Nile Tilapia Derived Antimicrobial Peptide TP4 Exerts Antineoplastic Activity Through Microtubule Disruption
Abstract
:1. Introduction
2. Results
2.1. Synthesized TP4 binds to Tubulin
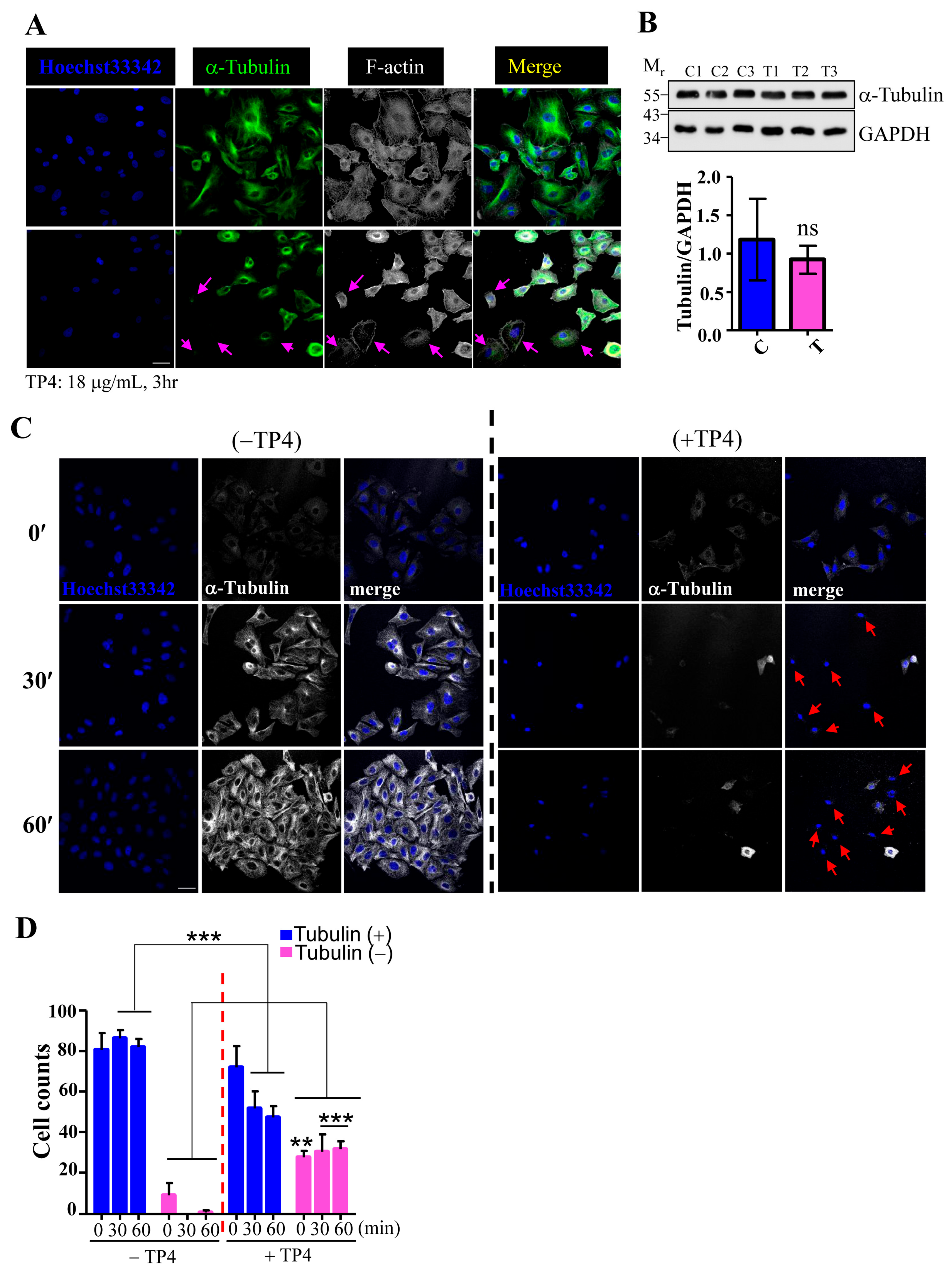
2.2. TP4 Disrupts the Microtubule Network in A549 Cells
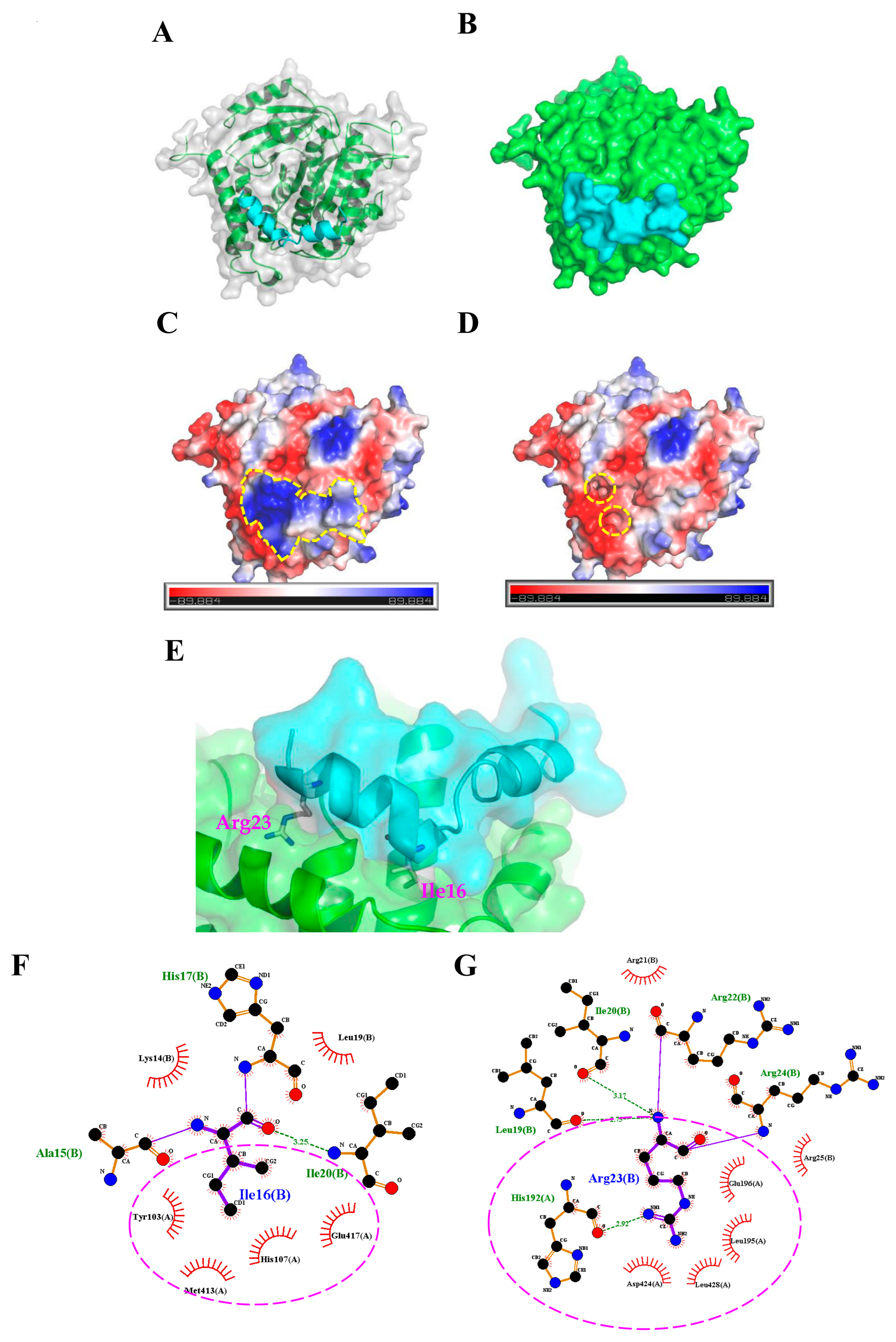
2.3. Molecular Docking Analysis of the TP4-Tubulin Interaction
2.4. TP4 Inhibits Microtubule Polymerization
2.5. TP4 Mutants Exhibit Diminished Cancer Cell Killing
3. Discussion
4. Material and Methods
4.1. Reagents and Peptide Sequence Analysis
4.2. Protein-Peptide Docking
4.3. Cell Culture and Cell Viability Assay
4.4. Co-Immunoprecipitation, LC-MS/MS Analysis, and Western Blot
4.5. Immunocytochemical and Immunohistochemical Studies
4.6. Microtubule Regrowth and In Vitro Tubulin Polymerization Assay
4.7. Statistical analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial Peptides and Their Therapeutic Potential for Bacterial Skin Infections and Wounds. Front. Pharmacol. 2018, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.N.; Chan, Y.L.; Wu, C.J.; Chen, J.Y. Tilapia Piscidin 4 (TP4) Stimulates Cell Proliferation and Wound Closure in MRSA-Infected Wounds in Mice. Mar. Drugs 2015, 13, 2813–2833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.N.; Pan, C.Y.; Chan, Y.L.; Chen, J.Y.; Wu, C.J. Use of the Antimicrobial Peptide Pardaxin (GE33) To Protect against Methicillin-Resistant Staphylococcus aureus Infection in Mice with Skin Injuries. Antimicrob. Agents Chemother. 2014, 58, 1538–1545. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.N.; Rajanbabu, V.; Pan, C.Y.; Chan, Y.L.; Wu, C.J.; Chen, J.Y. Use of the antimicrobial peptide Epinecidin-1 to protect against MRSA infection in mice with skin injuries. Biomaterials 2013, 34, 10319–10327. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Pan, C.Y.; Chen, J.Y. The antimicrobial peptide, epinecidin-1, mediates secretion of cytokines in the immune response to bacterial infection in mice. Peptides 2012, 36, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.; Pirri, G.; Bozzi, A.; Di Giulio, A.; Aschi, M.; Rinaldi, A.C. Antimicrobial peptides: natural templates for synthetic membrane-active compounds. Cell. Mol. Life Sci. 2008, 65, 2450–2460. [Google Scholar] [CrossRef] [PubMed]
- Scocchi, M.; Tossi, A.; Gennaro, R. Proline-rich antimicrobial peptides: converging to a non-lytic mechanism of action. Cell. Mol. Life Sci. 2011, 68, 2317–2330. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.M.; Tseng, C.C.; Chen, N.F.; Tai, M.H.; Hung, H.C.; Feng, C.W.; Cheng, S.Y.; Huang, S.Y.; Jean, Y.H.; Wen, Z.H. MSP-4, an Antimicrobial Peptide, Induces Apoptosis via Activation of Extrinsic Fas/FasL- and Intrinsic Mitochondria-Mediated Pathways in One Osteosarcoma Cell Line. Mar. Drugs 2018, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Ting, C.H.; Chen, Y.C.; Wu, C.J.; Chen, J.Y. Targeting FOSB with a cationic antimicrobial peptide, TP4, for treatment of triple-negative breast cancer. Oncotarget 2016, 7, 40329–40347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayana, J.L.; Huang, H.N.; Wu, C.J.; Chen, J.Y. Efficacy of the antimicrobial peptide TP4 against Helicobacter pylori infection: in vitro membrane perturbation via micellization and in vivo suppression of host immune responses in a mouse model. Oncotarget 2015, 6, 12936–12954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ting, C.H.; Huang, H.N.; Huang, T.C.; Wu, C.J.; Chen, J.Y. The mechanisms by which pardaxin, a, natural cationic antimicrobial peptide, targets the endoplasmic reticulum and induces c-FOS. Biomaterials 2014, 35, 3627–3640. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.; Hoskin, D.W.; Hilchie, A.L. Assessment of antimicrobial (host defense) peptides as anti-cancer agents. Methods Mol. Biol. 2014, 1088, 159–170. [Google Scholar]
- Huang, H.N.; Rajanbabu, V.; Pan, C.Y.; Chan, Y.L.; Wu, C.J.; Chen, J.Y. A cancer vaccine based on the marine antimicrobial peptide pardaxin (GE33) for control of bladder-associated tumors. Biomaterials 2013, 34, 10151–10159. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, D.; Veiga, A.S.; Castanho, M.A.R.B. From antimicrobial to anticancer peptides: A review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [PubMed]
- Hilchie, A.L.; Doucette, C.D.; Pinto, D.M.; Patrzykat, A.; Douglas, S.; Hoskin, D.W. Pleurocidin-family cationic antimicrobial peptides are cytolytic for breast carcinoma cells and prevent growth of tumor xenografts. Breast Cancer Res. 2011, 13, R102. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur. J. Pharmacol. 2009, 625, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.C.; Chen, J.Y. Proteomic analysis reveals that pardaxin triggers apoptotic signaling pathways in human cervical carcinoma HeLa cells: Cross talk among the UPR, c-Jun and ROS. Carcinogenesis 2013, 34, 1833–1842. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.C.; Lee, S.H.; Hour, A.L.; Pan, C.Y.; Lee, L.H.; Chen, J.Y. Five different piscidins from Nile tilapia, Oreochromis niloticus: Analysis of their expressions and biological functions. PloS One 2012, 7, e50263. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, Y.C.; Wang, K.S.; Tsai, S.H.; Chao, W.T.; Lung, F.D. Anticancer activities of an antimicrobial peptide derivative of Ixosin-B amide. Bioorg. Med. Chem. Lett. 2013, 23, 5744–5747. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Wu, Y.; Kang, S.; Ma, J.; Yang, J.; Zhang, F. CecropinXJ, a silkworm antimicrobial peptide, induces cytoskeleton disruption in esophageal carcinoma cells. Acta Biochim. Biophys. Sin. (Shanghai) 2014, 46, 867–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogales, E.; Wolf, S.G.; Khan, I.A.; Luduena, R.F.; Downing, K.H. Structure of Tubulin at 6.5 Angstrom and Location of the Taxol-Binding Site. Nature 1995, 375, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Krauss, N.E.; Heerding, J.M.; Swindell, C.S.; Ringel, I.; Orr, G.A.; Horwitz, S.B. 3′-(P-Azidobenzamido)Taxol Photolabels the N-Terminal 31 Amino-Acids of Beta-Tubulin. J. Biol. Chem. 1994, 269, 3132–3134. [Google Scholar] [PubMed]
- Rieder, C.L.; Maiato, H. Stuck in division or passing through: What happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 2004, 7, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Bekier, M.E.; Fischbach, R.; Lee, J.; Taylor, W.R. Length of mitotic arrest induced by microtubule-stabilizing drugs determines cell death after mitotic exit. Mol. Cancer Ther. 2009, 8, 1646–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gigant, B.; Wang, C.; Ravelli, R.B.G.; Roussi, F.; Steinmetz, M.O.; Curmi, P.A.; Sobel, A.; Knossow, M. Structural basis for the regulation of tubulin by vinblastine. Nature 2005, 435, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.S.; Wolff, J. Localization of the vinblastine-binding site on beta-tubulin. J. Biol. Chem. 1996, 271, 14707–14711. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.A.; Thrower, D.; Wilson, L. Mechanism of Inhibition of Cell-Proliferation by Vinca Alkaloids. Cancer Res. 1991, 51, 2212–2222. [Google Scholar] [PubMed]
- Ngan, V.K.; Bellman, K.; Hill, B.T.; Wilson, L.; Jordan, M.A. Mechanism of mitotic block and inhibition of cell proliferation by the semisynthetic vinca alkaloids vinorelbine and its newer derivative vinflunine. Mol. Pharmacol. 2001, 60, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Garland, D.L. Kinetics and Mechanism of Colchicine Binding to Tubulin: Evidence for Ligand-Induced Conformational Change. Biochemistry 1978, 17, 4266–4272. [Google Scholar] [CrossRef] [PubMed]
- Ravelli, R.B.G.; Gigant, B.; Curmi, P.A.; Jourdain, I.; Lachkar, S.; Sobel, A.; Knossow, M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 2004, 428, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.; Chaplin, D.J.; Rosenthal, D.S.; Boulares, A.H.; Li, L.Y.; Smulson, M.E. Induction of apoptosis in proliferating human endothelial cells by the tumor-specific antiangiogenesis agent combretastatin A-4. Cancer Rres. 1998, 58, 4510–4514. [Google Scholar]
- Zhou, J.; Giannakakou, P. Targeting microtubules for cancer chemotherapy. Curr. Med. Chem. Anticancer Agents 2005, 5, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.; Li, H.; Downing, K.H.; Nogales, E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J. Mol. Biol. 2001, 313, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Kikkawa, M.; Okada, Y.; Hirokawa, N. 15 A resolution model of the monomeric kinesin motor, KIF1A. Cell 2000, 100, 241–52. [Google Scholar] [CrossRef]
- Gigant, B.; Curmi, P.A.; Martin-Barbey, C.; Charbaut, E.; Lachkar, S.; Lebeau, L.; Siavoshian, S.; Sobel, A.; Knossow, M. The 4 A X-ray Structure of a Tubulin:Stathmin-like Domain Complex. Cell 2000, 102, 809–816. [Google Scholar] [CrossRef]
- Yang, C.P.H.; Verdier-Pinard, P.; Wang, F.; Lippaine-Horvath, E.; He, L.; Li, D.; Hofle, G.; Ojima, I.; Orr, G.A.; Horwitz, S.B. A highly epothilone B-resistant A549 cell line with mutations in tubulin that confer drug dependence. Mol. Cancer Ther. 2005, 4, 987–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, K.L.; Anders, K.R.; Nogales, E.; Schwartz, K.; Downing, K.H.; Botstein, D. Structure-function relationships in yeast tubulins. Mol. Biol. Cell 2000, 11, 1887–1903. [Google Scholar] [CrossRef] [PubMed]
- Grigoryan, H.; Lockridge, O. Nanoimages show disruption of tubulin polymerization by chlorpyrifos oxon: Implications for neurotoxicity. Toxicol. Appl. Pharmacol. 2009, 240, 143–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mencarelli, A.; Prontera, P.; Stangoni, G.; Mencaroni, E.; Principi, N.; Esposito, S. Epileptogenic Brain Malformations and Mutations in Tubulin Genes: A Case Report and Review of the Literature. Int. J. Mol. Sci. 2017, 18, 2273. [Google Scholar] [CrossRef] [PubMed]
- Thevenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tuffery, P. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tuffery, P. Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef] [PubMed]
- Lamiable, A.; Thevenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tuffery, P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Fiser, A.; Sali, A. Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol. 2003, 374, 461–491. [Google Scholar] [PubMed]
- Laskowski, R.A.; Macarthur, M.W.; Moss, D.S.; Thornton, J.M. Procheck: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Kurcinski, M.; Jamroz, M.; Blaszczyk, M.; Kolinski, A.; Kmiecik, S. CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 2015, 43, W419–W424. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand-Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.L.; Lin, Y.T.; Ting, C.H.; Lin-Chao, S.; Li, H.; Hsieh-Li, H.M. Stathmin, a microtubule-destabilizing protein, is dysregulated in spinal muscular atrophy. Hum. Mol. Genet. 2010, 19, 1766–1778. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ting, C.-H.; Liu, Y.-C.; Lyu, P.-C.; Chen, J.-Y. Nile Tilapia Derived Antimicrobial Peptide TP4 Exerts Antineoplastic Activity Through Microtubule Disruption. Mar. Drugs 2018, 16, 462. https://doi.org/10.3390/md16120462
Ting C-H, Liu Y-C, Lyu P-C, Chen J-Y. Nile Tilapia Derived Antimicrobial Peptide TP4 Exerts Antineoplastic Activity Through Microtubule Disruption. Marine Drugs. 2018; 16(12):462. https://doi.org/10.3390/md16120462
Chicago/Turabian StyleTing, Chen-Hung, Yi-Chung Liu, Ping-Chiang Lyu, and Jyh-Yih Chen. 2018. "Nile Tilapia Derived Antimicrobial Peptide TP4 Exerts Antineoplastic Activity Through Microtubule Disruption" Marine Drugs 16, no. 12: 462. https://doi.org/10.3390/md16120462
APA StyleTing, C.-H., Liu, Y.-C., Lyu, P.-C., & Chen, J.-Y. (2018). Nile Tilapia Derived Antimicrobial Peptide TP4 Exerts Antineoplastic Activity Through Microtubule Disruption. Marine Drugs, 16(12), 462. https://doi.org/10.3390/md16120462



