Cytotoxic, Anti-Migration, and Anti-Invasion Activities on Breast Cancer Cells of Angucycline Glycosides Isolated from a Marine-Derived Streptomyces sp.
Abstract
:1. Introduction
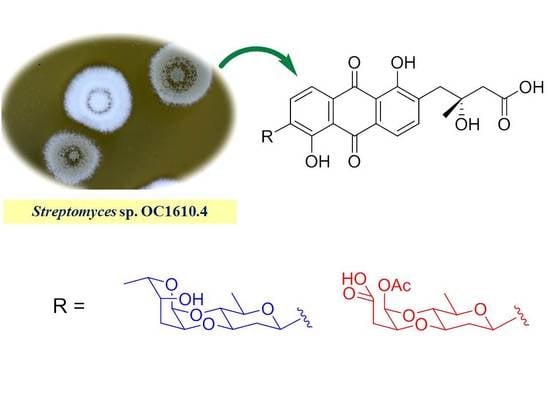
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Actinomycetes Strain
3.3. Fermentation, Extraction, and Isolation
3.4. Cell Culture
3.5. MTT Assay
3.6. Wound-Healing Assay
3.7. Transwell Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rohr, J.; Thiericke, R. Angucycline group antibiotics. Nat. Prod. Rep. 1992, 9, 103–137. [Google Scholar] [CrossRef] [PubMed]
- Kharel, M.K.; Pahari, P.; Shepherd, M.D.; Tibrewal, N.; Nybo, S.E.; Shaaban, K.A.; Rohr, J. Angucyclines: Biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 2012, 29, 264–325. [Google Scholar] [CrossRef]
- He, H.Y.; Ding, W.D.; Bernan, V.S.; Richardson, A.D.; Ireland, C.M.; Greenstein, M.; Ellestad, G.A.; Carter, G.T. Lomaiviticins A and B, potent antitumor antibiotics from Micromonospora lomaivitiensis. J. Am. Chem. Soc. 2001, 123, 5362–5363. [Google Scholar] [CrossRef]
- Zhang, W.J.; Liu, Z.; Li, S.M.; Lu, Y.Z.; Chen, Y.C.; Zhang, H.B.; Zhang, G.T.; Zhu, Y.G.; Zhang, G.Y.; Zhang, W.M.; et al. Fluostatins I–K from the south China sea-derived Micromonospora rosaria SCSIO N160. J. Nat. Prod. 2012, 75, 1937–1943. [Google Scholar] [CrossRef]
- Wu, C.S.; van der Heul, H.U.; Melnik, A.V.; Lubben, J.; Dorrestein, P.C.; Minnaard, A.J.; Choi, Y.H.; van Wezel, G.P. Lugdunomycin, an angucycline-derived molecule with unprecedented chemical architecture. Angew. Chem. Int. Edit. 2019, 58, 2809–2814. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, K.A.; Ahmed, T.A.; Leggas, M.; Rohr, J. Saquayamycins G-K, cytotoxic angucyclines from Streptomyces sp. including two analogues bearing the aminosugar rednose. J. Nat. Prod. 2012, 75, 1383–1392. [Google Scholar] [CrossRef]
- Vicente, J.; Stewart, A.K.; van Wagoner, R.M.; Elliott, E.; Bourdelais, A.J.; Wright, J.L.C. Monacyclinones, new angucyclinone metabolites isolated from Streptomyces sp. M7_15 associated with the Puerto Rican sponge Scopalina ruetzleri. Mar. Drugs 2015, 13, 4682–4700. [Google Scholar] [CrossRef]
- Zhu, X.C.; Duan, Y.W.; Cui, Z.M.; Wang, Z.; Li, Z.X.; Zhang, Y.; Ju, J.H.; Huang, H.B. Cytotoxic rearranged angucycline glycosides from deep sea-derived Streptomyces lusitanus SCSIO LR32. J. Antibiot. 2017, 70, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Korynevska, A.; Heffeter, P.; Matselyukh, B.; Elbling, L.; Micksche, M.; Stoika, R.; Berger, W. Mechanisms underlying the anticancer activities of the angucycline landomycin E. Biochem Pharmacol. 2007, 74, 1713–1726. [Google Scholar] [CrossRef] [PubMed]
- Panchuk, R.R.; Lehka, L.V.; Terenzi, A.; Matselyukh, B.P.; Rohr, J.; Jha, A.K.; Downey, T.; Kril, I.J.; Herbacek, I.; van Schoonhoven, S.; et al. Rapid generation of hydrogen peroxide contributes to the complex cell death induction by the angucycline antibiotic landomycin E. Free Radical Bio. Med. 2017, 106, 134–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, S.R.; Blundon, H.L.; Ladda, M.A.; Robertson, A.W.; Martinez-Farina, C.F.; Jakeman, D.L.; Goralski, K.B. Jadomycin breast cancer cytotoxicity is mediated by a copper-dependent, reactive oxygen species-inducing mechanism. Pharmacol. Res. Perspect. 2015, 3, e00110. [Google Scholar] [CrossRef]
- Hall, S.R.; Toulany, J.; Bennett, L.G.; Martinez-Farina, C.F.; Robertson, A.W.; Jakeman, D.L.; Goralski, K.B. Jadomycins inhibit type II topoisomerases and promote DNA damage and apoptosis in multidrug-resistant triple-negative breast cancer cells. J. Pharmacol. Exp. Ther. 2017, 363, 196–210. [Google Scholar] [CrossRef]
- Woo, C.M.; Beizer, N.E.; Janso, J.E.; Herzon, S.B. The cytotoxicity of (-)-lomaiviticin A arises from induction of double-strand breaks in DNA. Nat. Chem. 2014, 6, 504–510. [Google Scholar]
- Colis, L.C.; Hegan, D.C.; Kaneko, M.; Glazer, P.M.; Herzon, S.B. Mechanism of action studies of lomaiviticin A and the monomeric lomaiviticin aglycon. Selective and potent activity toward DNA double-strand break repair-deficient cell lines. J. Am. Chem. Soc. 2015, 137, 5741–5747. [Google Scholar] [CrossRef]
- Herzon, S.B. The mechanism of action of (-)-lomaiviticin A. Accounts Chem. Res. 2017, 50, 2577–2588. [Google Scholar] [CrossRef]
- Guo, L.; Xie, Z.P.; Yang, Q.; Feng, L.L.; Zhang, L.; Zhang, Y.Z.; Li, X.N.; Pescitelli, G.; Zhang, S.M. Kiamycins B and C, unusual bridged angucyclinones from a marine sediment-derived Streptomyces sp. Tetrahedron Lett. 2018, 59, 2176–2180. [Google Scholar] [CrossRef]
- Jin, J.; Yang, X.Y.; Liu, T.; Xiao, H.; Wang, G.Y.; Zhou, M.J.; Liu, F.W.; Zhang, Y.T.; Liu, D.; Chen, M.H.; et al. Fluostatins M-Q featuring a 6-5-6-6 ring skeleton and high oxidized A-rings from marine Streptomyces sp. PKU-MA00045. Mar. Drugs 2018, 16, 87. [Google Scholar] [CrossRef]
- Hu, Z.J.; Qin, L.L.; Wang, Q.Q.; Ding, W.J.; Chen, Z.; Ma, Z.J. Angucycline antibiotics and its derivatives from marine-derived actinomycete Streptomyces sp. A6H. Nat. Prod. Res. 2016, 30, 2551–2558. [Google Scholar] [CrossRef]
- Yang, L.; Hou, L.K.; Li, H.Y.; Li, W.L. Antibiotic angucycline derivatives from the deepsea-derived Streptomyces lusitanus. Nat. Prod. Res. 2019, 1–7. [Google Scholar] [CrossRef]
- Gui, C.; Liu, Y.N.; Zhou, Z.B.; Zhang, S.W.; Hu, Y.F.; Gu, Y.C.; Huang, H.B.; Ju, J.H. Angucycline glycosides from mangrove-derived Streptomyces diastaticus subsp. SCSIO GJ056. Mar. Drugs 2018, 16, 185. [Google Scholar] [CrossRef]
- Peng, A.H.; Qu, X.Y.; Liu, F.Y.; Li, X.; Li, E.W.; Xie, W.D. Angucycline glycosides from an intertidal sediments strain Streptomyces sp. and their cytotoxic activity against hepatoma carcinoma cells. Mar. Drugs 2018, 16, 470. [Google Scholar] [CrossRef]
- Danishefsky, S.J.; Biing, J.U.; Quallich, G. Total synthesis of vineomycin-B2 aglycon. J. Am. Chem. Soc. 1984, 106, 2453–2455. [Google Scholar] [CrossRef]
- Maskey, R.P.; Helmke, E.; Laatsch, H. Himalomycin A and B: Isolation and structure elucidation of new fridamycin type antibiotics from a marine Streptomyces isolate. J. Antibiot. 2003, 56, 942–949. [Google Scholar] [CrossRef]
- Abdelfattah, M.S.; Kharel, M.K.; Hitron, J.A.; Baig, I.; Rohr, J. Moromycins A and B, isolation and structure elucidation of C-glycosylangucycline-type antibiotics from Streptomyces sp. KY002. J. Nat. Prod. 2008, 71, 1569–1573. [Google Scholar] [CrossRef]
- Uchida, T.; Imoto, M.; Watanabe, Y.; Miura, K.; Dobashi, T.; Matsuda, N.; Sawa, T.; Naganawa, H.; Hamada, M.; Takeuchi, T.; et al. Saquayamycins, new aquayamycin-group antibiotics. J. Antibiot. 1985, 38, 1171–1181. [Google Scholar] [CrossRef]
- Huang, H.B.; Yang, T.T.; Ren, X.M.; Liu, J.; Song, Y.X.; Sun, A.J.; Ma, J.Y.; Wang, B.; Zhang, Y.; Huang, C.G.; et al. Cytotoxic angucycline class glycosides from the deep sea actinomycete Streptomyces lusitanus SCSIO LR32. J. Nat. Prod. 2012, 75, 202–208. [Google Scholar] [CrossRef]
- Kawashima, A.; Yoshimura, Y.; Goto, J.; Nakaike, S.; Mizutani, T.; Hanada, K.; Omura, S. Pi-083, a new platelet-aggregation inhibitor. J. Antibiot. 1988, 41, 1913–1914. [Google Scholar] [CrossRef]
- Lafourcade, A.; His, M.; Baglietto, L.; Boutron-Ruault, M.C.; Dossus, L.; Rondeau, V. Factors associated with breast cancer recurrences or mortality and dynamic prediction of death using history of cancer recurrences: The French E3N cohort. Bmc Cancer 2018, 18, 171. [Google Scholar] [CrossRef]
- Narod, S.A.; Sopik, V. Is invasion a necessary step for metastases in breast cancer? Breast Cancer Res. Tr. 2018, 169, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Zheng, B.B.; Wu, L.H.; Ma, L.S.; Liu, S.S.; Li, L.; Xie, W.D.; Li, X. Telekin induces apoptosis associated with the mitochondria-mediated pathway in human hepatocellular carcinoma cells. Biol. Pharm. Bull. 2013, 36, 1118–1125. [Google Scholar] [CrossRef]
- Ma, J.H.; Qi, J.; Liu, FY.; Lin, S.Q.; Zhang, C.Y.; Xie, W.D.; Zhang, H.Y.; Li, X. Ivalin inhibits proliferation, migration and invasion by suppressing epithelial mesenchymal transition in breast cancer cells. Nutr. Cancer 2018, 70, 1330–1338. [Google Scholar] [CrossRef] [PubMed]




| No. | 1 b | 2 c | ||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1 | 176.4 s | - | 173.8 s | - |
| 2 | 46.7 t | 2.48, d (15.5) 2.51, d (15.5) | 45.5 t | 2.56, d (15.2) 2.59, d (15.2) |
| 3 | 73.0 s | - | 72.1 s | - |
| 4 | 41.2 t | 3.01, d (13.1) 3.08, d (13.1) | 40.8 t | 3.10, brs |
| 4a | 136.3 s | - | 136.2 s | - |
| 5 | 140.8 d | 7.70, mc | 140.8 | 7.85, d (5.6) |
| 6 | 119.6 d | 7.68, mc | 119.3 d | 7.78, d (5.6) |
| 6a | 132.9 s | - | 132.5 s | - |
| 7 | 189.2 s | - | 189.1 s | - |
| 7a | 116.6 s | - | 116.3 s | - |
| 8 | 159.7 s | - | 159.6 s | - |
| 9 | 139.2 s | - | 138.8 s | - |
| 10 | 134.4 d | 7.84, d (7.3) | 134.2 d | 7.96, d (6.6) |
| 11 | 120.1 d | 7.72, d (7.3) | 119.8 d | 7.84, d (6.6) |
| 11a | 133.3 s | - | 133.0 s | - |
| 12 | 189.3 s | - | 189.2 s | - |
| 12a | 116.6 s | - | 116.4 s | - |
| 12b | 162.4 s | - | 162.3 s | - |
| 13 | 27.1 q | 1.27, s | 27.3 q | 1.31, s |
| Sugar A, β-d-olivose | ||||
| 1A | 72.7 d | 4.92, brd (11.4) | 72.2 d | 5.03, brd (9.1) |
| 2A | 37.9 t | 1.50, m2.38, m | 37.3 t | 1.56, m2.43, m |
| 3A | 78.0 d | 3.68, m | 77.5 d | 3.89, m |
| 4A | 75.1 d | 3.47, dd (9.3, 9.3) | 75.3 d | 3.46, dd (7.6, 7.8) |
| 5A | 75.6 d | 3.56, m | 74.6 d | 3.67, m |
| 6A | 17.9 q | 1.31, d (5.8) | 17.6 q | 1.26, d (5.6) |
| Sugar B | ||||
| 1B | 91.0 s | 4.97, brs | 90.5 d | 6.06, brs |
| 2B | 73.7 d | 3.88, m | 74.4 d | 4.34, m |
| 3B | 32.6 t | 1.92, m1.96, m | 35.9 d | 2.54, m |
| 4B | 64.1 d | 4.12, m | 171.4 s | - |
| 5B | 74.7 d | 4.24, m | 169.8 s | - |
| 6B | 11.5 q | 1.26, d (6.9) | 20.8 q | 2.17, s |
| Compounds | Cell lines | ||
|---|---|---|---|
| MCF-7 | MDA-MB-231 | BT-474 | |
| 1 | 6.07 ± 0.09 | 7.72 ± 0.76 | 4.27 ± 2.09 |
| 2 | >20 | >20 | >20 |
| 3 | >20 | >20 | >20 |
| 4 | >20 | >20 | >20 |
| 5 | 7.58 ± 1.19 | 8.01 ± 0.55 | 6.46 ± 1.92 |
| 6 | >20 | >20 | >20 |
| 7 | 0.42 ± 0.03 | 0.35 ± 0.03 | 0.67 ± 0.09 |
| 8 | 0.24 ± 0.01 | 0.16 ± 0.02 | 0.28 ± 0.09 |
| 9 | 0.40 ± 0.01 | 0.38 ± 0.04 | 0.41 ± 0.15 |
| 10 | >20 | >20 | >20 |
| 11 | >20 | >20 | >20 |
| Doxorubicin | 0.86 ± 0.64 | 1.30 ± 0.25 | 0.39 ± 0.06 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, X.-Y.; Ren, J.-W.; Peng, A.-H.; Lin, S.-Q.; Lu, D.-D.; Du, Q.-Q.; Liu, L.; Li, X.; Li, E.-W.; Xie, W.-D. Cytotoxic, Anti-Migration, and Anti-Invasion Activities on Breast Cancer Cells of Angucycline Glycosides Isolated from a Marine-Derived Streptomyces sp. Mar. Drugs 2019, 17, 277. https://doi.org/10.3390/md17050277
Qu X-Y, Ren J-W, Peng A-H, Lin S-Q, Lu D-D, Du Q-Q, Liu L, Li X, Li E-W, Xie W-D. Cytotoxic, Anti-Migration, and Anti-Invasion Activities on Breast Cancer Cells of Angucycline Glycosides Isolated from a Marine-Derived Streptomyces sp. Marine Drugs. 2019; 17(5):277. https://doi.org/10.3390/md17050277
Chicago/Turabian StyleQu, Xin-Ying, Jin-Wei Ren, Ai-Hong Peng, Shi-Qi Lin, Dan-Dan Lu, Qian-Qian Du, Ling Liu, Xia Li, Er-Wei Li, and Wei-Dong Xie. 2019. "Cytotoxic, Anti-Migration, and Anti-Invasion Activities on Breast Cancer Cells of Angucycline Glycosides Isolated from a Marine-Derived Streptomyces sp." Marine Drugs 17, no. 5: 277. https://doi.org/10.3390/md17050277
APA StyleQu, X. -Y., Ren, J. -W., Peng, A. -H., Lin, S. -Q., Lu, D. -D., Du, Q. -Q., Liu, L., Li, X., Li, E. -W., & Xie, W. -D. (2019). Cytotoxic, Anti-Migration, and Anti-Invasion Activities on Breast Cancer Cells of Angucycline Glycosides Isolated from a Marine-Derived Streptomyces sp. Marine Drugs, 17(5), 277. https://doi.org/10.3390/md17050277