Rational Design of Alginate Lyase from Microbulbifer sp. Q7 to Improve Thermal Stability
Abstract
:1. Introduction
2. Results
2.1. Selection of Potential Disulfide Bonds in cAlyM
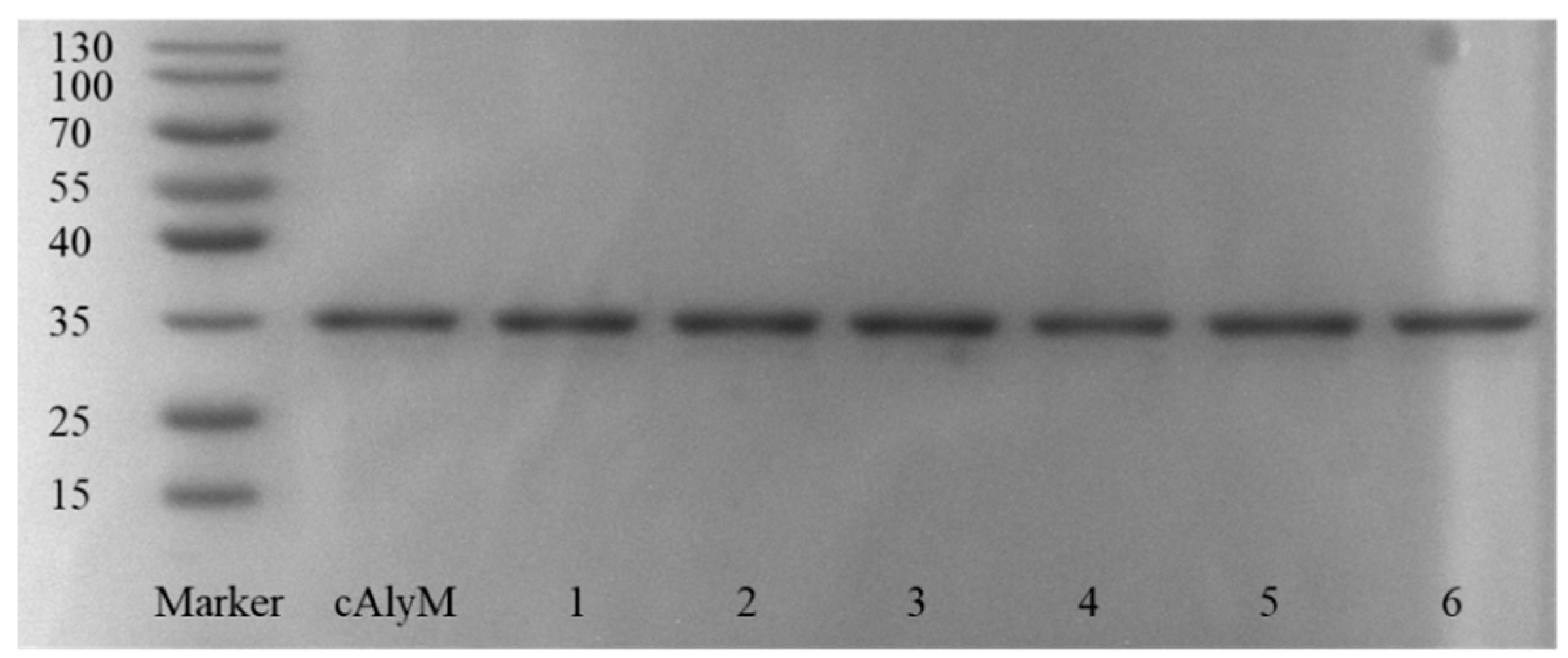
2.2. Determination of Disulfide Bonds of Enzymes
2.3. Enzymatic Properties of the Enzymes
2.4. Analysis of the Molecular Structure of cAlyM Mutants
3. Discussion
4. Materials and Methods
4.1. Strains, Media, and Chemicals
4.2. Computational Analysis of Enzymes
4.3. Construction, Expression, and Purification of cAlyM and Its Mutants
4.4. Determination of Disulfide Bonds Formation
4.5. Enzyme Activity Assays
4.6. Properties of cAlyM and Its Mutant
4.7. Circular Dichroism (CD) Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chen, X.L.; Sheng, D.; Fei, X.; Fang, D.; Li, P.Y.; Zhang, X.Y.; Zhou, B.C.; Zhang, Y.Z.; Xie, B.B. Characterization of a new cold–adapted and salt–activated polysaccharide lyase family 7 alginate lyase from pseudoalteromonas sp. SM0524. Front. Microbiol. 2016, 7, e30105. [Google Scholar] [CrossRef] [PubMed]
- Preiss, J.; Ashwell, G. Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosac–charides and 4–deoxy–l–erythro–5–hexoseulose uronic acid. J. Biol. Chem. 1962, 237, 309–316. [Google Scholar] [PubMed]
- Chen, J.; Hu, Y.; Zhang, L.; Wang, Y.; Wang, S.; Zhang, Y.; Guo, H.; Ji, D.; Wang, Y. Alginate oligosaccharide DP5 exhibits antitumor effects in osteosarcoma patients following surgery. Front. Pharmacol. 2017, 8, 623. [Google Scholar] [CrossRef] [PubMed]
- Wargacki, A.J.; Leonard, E.; Win, M.N.; Regitsky, D.D.; Santos, C.N.S.; Kim, P.B.; Cooper, S.R.; Raisner, R.M.; Herman, A.; Sivitz, A.B.; et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 2012, 335, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Shi, X.; Gao, Y.; Cai, N.; Jiang, Z.; Xu, X. Anti-inflammatory activity of guluronate oligosaccharides obtained by oxidative degradation from alginate in lipopolysaccharide-activated murine macrophage RAW 264.7 cells. J. Agric. Food Chem. 2015, 63, 160–168. [Google Scholar] [CrossRef]
- Miyake, O.; Moriwaki, S.; Murata, K.; Yamasaki, M.; Mikami, B.; Hashimoto, W. Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL7: A novel alginate lyase with a beta–sandwich fold. J. Biol. Chem. 2004, 279, 31863–31872. [Google Scholar]
- Rahman, M.M.; Wang, L.; Inoue, A.; Ojima, T. cDNA cloning and bacterial expression of a PL–14 alginate lyase from a herbivorous marine snail littorina brevicula. Carbohydr. Res. 2012, 360, 69–77. [Google Scholar] [CrossRef]
- Zhu, B.; Tan, H.; Qin, Y.; Xu, Q.; Du, Y.; Yin, H. Characterization of a new endo-type alginate lyase from vibrio sp. W13. Int. J. Biol. Macromol. 2015, 75, 330–337. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, L.; Jia, L.; Gui, S.; Fu, Y.; Zheng, D.; Guo, W.; Lu, F.P. Improvement of the acid stability of Bacillus licheniformis alpha amylase by site–directed mutagenesis. Process Biochem. 2017, 58, 174–180. [Google Scholar] [CrossRef]
- Couturier, M.; Féliu, J.; Bozonnet, S.; Roussel, A.; Berrin, J.G. Molecular engineering of fungal GH5 and GH26 beta-(1,4)-mannanases toward improvement of enzyme activity. PLoS ONE 2013, 8, e79800. [Google Scholar] [CrossRef]
- Rha, E.; Kim, S.; Choi, S.L.; Hong, S.P.; Sung, M.H.; Song, J.J.; Lee, S.G. Simultaneous improvement of catalytic activity and thermal stability of tyrosine phenol–lyase by directed evolution. FEBS J. 2010, 276, 6187–6194. [Google Scholar] [CrossRef]
- Wu, X.; Tian, Z.; Jiang, X.; Zhang, Q.; Wang, L. Enhancement in catalytic activity of Aspergillus niger XynB by selective site-directed mutagenesis of active site amino acids. Appl. Microbiol. Biotechnol. 2017, 102, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.H.; Piao, Y.L.; Huang, X.; Yoon, E.J.; Park, S.H.; Lee, K.; Zhan, C.G.; Cho, H. Modeling and re–engineering of Azotobacter vinelandii alginate lyase to enhance its catalytic efficiency for accelerating biofilm degradation. PLoS ONE 2016, 11, e0156197. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Xu, T.; Selvaraj, J.N.; Yi, L.; Zhang, G. Improving the specific activity and thermo–stability of alkaline pectate lyase from Bacillus subtilis 168 for bioscouring. Biochem. Eng. J. 2018, 129, 74–83. [Google Scholar] [CrossRef]
- Wu, T.H.; Chen, C.C.; Cheng, Y.S.; Ko, T.P.; Lin, C.Y.; Lai, H.L.; Huang, T.Y.; Liu, J.R.; Guo, R.T. Improving specific activity and thermostability of Escherichia coli phytase by structure–based rational design. J. Biotechnol. 2014, 175, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Zhang, K.; Liu, X.; Liu, W.; Lyu, Q.; Ji, A. Characterization of a novel polyM-preferred alginate lyase from marine Vibrio splendidus OU02. Mar. Drugs 2018, 16, 295. [Google Scholar] [CrossRef] [PubMed]
- Mansfeld, J.; Vriend, G.; Dijkstra, B.W.; Veltman, O.R.; Van den Burg, B.; Venema, G.; Ulbrich-Hofmann, R.; Eijsink, V.G. Extreme stabilization of a thermolysin-like protease by an engineered disulfide bond. J. Biol. Chem. 1997, 272, 11152–11156. [Google Scholar] [CrossRef]
- Liu, T.; Wang, Y.; Luo, X.; Li, J.; Reed, S.A.; Xiao, H.; Young, T.S.; Schultz, P.G. Enhancing protein stability with extended disulfide bonds. Proc. Nati. Acad. Sci. USA 2016, 113, 5910. [Google Scholar] [CrossRef]
- Le, Q.A.T.; Joo, J.C.; Yoo, Y.J.; Yong, H.K. Development of thermostable Candida antarctica lipase B through novel in silico design of disulfide bridge. Biotechnol. Bioeng. 2012, 109, 867–876. [Google Scholar] [CrossRef]
- Tan, H.; Miao, R.; Liu, T.; Cao, X.; Wu, X.; Xie, L.; Huang, Z.; Peng, W.; Gan, B. Enhancing thermal resistance of a novel acidobacteria–derived phytase by engineering of disulfide bridges. J. Microbiol. Biotechnol. 2016, 26, 1717–1722. [Google Scholar] [CrossRef]
- Liu, L.; Deng, Z.; Yang, H.; Li, J.; Shin, H.D.; Chen, R.R.; Chen, J. In silico rational design and systems engineering of disulfide bridges in the catalytic domain of an alkaline α–amylase from Alkalimonas amylolytica to improve thermostability. Appl. Environ. Microbiol. 2014, 80, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, N.N.; Yang, S.X.; Yu, Y.; Han, Z.L.; Li, L.; Mou, H.J. Study on expression and action mode of recombinant alginate lyases based on conserved domains reconstruction. Appl. Microbiol. Biotechnol. 2018, 103, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef]
- Miller, G. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Niu, C.; Zhu, L.; Xu, X.; Li, Q. Rational design of disulfide bonds increases thermostability of a mesophilic 1,3-1,4-β-glucanase from Bacillus terquilensis. PLoS ONE 2016, 11, e0154036. [Google Scholar] [CrossRef]
- Yang, M.; Yu, Y.; Yang, S.X.; Shi, X.H.; Mou, H.J.; Li, L. Expression and characterization of a new polyG-specific alginate lyase from marine bacterium Microbulbifer sp. Q7. Front. Microbiol. 2018, 9, 2894. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Inoue, A.; Mashino, C.; Uji, T.; Saga, N.; Mikami, K.; Ojima, T. Characterization of an eukaryotic PL7 alginate lyase in the marine red alga Pyropia yezoensis. Curr. Biotechnol. 2015, 4, 240–248. [Google Scholar] [CrossRef]
- Yagi, H.; Fujise, A.; Itabashi, N.; Ohshiro, T. Purification and characterization of a novel alginate lyase from the marine bacterium Cobetia sp. NAP1 isolated from brown algae. Biosci. Biotechno. Biochem. 2016, 80, 2338–2346. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Zhang, Y.; Chen, L. High–level expression of a thermally stable alginate lyase using pichia pastoris, characterization and application in producing brown alginate oligosaccharide. Mar. Drugs 2018, 16, 158. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Chang, Y.; Shen, J. Cloning, expression and characterization of an endo-acting bifunctional alginate lyase of marine bacterium Wenyingzhuangia fucanilytica. Protein Express. Purify. 2019, 154, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, L.; Chen, X.; Zhao, W.; Sun, M.; Han, Y. Cloning, expression, and biochemical characterization of two new oligoalginate lyases with synergistic degradation capability. Mar. Biotechnol. 2018, 20, 75–86. [Google Scholar] [CrossRef]
- Wang, L.; Li, S.; Yu, W.; Gong, Q. Cloning, overexpression and characterization of a new oligoalginate lyase from a marine bacterium Shewanella sp. Biotechnol. Lett. 2015, 37, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tsai, C.J.; Ma, B.; Nussinov, R. Contribution of salt bridges toward protein thermostability. J. Biomol. Struct. Dyn. 2000, 17, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Mamonova, T.B.; Glyakina, A.V.; Galzitskaya, O.V.; Kurnikova, M.G. Stability and rigidity/flexibility-two sides of the same coin? Biochim. Biophys. Acta 2013, 1834, 854–866. [Google Scholar] [CrossRef]
- Li, P.Y.; Chen, X.L.; Ji, P.; Li, C.Y.; Wang, P.; Zhang, Y.; Xie, B.B.; Qin, Q.L.; Su, H.N.; Zhou, B.C.; et al. Interdomain hydrophobic interactions modulate the thermostability of microbial esterases from the hormone–sensitive lipase family. J. Biol. Chem. 2015, 290, 11188–11198. [Google Scholar] [CrossRef]
- Badieyan, S.; Bevan, D.R.; Zhang, C. Study and design of stability in GH5 cellulases. Biotechnol. Bioeng. 2012, 109, 31–44. [Google Scholar] [CrossRef]
- Goedegebuur, F.; Dankmeyer, L.; Gualfetti, P.; Karkehabadi, S.; Hansson, H.; Jana, S.; Huynh, V.; Kelemen, B.R.; Kruithof, P.; Larenas, E.A.; et al. Improving the thermal stability of cellobiohydrolase Cel7A from Hypocrea jecorina by directed evolution. J. Biol. Chem. 2017, 292, 17418–17430. [Google Scholar] [CrossRef]
- Qin, H.M.; Miyakawa, T.; Inoue, A.; Nishiyama, R.; Nakamura, A.; Asano, A.; Ojima, T.; Tanokura, M. Structural basis for controlling the enzymatic properties of polymannuronate preferred alginate lyase FlAlyA from the PL-7 family. Chem. Commun. 2018, 54, 555–558. [Google Scholar] [CrossRef] [Green Version]
- Inoue, A.; Anraku, M.; Nakagawa, S.; Ojima, T. Discovery of a novel alginate lyase from Nitratiruptor sp. SB155-2 thriving at deep-sea hydrothermal vents and identification of the residues responsible for its heat stability. J. Biol. Chem. 2016, 291, 5551–15563. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, M.; Ogura, K.; Hashimoto, W. A structural basis for depolymerization of alginate by polysaccharide lyase family-7. J. Mol. Biol. 2005, 352, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.T.N.; Jonson, P.H.; Petersen, S.B. Amino acid neighbours and detailed conformational analysis of cysteines in proteins. Protein Eng. 1999, 12, 535–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, X.; Zhang, Y.; Yuan, T. Enhanced thermostability of glucose oxidase through computer-aided molecular design. Int. J. Mol. Sci. 2018, 19, 425. [Google Scholar] [CrossRef] [PubMed]
- Radestock, S.; Gohlke, H. Protein rigidity and thermophilic adaptation. Proteins 2015, 79, 1089–1108. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Huang, H. Engineering proteins for thermostability through rigidifying flexible sites. Biotechnol. Adv. 2014, 32, 308–315. [Google Scholar] [CrossRef]




| Mutants | Average Overall RMSD | Mutation Sites RMSF | ||||
|---|---|---|---|---|---|---|
| cAlyM | Mutants | RMSD (cAlyM -Mutants) | cAlyM | Mutants | RMSF (cAlyM -Mutants) | |
| V59C-Y86C | 1.022 | 0.925 | 0.097 | 0.965 | 0.921 | 0.044 |
| D102C-A300C | 1.004 | 0.018 | 1.277 | 1.204 | 0.073 | |
| G103C-T113C | 0.996 | 0.026 | 0.868 | 0.994 | −0.126 | |
| R122C-N136C | 1.003 | 0.019 | 0.921 | 0.869 | 0.052 | |
| S173C-S229C | 0.989 | 0.033 | 1.772 | 1.201 | 0.571 | |
| V207C-I224C | 0.962 | 0.060 | 0.856 | 0.823 | 0.033 | |
| Enzymes | Km (mg/mL) | Vmax (U/mg) | Kcat (s−1) | Kcat/Km (mL/s/mg) | Tm (°C) |
|---|---|---|---|---|---|
| cAlyM | 0.37 ± 0.09 | 1386.3 ± 23.5 | 762.4 ± 19.6 | 2060.7 | 57.2 |
| V59C-Y86C | 0.95 ± 0.21 | 1166.9 ± 19.2 | 641.8 ± 18.5 | 675.6 | 54.3 |
| D102C-A300C | 0.28 ± 0.04 | 1567.6 ± 52.4 | 862.2 ± 30.3 | 3079.2 | 58.9 |
| G103C-T113C | 1.26 ± 0.32 | 754.7 ± 16.8 | 415.1 ± 15.5 | 329.4 | 57.6 |
| R122C-N136C | 1.94 ± 0.28 | 521.6 ± 17.2 | 286.9 ± 14.4 | 147.9 | 55.6 |
| S173C-S229C | 0.31 ± 0.08 | 1455.1 ± 42.8 | 800.33 ± 32.3 | 2581.7 | 55.4 |
| V207C-I224C | 0.21 ± 0.07 | 1675.4 ± 37.2 | 921.5 ± 28.6 | 4387.9 | 55.4 |
| Mutants | cAlyM | V59C-Y86C | D102C-A300C | G103C-T113C | R122C-N136C | S173C-S229C | V207C-I224C |
|---|---|---|---|---|---|---|---|
| H-bond | 281 | 282 | 281 | 280 | 275 | 281 | 281 |
| Salt-bond | 26 | 26 | 25 | 26 | 24 | 26 | 26 |
| Hydrophobic interaction | 120 | 117 | 120 | 126 | 121 | 120 | 118 |
| Primer Name | Sequence (5’-3’) |
|---|---|
| V59C-F | GGTACCTGAGCTGTCCTACCGACAAC |
| V59C-R | GTTGTCGGTAGGACAGCTCAGGTACC |
| Y86C-F | CAGATGGCACCTGCTTCTATACTGCTG |
| Y86C-R | CAGCAGTATAGAAGCAGGTGCCATCTG |
| D102C-F | GCTGCCCGATCTGTGGCTATAAAAC |
| D102C-R | GTTTTATAGCCACAGATCGGGCAGC |
| A300C-F | CACCGGCAATTGCAGTGACTATGTC |
| A300C-R | GACATAGTCACTGCAATTGCCGGTG |
| G103C-F | GCCCGATCGATTGCTATAAAACATCG |
| G103C-R | CGATGTTTTATAGCAATCGATCGGGC |
| T113C-F | CACGTCCTATTGCCGCACCGAGCTG |
| T113C-R | CAGCTCGGTGCGGCAATAGGACGTG |
| R122C-F | CGCGAGATGCTATGTCGTGGCGACACC |
| R122C-R | GGTGTCGCCACGACATAGCATCTCGCG |
| N136C-F | GGGTCAATGGATGCAACTGGGTATTCG |
| N136C-R | CGAATACCCAGTTGCATCCATTGACCC |
| S173C-F | CTACCGGAGATTGCGGCCAGGTTGGAC |
| S173C-R | GTCCAACCTGGCCGCAATCTCCGGTAG |
| S229C-F | GGCAGCCGTTCCTGCAGCGCCTCGGAC |
| S229C-R | GTCCGAGGCGCTGCAGGAACGGCTGC |
| V207C-F | GCAAAGGTTCTTGCTATATCGCCCATG |
| V207C-R | CATGGGCGATATAGCAAGAACCTTTGC |
| I224C-F | GGTACGACATGTGTGGCAGCCGTTCC |
| I224C-R | GGAACGGCTGCCACACATGTCGTACC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Yang, S.-X.; Liu, Z.-M.; Li, N.-N.; Li, L.; Mou, H.-J. Rational Design of Alginate Lyase from Microbulbifer sp. Q7 to Improve Thermal Stability. Mar. Drugs 2019, 17, 378. https://doi.org/10.3390/md17060378
Yang M, Yang S-X, Liu Z-M, Li N-N, Li L, Mou H-J. Rational Design of Alginate Lyase from Microbulbifer sp. Q7 to Improve Thermal Stability. Marine Drugs. 2019; 17(6):378. https://doi.org/10.3390/md17060378
Chicago/Turabian StyleYang, Min, Su-Xiao Yang, Zhe-Min Liu, Nan-Nan Li, Li Li, and Hai-Jin Mou. 2019. "Rational Design of Alginate Lyase from Microbulbifer sp. Q7 to Improve Thermal Stability" Marine Drugs 17, no. 6: 378. https://doi.org/10.3390/md17060378
APA StyleYang, M., Yang, S.-X., Liu, Z.-M., Li, N.-N., Li, L., & Mou, H.-J. (2019). Rational Design of Alginate Lyase from Microbulbifer sp. Q7 to Improve Thermal Stability. Marine Drugs, 17(6), 378. https://doi.org/10.3390/md17060378





