
- Azam, L.; McIntosh, J.M. Molecular basis for the differential sensitivity of rat and human alpha9alpha10 nAChRs to alpha-conotoxin RgIA. J. Neurochem. 2012, 122, 1137–1144. [Google Scholar] [CrossRef]
- Marx, A.; O’Connor, R.; Tzartos, S.; Kalies, I.; Kirchner, T.; Muller-Hermelink, H.K. Acetylcholine receptor epitope in proteins of myasthenia gravis-associated thymomas and non-thymic tissues. Thymus 1989, 14, 171–178. [Google Scholar]
- Navaneetham, D.; Penn, A.S.; Howard, J.F., Jr.; Conti-Fine, B.M. Human thymuses express incomplete sets of muscle acetylcholine receptor subunit transcripts that seldom include the delta subunit. Muscle Nerve 2001, 24, 203–210. [Google Scholar] [CrossRef]
- Horton, R.M.; Manfredi, A.A.; Conti-Tronconi, B.M. The ‘embryonic’ gamma subunit of the nicotinic acetylcholine receptor is expressed in adult extraocular muscle. Neurology 1993, 43, 983–986. [Google Scholar] [CrossRef]
- Gattenlohner, S.; Schneider, C.; Thamer, C.; Klein, R.; Roggendorf, W.; Gohlke, F.; Niethammer, C.; Czub, S.; Vincent, A.; Muller-Hermelink, H.K.; et al. Expression of foetal type acetylcholine receptor is restricted to type 1 muscle fibres in human neuromuscular disorders. Brain 2002, 125, 1309–1319. [Google Scholar] [CrossRef]
- Gattenloehner, S.; Vincent, A.; Leuschner, I.; Tzartos, S.; Muller-Hermelink, H.K.; Kirchner, T.; Marx, A. The fetal form of the acetylcholine receptor distinguishes rhabdomyosarcomas from other childhood tumors. Am. J. Pathol. 1998, 152, 437–444. [Google Scholar]
- Gattenloehner, S.; Dockhorn-Dworniczak, B.; Leuschner, I.; Vincent, A.; Muller-Hermelink, H.K.; Marx, A. A comparison of MyoD1 and fetal acetylcholine receptor expression in childhood tumors and normal tissues: Implications for the molecular diagnosis of minimal disease in rhabdomyosarcomas. J. Mol. Diagn. 1999, 1, 23–31. [Google Scholar] [CrossRef]
- Gu, Y.; Hall, Z.W. Characterization of acetylcholine receptor subunits in developing and in denervated mammalian muscle. J. Biol. Chem. 1988, 263, 12878–12885. [Google Scholar]
- Witzemann, V.; Barg, B.; Criado, M.; Stein, E.; Sakmann, B. Developmental regulation of five subunit specific mRNAs encoding acetylcholine receptor subtypes in rat muscle. FEBS Lett. 1989, 242, 419–424. [Google Scholar] [CrossRef]
- Sine, S.M.; Claudio, T. Gamma- and delta-subunits regulate the affinity and the cooperativity of ligand binding to the acetylcholine receptor. J. Biol. Chem. 1991, 266, 19369–19377. [Google Scholar]
- Changeux, J.P. The TiPS lecture. The nicotinic acetylcholine receptor: An allosteric protein prototype of ligand-gated ion channels. Trends Pharmacol. Sci. 1990, 11, 485–492. [Google Scholar] [CrossRef]
- Arias, H.R.; Blanton, M.P. Alpha-conotoxins. Int. J. Biochem. Cell. Biol. 2000, 32, 1017–1028. [Google Scholar] [CrossRef]
- Jackson, M.B. Perfection of a synaptic receptor: Kinetics and energetics of the acetylcholine receptor. Proc. Natl. Acad. Sci. USA 1989, 86, 2199–2203. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, J.; Auerbach, A. Activation of recombinant mouse acetylcholine receptors by acetylcholine, carbamylcholine and tetramethylammonium. J. Physiol. 1995, 486, 189–206. [Google Scholar]
- Sine, S.M.; Claudio, T.; Sigworth, F.J. Activation of Torpedo acetylcholine receptors expressed in mouse fibroblasts. Single channel current kinetics reveal distinct agonist binding affinities. J. Gen. Physiol. 1990, 96, 395–437. [Google Scholar] [CrossRef]
- Khalid, M.A.A. Membrane Electrochemistry: Electrochemical Processes in Bilayer Lipid Membrane. Electrochemistry; Mohammed, A.A.K., Ed.; InTech: Rijeka, Croatia, 2013. Available online: http://www.intechopen.com/books/electrochemistry/membrane-electrochemistry-electrochemical-processes-in-bilayer-lipid-membrane (accessed on 21 April 2014). [CrossRef]
- Kreienkamp, H.J.; Sine, S.M.; Maeda, R.K.; Taylor, P. Glycosylation sites selectively interfere with alpha-toxin binding to the nicotinic acetylcholine receptor. J. Biol. Chem. 1994, 269, 8108–8114. [Google Scholar]
- Groebe, D.R.; Dumm, J.M.; Levitan, E.S.; Abramson, S.N. alpha-Conotoxins selectively inhibit one of the two acetylcholine binding sites of nicotinic receptors. Mol. Pharmacol. 1995, 48, 105–111. [Google Scholar]
- Hann, R.M.; Pagan, O.R.; Eterovic, V.A. The alpha-conotoxins GI and MI distinguish between the nicotinic acetylcholine receptor agonist sites while SI does not. Biochemistry 1994, 33, 14058–14063. [Google Scholar] [CrossRef]
- Utkin, Y.N.; Kobayashi, Y.; Hucho, F.; Tsetlin, V.I. Relationship between the binding sites for an alpha-conotoxin and snake venom neurotoxins in the nicotinic acetylcholine receptor from Torpedo californica. Toxicon 1994, 32, 1153–1157. [Google Scholar] [CrossRef]
- Sine, S.M.; Kreienkamp, H.J.; Bren, N.; Maeda, R.; Taylor, P. Molecular dissection of subunit interfaces in the acetylcholine receptor: Identification of determinants of alpha-conotoxin M1 selectivity. Neuron 1995, 15, 205–211. [Google Scholar] [CrossRef]
- Gu, Y.; Camacho, P.; Gardner, P.; Hall, Z.W. Identification of two amino acid residues in the epsilon subunit that promote mammalian muscle acetylcholine receptor assembly in COS cells. Neuron 1991, 6, 879–887. [Google Scholar] [CrossRef]
- Gehrmann, J.; Alewood, P.F.; Craik, D.J. Structure determination of the three disulfide bond isomers of alpha-conotoxin GI: A model for the role of disulfide bonds in structural stability. J. Mol. Biol. 1998, 278, 401–415. [Google Scholar] [CrossRef]
- Groebe, D.R.; Gray, W.R.; Abramson, S.N. Determinants involved in the affinity of alpha-conotoxins GI and SI for the muscle subtype of nicotinic acetylcholine receptors. Biochemistry 1997, 36, 6469–6474. [Google Scholar] [CrossRef]
- Hann, R.M.; Pagan, O.R.; Gregory, L.M.; Jacome, T.; Eterovic, V.A. The 9-arginine residue of alpha-conotoxin GI is responsible for its selective high affinity for the alphagamma agonist site on the electric organ acetylcholine receptor. Biochemistry 1997, 36, 9051–9056. [Google Scholar] [CrossRef]
- Martinez, J.S.; Olivera, B.M.; Gray, W.R.; Craig, A.G.; Groebe, D.R.; Abramson, S.N.; McIntosh, J.M. alpha-Conotoxin EI, a new nicotinic acetylcholine receptor antagonist with novel selectivity. Biochemistry 1995, 34, 14519–14526. [Google Scholar] [CrossRef]
- Hu, S.H.; Loughnan, M.; Miller, R.; Weeks, C.M.; Blessing, R.H.; Alewood, P.F.; Lewis, R.J.; Martin, J.L. The 1.1 A resolution crystal structure of [Tyr15]EpI, a novel alpha-conotoxin from Conus episcopatus, solved by direct methods. Biochemistry 1998, 37, 11425–11433. [Google Scholar] [CrossRef]
- Jacobsen, R.B.; DelaCruz, R.G.; Grose, J.H.; McIntosh, J.M.; Yoshikami, D.; Olivera, B.M. Critical residues influence the affinity and selectivity of alpha-conotoxin MI for nicotinic acetylcholine receptors. Biochemistry 1999, 38, 13310–13315. [Google Scholar]
- Lopez-Vera, E.; Aguilar, M.B.; Schiavon, E.; Marinzi, C.; Ortiz, E.; Restano Cassulini, R.; Batista, C.V.; Possani, L.D.; Heimer de la Cotera, E.P.; Peri, F.; et al. Novel alpha-conotoxins from Conus spurius and the alpha-conotoxin EI share high-affinity potentiation and low-affinity inhibition of nicotinic acetylcholine receptors. FEBS J. 2007, 274, 3972–3985. [Google Scholar] [CrossRef]
- Lluisma, A.O.; Lopez-Vera, E.; Bulaj, G.; Watkins, M.; Olivera, B.M. Characterization of a novel psi-conotoxin from Conus parius Reeve. Toxicon 2008, 51, 174–180. [Google Scholar] [CrossRef]
- Shon, K.J.; Grilley, M.; Jacobsen, R.; Cartier, G.E.; Hopkins, C.; Gray, W.R.; Watkins, M.; Hillyard, D.R.; Rivier, J.; Torres, J.; et al. A noncompetitive peptide inhibitor of the nicotinic acetylcholine receptor from Conus purpurascens venom. Biochemistry 1997, 36, 9581–9587. [Google Scholar] [CrossRef]
- Teichert, R.W.; Rivier, J.; Torres, J.; Dykert, J.; Miller, C.; Olivera, B.M. A uniquely selective inhibitor of the mammalian fetal neuromuscular nicotinic acetylcholine receptor. J. Neurosci. 2005, 25, 732–736. [Google Scholar] [CrossRef]
- Han, K.H.; Hwang, K.J.; Kim, S.M.; Kim, S.K.; Gray, W.R.; Olivera, B.M.; Rivier, J.; Shon, K.J. NMR structure determination of a novel conotoxin, [Pro 7,13] alpha A-conotoxin PIVA. Biochemistry 1997, 36, 1669–1677. [Google Scholar]
- Lebbe, E.K.; Peigneur, S.; Maiti, M.; Devi, P.; Ravichandran, S.; Lescrinier, E.; Ulens, C.; Waelkens, E.; D’Souza, L.; Herdewijn, P.; et al. Structure-Function Elucidation of a New alpha-conotoxin, Lo1a, from Conus longurionis. J. Biol. Chem. 2014, 289, 9573–9583. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lebbe, E.K.M.; Peigneur, S.; Wijesekara, I.; Tytgat, J. Conotoxins Targeting Nicotinic Acetylcholine Receptors: An Overview. Mar. Drugs 2014, 12, 2970-3004. https://doi.org/10.3390/md12052970
Lebbe EKM, Peigneur S, Wijesekara I, Tytgat J. Conotoxins Targeting Nicotinic Acetylcholine Receptors: An Overview. Marine Drugs. 2014; 12(5):2970-3004. https://doi.org/10.3390/md12052970
Chicago/Turabian StyleLebbe, Eline K. M., Steve Peigneur, Isuru Wijesekara, and Jan Tytgat. 2014. "Conotoxins Targeting Nicotinic Acetylcholine Receptors: An Overview" Marine Drugs 12, no. 5: 2970-3004. https://doi.org/10.3390/md12052970
APA StyleLebbe, E. K. M., Peigneur, S., Wijesekara, I., & Tytgat, J. (2014). Conotoxins Targeting Nicotinic Acetylcholine Receptors: An Overview. Marine Drugs, 12(5), 2970-3004. https://doi.org/10.3390/md12052970




