The Antibacterial and Anti-Eukaryotic Type VI Secretion System MIX-Effector Repertoire in Vibrionaceae
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identifying MIX-Effectors in Vibrionaceae
2.2. Vibrionaceae MIX-Effectors Carry Diverse C-Terminal Toxin Domains
2.3. Predicted Anti-Eukaryotic MIX-Effector Families
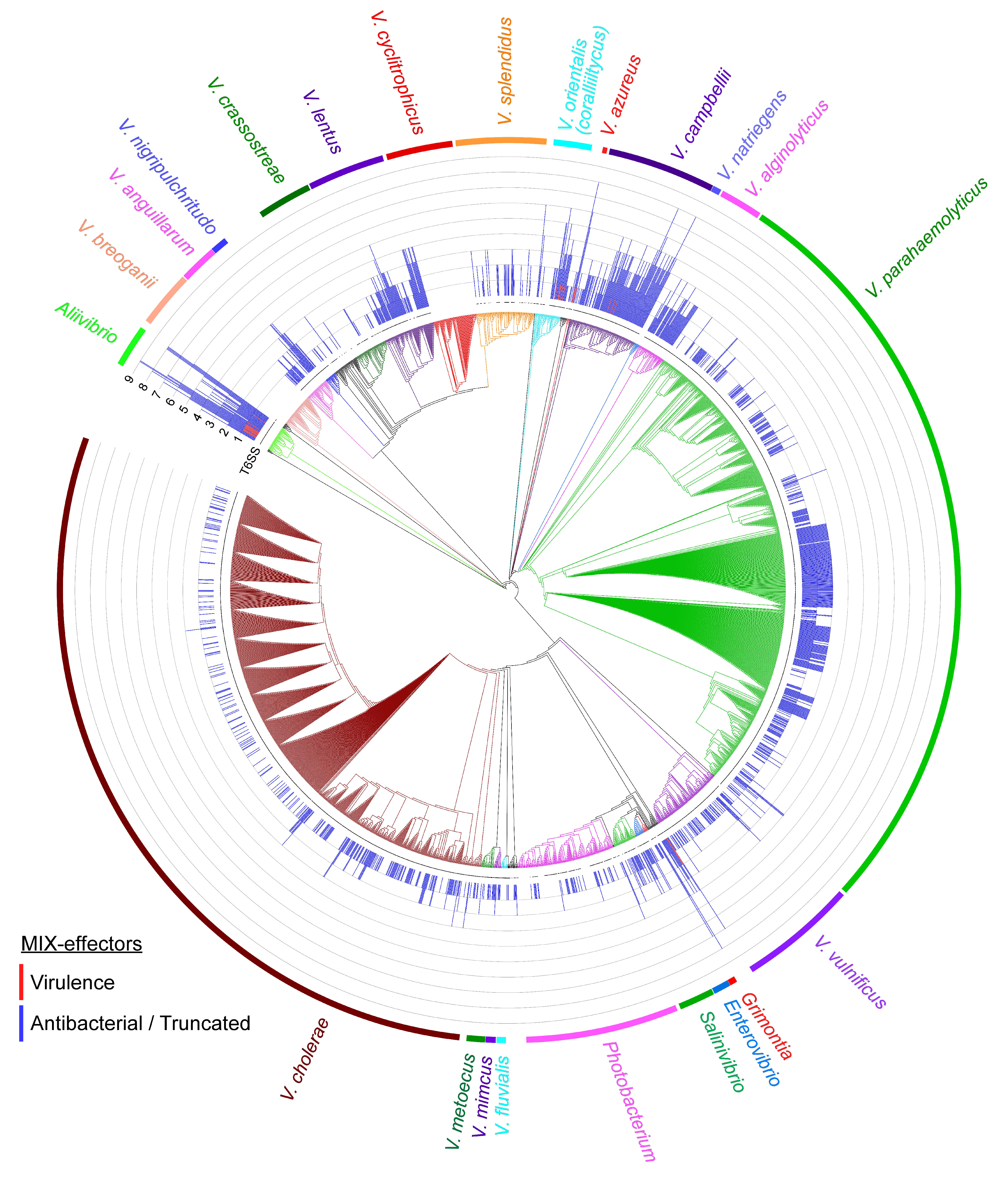
2.4. MIX-Effectors Are Unevenly Distributed Among Vibrionaceae Species
2.5. Concluding Remarks
3. Materials and Methods
3.1. Construction of Position-Specific Scoring Matrices (PSSMs) of Various MIX Domains
3.2. Identification of MIX-Effectors in Vibrionaceae
3.3. Construction of the Phylogenetic Tree
3.4. Strains and Media
3.5. Plasmids
3.6. Bacterial Growth Assays
3.7. Yeast Toxicity
3.8. Protein Expression
3.9. Multiple Sequence Alignments and Secondary Structure Predictions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boyd, E.F.; Carpenter, M.R.; Chowdhury, N.; Cohen, A.L.; Haines-Menges, B.L.; Kalburge, S.S.; Kingston, J.J.; Lubin, J.B.B.; Ongagna-Yhombi, S.Y.; Whitaker, W.B. Post-Genomic Analysis of Members of the Family. Vibrionaceae 2015, 3, 1–26. [Google Scholar] [CrossRef]
- Horseman, M.A.; Bray, R.; Lujan-Francis, B.; Matthew, E. Infections Caused by Vibrionaceae. Infect. Dis. Clin. Pract. 2013, 21, 222–232. [Google Scholar] [CrossRef]
- Huehn, S.; Eichhorn, C.; Urmersbach, S.; Breidenbach, J.; Bechlars, S.; Bier, N.; Alter, T.; Bartelt, E.; Frank, C.; Oberheitmann, B.; et al. Pathogenic Vibrios in Environmental, Seafood and Clinical Sources in Germany. Int. J. Med. Microbiol. 2014, 304, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Romalde, J.L.; Dieguez, A.L.; Lasa, A.; Balboa, S. New Vibrio Species Associated to Molluscan Microbiota: A review. Front. Microbiol. 2014, 4, 413. [Google Scholar] [CrossRef] [PubMed]
- Nyholm, S.V.; McFall-Ngai, M. The Winnowing: Establishing the Squid–Vibrio Symbiosis. Nat. Rev. Microbiol. 2004, 2, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Vezzulli, L.; Grande, C.; Reid, P.C.; Hélaouët, P.; Edwards, M.; Höfle, M.G.; Brettar, I.; Colwell, R.R.; Pruzzo, C. Climate Influence on Vibrio and Associated Human Diseases During the Past Half-Century in the Coastal North Atlantic. Proc. Natl. Acad. Sci. USA 2016, 113, E5062–E5071. [Google Scholar] [CrossRef] [PubMed]
- Lemire, A.; Goudenège, D.; Versigny, T.; Petton, B.; Calteau, A.; Labreuche, Y.; Le Roux, F. Populations, Not Clones, are the Unit of Vibrio Pathogenesis in Naturally Infected Oysters. ISME J. 2015, 9, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.N. Fitness Factors in Vibrios: A Mini-review. Microb. Ecol. 2013, 65, 826–851. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Orth, K. Virulence Determinants for Vibrio parahaemolyticus Infection. Curr. Opin. Microbiol. 2013, 16, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Boyer, F.; Fichant, G.; Berthod, J.; Vandenbrouck, Y.; Attree, I. Dissecting the Bacterial Type VI Secretion System By a Genome Wide In Silico Analysis: What Can Be Learned From Available Microbial Genomic Resources? BMC Genom. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Mougous, J.D.; Cuff, M.E.; Raunser, S.; Shen, A.; Zhou, M.; Gifford, C.A.; Goodman, A.L.; Joachimiak, G.; Ordoñez, C.L.; Lory, S.; et al. A Virulence Locus of Pseudomonas aeruginosa Encodes a Protein Secretion Apparatus. Science 2006, 312, 1526–1530. [Google Scholar] [CrossRef] [PubMed]
- Pukatzki, S.; Ma, A.T.; Sturtevant, D.; Krastins, B.; Sarracino, D.; Nelson, W.C.; Heidelberg, J.F.; Mekalanos, J.J. Identification of a Conserved Bacterial Protein Secretion System in Vibrio cholerae Using the Dictyostelium Host Model System. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Durand, E.; Cambillau, C.; Cascales, E.; Journet, L. VgrG, Tae, Tle, and beyond: The Versatile Arsenal of Type VI Secretion Effectors. Trends Microbiol. 2014, 22, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Pukatzki, S.; Ma, A.T.; Revel, A.T.; Sturtevant, D.; Mekalanos, J.J. Type VI Secretion System Translocates a Phage Tail Spike-like Protein Into Target Cells Where it Cross-links Actin. Proc. Natl. Acad. Sci. USA 2007, 104, 15508–15513. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Hood, R.D.; Bui, N.K.; Leroux, M.; Vollmer, W.; Mougous, J.D. Type VI Secretion Delivers Bacteriolytic Effectors to Target Cells. Nature 2011, 475, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Schwartz, N.; de Souza Santos, M.; Zhang, J.; Orth, K.; Salomon, D. Type VI Secretion System MIX-effectors Carry Both Antibacterial and Anti-eukaryotic Activities. EMBO Rep. 2017, e201744226. [Google Scholar] [CrossRef] [PubMed]
- Cianfanelli, F.R.; Monlezun, L.; Coulthurst, S.J. Aim, Load, Fire: The Type VI Secretion System, a Bacterial Nanoweapon. Trends Microbiol. 2016, 24, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Singh, P.; Brittnacher, M.; Bui, N.K.; Hood, R.D.; Carl, M.A.; Agnello, D.M.; Schwarz, S.; Goodlett, D.R.; Vollmer, W.; et al. A Widespread Bacterial Type VI Secretion Effector Superfamily Identified Using a Heuristic Approach. Cell Host Microbe 2012, 11, 538–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacIntyre, D.L.; Miyata, S.T.; Kitaoka, M.; Pukatzki, S. The Vibrio cholerae Type VI Secretion System Displays Antimicrobial Properties. Proc. Natl. Acad. Sci. USA 2010, 107, 19520–19524. [Google Scholar] [CrossRef] [PubMed]
- Salomon, D.; Gonzalez, H.; Updegraff, B.L.; Orth, K. Vibrio parahaemolyticus Type VI Secretion System 1 Is Activated in Marine Conditions to Target Bacteria, and Is Differentially Regulated from System 2. PLoS ONE 2013, 8, e61086. [Google Scholar] [CrossRef] [PubMed]
- Salomon, D.; Klimko, J.A.; Trudgian, D.C.; Kinch, L.N.; Grishin, N.V.; Mirzaei, H.; Orth, K. Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria. PLoS Pathog. 2015, 11, e1005128. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Du, P.; Zhao, M.; Liu, W.; Du, Y.; Diao, B.; Li, J.; Kan, B.; Liang, W. Functional Characterization and Conditional Regulation of the Type VI Secretion System in Vibrio fluvialis. Front. Microbiol. 2017, 8, 528. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yue, S.; Li, G.-Y.; Li, J.; Wang, X.-R.; Li, S.-F.; Mo, Z.-L. Expression, Secretion and Bactericidal Activity of Type VI Secretion System in Vibrio anguillarum. Arch. Microbiol. 2016, 198, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Speare, L.; Cecere, A.G.; Guckes, K.R.; Smith, S.; Wollenberg, M.S.; Mandel, M.J.; Miyashiro, T.; Septer, A.N. Bacterial Symbionts Use a Type VI Secretion System to Eliminate Competitors in Their Natural Host. Proc. Natl. Acad. Sci. USA 2018, 115, E8528–E8537. [Google Scholar] [CrossRef] [PubMed]
- Church, S.R.; Lux, T.; Baker-Austin, C.; Buddington, S.P.; Michell, S.L. Vibrio vulnificus Type 6 Secretion System 1 Contains Anti-bacterial Properties. PLoS ONE 2016, 11, e0165500. [Google Scholar] [CrossRef] [PubMed]
- Salomon, D.; Kinch, L.N.; Trudgian, D.C.; Guo, X.; Klimko, J.A.; Grishin, N.V.; Mirzaei, H.; Orth, K. Marker for Type VI Secretion System Effectors. Proc. Natl. Acad. Sci. USA 2014, 111, 9271–9276. [Google Scholar] [CrossRef] [PubMed]
- Brooks, T.M.; Unterweger, D.; Bachmann, V.; Kostiuk, B.; Pukatzki, S. Lytic Activity of the Vibrio cholerae Type VI Secretion Toxin VgrG-3 Is Inhibited by the Antitoxin TsaB. J. Biol. Chem. 2013, 288, 7618–7625. [Google Scholar] [CrossRef] [PubMed]
- Altindis, E.; Dong, T.; Catalano, C.; Mekalanos, J. Secretome Analysis of Vibrio cholerae Type VI Secretion System Reveals a New Effector-Immunity Pair. MBio 2015, 6, e00075. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.T.; Unterweger, D.; Rudko, S.P.; Pukatzki, S. Dual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic Vibrio cholerae. PLoS Pathog. 2013, 9, e1003752. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.T.; Kitaoka, M.; Brooks, T.M.; McAuley, S.B.; Pukatzki, S. Vibrio cholerae Requires the Type VI Secretion System Virulence Factor VasX to Kill Dictyostelium Discoideum. Infect. Immun. 2011, 79, 2941–2949. [Google Scholar] [CrossRef] [PubMed]
- Unterweger, D.; Miyata, S.T.; Bachmann, V.; Brooks, T.M.; Mullins, T.; Kostiuk, B.; Provenzano, D.; Pukatzki, S. The Vibrio cholerae Type VI Secretion System Employs Diverse Effector Modules for Intraspecific Competition. Nat. Commun. 2014, 5, 3549. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Kinch, L.N.; Ray, A.; Dalia, A.B.; Cong, Q.; Nunan, L.M.; Camilli, A.; Grishin, N.V.; Salomon, D.; Orth, K. Acute Hepatopancreatic Necrosis Disease-Causing Vibrio parahaemolyticus Strains Maintain an Antibacterial Type VI Secretion System with Versatile Effector Repertoires. Appl. Environ. Microbiol. 2017, 83, e00737-17. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Anderson, J.B.; Derbyshire, M.K.; DeWeese-Scott, C.; Gonzales, N.R.; Gwadz, M.; Hao, L.; He, S.; Hurwitz, D.I.; Jackson, J.D.; et al. CDD: A Conserved Domain Database for Interactive Domain Family Analysis. Nucleic Acids Res. 2007, 35, D237–D240. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Lakey, J.H.; Slatin, S.L. Pore-forming Colicins and their Relatives. Curr. Top. Microbiol. Immunol. 2001, 257, 131–161. [Google Scholar] [PubMed]
- Russell, A.B.; Leroux, M.; Hathazi, K.; Agnello, D.M.; Ishikawa, T.; Wiggins, P.A.; Wai, S.N.; Mougous, J.D. Diverse Type VI Secretion Phospholipases are Functionally Plastic Antibacterial Effectors. Nature 2013, 496, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Frickey, T.; Lupas, A. CLANS: A Java Application for Visualizing Protein Families Based on Pairwise Similarity. Bioinformatics 2004, 20, 3702–3704. [Google Scholar] [CrossRef] [PubMed]
- Klimpel, K.R.; Arora, N.; Leppla, S.H. Anthrax Toxin Lethal factor contains a zinc Metalloprotease Consensus Sequence Which is Required for Lethal Toxin Activity. Mol. Microbiol. 1994, 13, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Dalkas, G.A.; Chasapis, C.T.; Gkazonis, P.V.; Bentrop, D.; Spyroulias, G.A. Conformational Dynamics of the Anthrax Lethal Factor Catalytic Center. Biochemistry 2010, 49, 10767–10769. [Google Scholar] [CrossRef] [PubMed]
- Prochazkova, K.; Shuvalova, L.A.; Minasov, G.; Voburka, Z.; Anderson, W.F.; Satchell, K.J.F. Structural and Molecular Mechanism for Autoprocessing of MARTX Toxin of Vibrio cholerae at Multiple Sites. J. Biol. Chem. 2009, 284, 26557–26568. [Google Scholar] [CrossRef] [PubMed]
- Egerer, M.; Giesemann, T.; Jank, T.; Satchell, K.J.F.; Aktories, K. Auto-catalytic Cleavage of Clostridium difficile Toxins A and B Depends on Cysteine Protease Activity. J. Biol. Chem. 2007, 282, 25314–25321. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, X.; Yang, C.; Wu, Z.; Xu, H.; Shen, Y. Crystal Structure of Outer Membrane Protein NMB0315 from Neisseria meningitidis. PLoS ONE 2011, 6, e26845. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; de Souza, R.F.; Anantharaman, V.; Iyer, L.M.; Aravind, L. Polymorphic Toxin Systems: Comprehensive Characterization of Trafficking Modes, Processing, Mechanisms of Action, Immunity and Ecology Using Comparative Genomics. Biol. Direct 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, G.; Dowling, A.J.; Gerike, U.; ffrench-Constant, R.H.; Waterfield, N.R. Photorhabdus Virulence Cassettes Confer Injectable Insecticidal Activity against the Wax Moth. J. Bacteriol. 2006, 188, 2254–2261. [Google Scholar] [CrossRef] [PubMed]
- Kimes, N.E.; Grim, C.J.; Johnson, W.R.; Hasan, N.A.; Tall, B.D.; Kothary, M.H.; Kiss, H.; Munk, A.C.; Tapia, R.; Green, L.; et al. Temperature Regulation of Virulence Factors in the Pathogen Vibrio coralliilyticus. ISME J. 2012, 6, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Salomon, D.; Dar, D.; Sreeramulu, S.; Sessa, G. Expression of Xanthomonas campestris pv. vesicatoria Type III Effectors in Yeast Affects Cell Growth and Viability. Mol. Plant. Microbe Interact. 2011, 24, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Bosis, E.; Salomon, D.; Sessa, G. A Simple Yeast-based Strategy to Identify Host Cellular Processes Targeted By Bacterial Effector Proteins. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Salomon, D.; Bosis, E.; Dar, D.; Nachman, I.; Sessa, G. Expression of Pseudomonas syringae Type III Effectors in Yeast Under Stress Conditions Reveals that HopX1 Attenuates Activation of the High Osmolarity Glycerol MAP Kinase Pathway. Microbiology 2012, 158, 2859–2869. [Google Scholar] [CrossRef] [PubMed]
- Siggers, K.A.; Lesser, C.F. The Yeast Saccharomyces cerevisiae: A Versatile Model System for the Identification and Characterization of Bacterial Virulence Proteins. Cell Host Microbe 2008, 4, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Slagowski, N.L.; Kramer, R.W.; Morrison, M.F.; LaBaer, J.; Lesser, C.F. A Functional Genomic Yeast Screen to Identify Pathogenic Bacterial Proteins. PLoS Pathog. 2008, 4, e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, R.W.; Slagowski, N.L.; Eze, N.A.; Giddings, K.S.; Morrison, M.F.; Siggers, K.A.; Starnbach, M.N.; Lesser, C.F. Yeast Functional Genomic Screens Lead to Identification of a Role for a Bacterial Effector in Innate Immunity Regulation. PLoS Pathog. 2007, 3, e21. [Google Scholar] [CrossRef] [PubMed]
- Haldar, S.; Chatterjee, S.; Sugimoto, N.; Das, S.; Chowdhury, N.; Hinenoya, A.; Asakura, M.; Yamasaki, S. Identification of Vibrio campbellii Isolated From Diseased Farm-shrimps From South India and Establishment of its Pathogenic Potential in an Artemia Model. Microbiology 2011, 157, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Ben-Haim, Y.; Zicherman-Keren, M.; Rosenberg, E. Temperature-regulated Bleaching and Lysis of the Coral Pocillopora damicornis by the Novel Pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 2003, 69, 4236–4242. [Google Scholar] [CrossRef] [PubMed]
- Lunder, T.; Sørum, H.; Holstad, G.; Steigerwalt, A.G.; Mowinckel, P.; Brenner, D.J. Phenotypic and Genotypic Characterization of Vibrio viscosus sp. nov. and Vibrio wodanis sp. nov. Isolated From Atlantic salmon (Salmo salar) with “Winter Ulcer”. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 2, 427–450. [Google Scholar] [CrossRef] [PubMed]
- López, J.; Lorenzo, L.; Alcantara, R.; Navas, J. Characterization of Aliivibrio fischeri Strains Associated with Disease Outbreak in Brill Scophthalmus rhombus. Dis. Aquat. Organ. 2017, 124, 215–222. [Google Scholar] [CrossRef] [PubMed]
- McFall-Ngai, M.J.; Ruby, E.G. Symbiont Recognition and Subsequent Morphogenesis as Early Events in an Animal-bacterial Mutualism. Science 1991, 254, 1491–1494. [Google Scholar] [CrossRef] [PubMed]
- Hachani, A.; Wood, T.E.; Filloux, A. Type VI Secretion and Anti-host Effectors. Curr. Opin. Microbiol. 2016, 29, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein Domain Annotations on the Fly. Nucleic Acids Res. 2004, 32. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2017. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The Neighbor-joining Method: A New Nethod for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for Inferring Very Large Phylogenies by Using the Neighbor-joining Method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic Sssembly of DNA Molecules Up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Salomon, D.; Sessa, G. Identification of Growth Inhibition Phenotypes Induced by Expression of Bacterial Type III Effectors in Yeast. J. Vis. Exp. 2010, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A Protein Secondary Structure Prediction Server. Nucleic Acids Res. 2015, 43. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2--a Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]


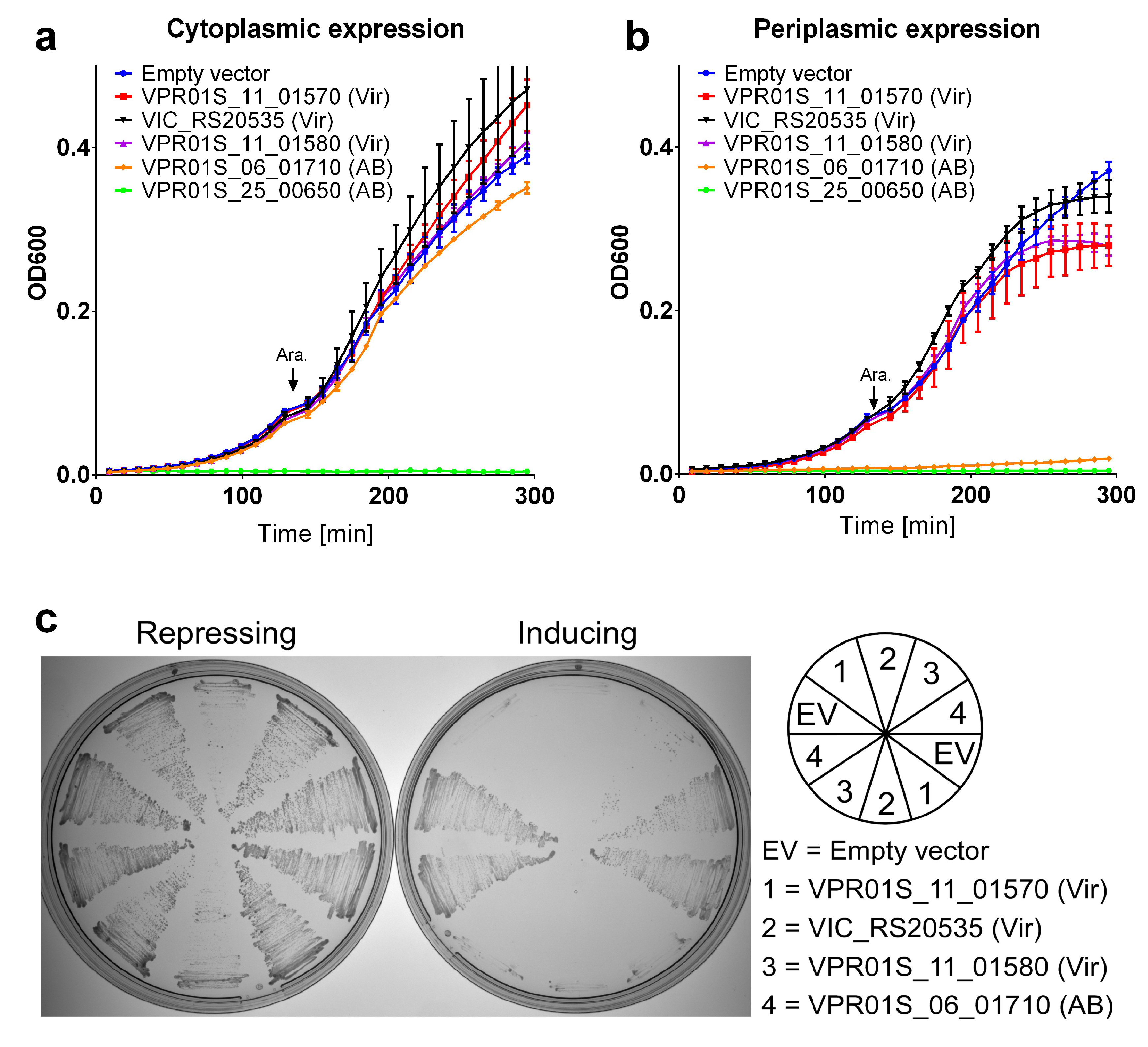

| Predicted Toxic Activity | Occurrences | MIX Clans | Predicted Antibacterial (AB)/Virulence (Vir) |
|---|---|---|---|
| Pore-forming | 835 | I, II, IV, IV + II, V + I, V + IV | AB |
| VP1388 homologs | 800 | I | AB |
| Nuclease | 331 | V, V + I | AB |
| Truncated (no C-terminal toxin domain) | 124 | I, IV, V, V + IV | - |
| DUF2335 (phospholipase) | 121 | V, V + I | AB |
| Peptidoglycan hydrolase | 35 | V | AB |
| Peptidase | 6 | V | AB |
| Nucleotide deaminase | 2 | I | AB |
| Unknown | 61 | I, IV, V, V + IV, V + I + IV | AB |
| Protease | 15 | V | Vir |
| Glycosyltransferase | 5 | V | Vir |
| Pore-forming | 2 | I | Vir |
| CNF1 | 1 | V | Vir |
| Unknown | 4 | V | Vir |
| Cluster | Accessions | Bacterial Species | Predicted Activity/Toxin Domain |
|---|---|---|---|
| 83 | WP_040902815.1 | V. proteolyticus | CNF1 deamidase |
| 27 | WP_065611703.1, WP_061012685.1, WP_060992952.1, WP_017020872.1, WP_061036948.1, WP_061012685.1, WP_105064022.1, WP_061029312.1, WP_061004481.1, WP_023604334.1 | A. wodanis, A. logei, A. sifiae | Protease |
| 55 | WP_005429108.1, WP_052438057.1 | V. campbellii | Cysteine protease |
| 53 | WP_012535377.1, WP_063646315.1 | A. fischeri | Peptidase |
| 58 | WP_021709833.1, WP_052035761.1 | V. proteolyticus, V. azeurus | Unknown |
| 38 | WP_006962196.1, WP_043008000.1, WP_095560114.1, WP_099609290.1, WP_064487344.1 | V. coralliilyticus | Glycosyltransferase |
| 72 | WP_073603189.1 | V. aerogenes | Pore-forming |
| 76 | WP_073605246.1 | V. aerogenes | Pore-forming |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dar, Y.; Salomon, D.; Bosis, E. The Antibacterial and Anti-Eukaryotic Type VI Secretion System MIX-Effector Repertoire in Vibrionaceae. Mar. Drugs 2018, 16, 433. https://doi.org/10.3390/md16110433
Dar Y, Salomon D, Bosis E. The Antibacterial and Anti-Eukaryotic Type VI Secretion System MIX-Effector Repertoire in Vibrionaceae. Marine Drugs. 2018; 16(11):433. https://doi.org/10.3390/md16110433
Chicago/Turabian StyleDar, Yasmin, Dor Salomon, and Eran Bosis. 2018. "The Antibacterial and Anti-Eukaryotic Type VI Secretion System MIX-Effector Repertoire in Vibrionaceae" Marine Drugs 16, no. 11: 433. https://doi.org/10.3390/md16110433
APA StyleDar, Y., Salomon, D., & Bosis, E. (2018). The Antibacterial and Anti-Eukaryotic Type VI Secretion System MIX-Effector Repertoire in Vibrionaceae. Marine Drugs, 16(11), 433. https://doi.org/10.3390/md16110433






