Two Highly Similar Chitinases from Marine Vibrio Species have Different Enzymatic Properties
Abstract
:1. Introduction
2. Results
2.1. Amino Acid Sequences Analysis of Chi1557 and Chi4668
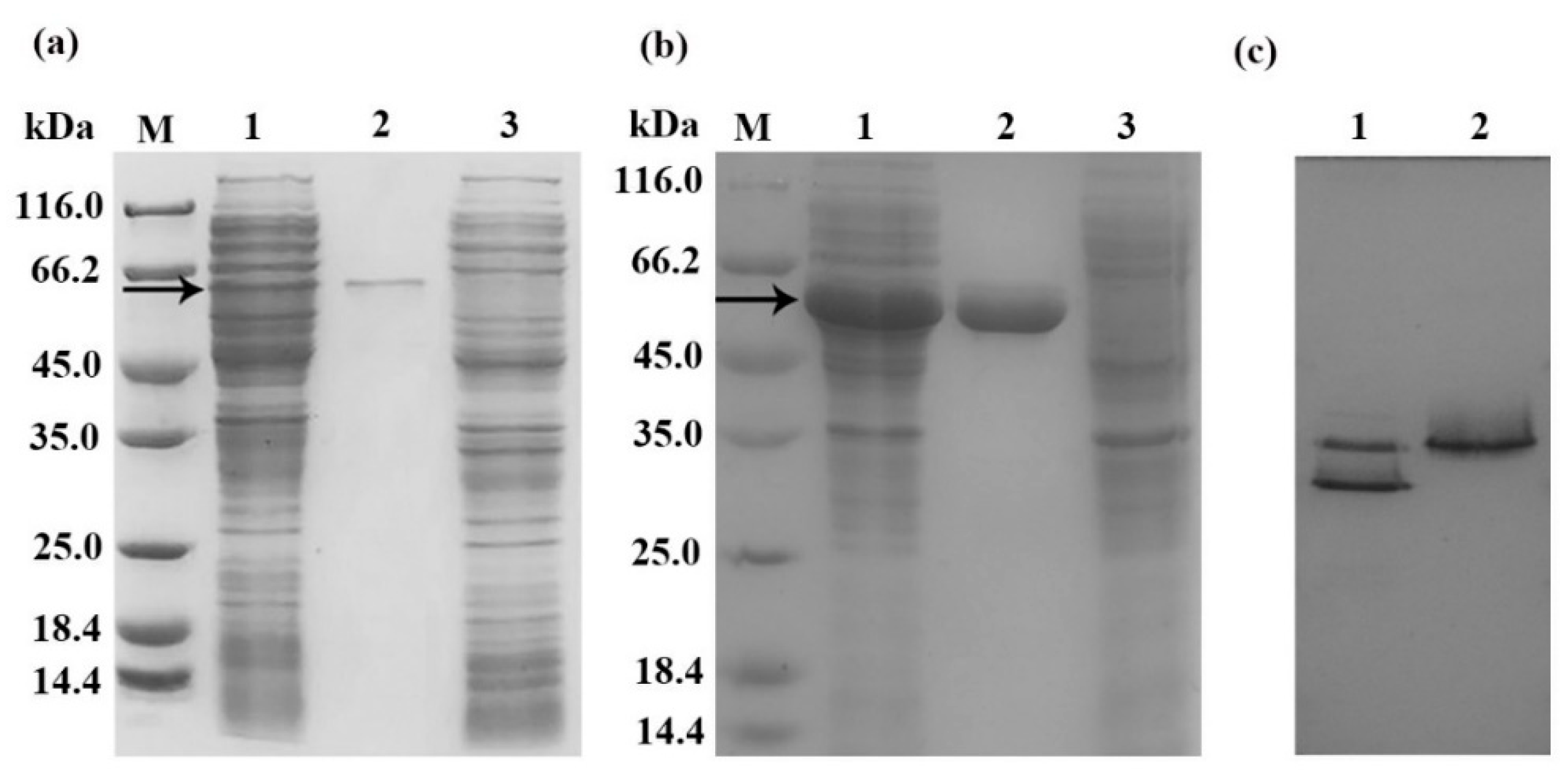
2.2. Expression, Purification, and Activity Detection of Recombinant Chitinases
2.3. The Activity and Stability of Recombinant Chitinases for Temperature and pH
2.4. Metal Ions and Reductants on the Activity of Recombinant Chitinases
2.5. The Kinetic Parameters and Hydrolysis Property of Recombinant Chitinases
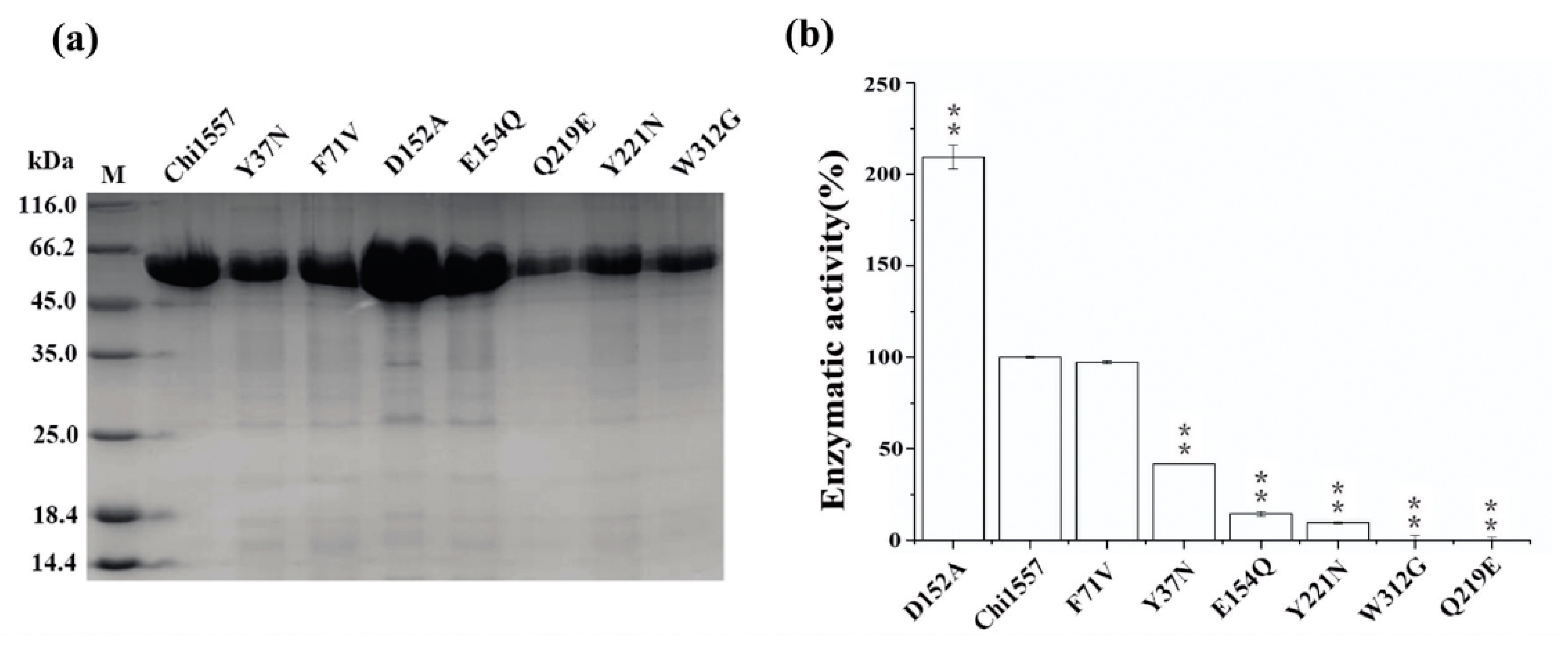
2.6. Site-Directed Mutagenesis of Chi1557
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Media and Growth Conditions
4.2. Sequence Analysis of Chitinase Genes
4.3. Preparation of Chitin Colloids
4.4. Expression and Purification of Recombinant Chitinases
4.5. Chitinase Activity Assay and Protein Quantification
4.6. Characterization of Purified Chitinases
4.7. Kinetic Parameters and Hydrolytic Properties of Chitinases
4.8. Site-Directed Mutagenesis of Chi1557
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Hamed, I.; Özogul, F.; Regenstein, J.M. Industrial applications of crustacean by-products (chitin, chitosan, and chitooligosaccharides): A review. Trends Food Sci. Technol. 2016, 48, 40–50. [Google Scholar] [CrossRef]
- Beier, S.; Bertilsson, S. Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front Microbiol. 2013, 4, 149. [Google Scholar] [CrossRef] [Green Version]
- Kirchman, D.L.; White, J. Hydrolysis and mineralization of chitin in the Delaware Estuary. Aquat. Microb. Ecol. 1999, 18, 187–196. [Google Scholar] [CrossRef]
- Howard, M.B.; Ekborg, N.A.; Weiner, R.M.; Hutcheson, S.W. Detection and characterization of chitinases and other chitin-modifying enzymes. J. Ind. Microbiol. Biotechnol. 2003, 30, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.P.; Almeida, B.C.; Colwell, R.R.; Rivera, I.N. The importance of chitin in the marine environment. Mar. Biotechnol. 2011, 13, 823. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, N.; Tewari, R.; Hoondal, G.S. Biotechnological aspects of chitinolytic enzymes: A review. Appl. Microbiol. Biotechnol. 2006, 71, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Karthik, N.; Binod, P.; Pandey, A. Purification and characterisation of an acidic and antifungal chitinase produced by a Streptomyces sp. Bioresour. Technol. 2015, 188, 195–201. [Google Scholar] [CrossRef]
- Stoykov, Y.M.; Pavlov, A.I.; Krastanov, A.I. Chitinase biotechnology: Production, purification, and application. Eng. Life Sci. 2015, 15, 30–38. [Google Scholar] [CrossRef]
- Tarentino, A.L.; Maley, F. Purification and properties of an endo-β-N-acetylglucosaminidase from Streptomyces griseus. J. Biol. Chem. 1974, 249, 811–817. [Google Scholar]
- Honda, Y.; Taniguchi, H.; Kitaoka, M. A reducing-end-acting chitinase from Vibrio proteolyticus belonging to glycoside hydrolase family 19. Appl. Microbiol. Biotechnol. 2008, 78, 627–634. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Horn, S.J.; Sørlie, M.; Eijsink, V.G. The chitinolytic machinery of Serratia marcescens-a model system for enzymatic degradation of recalcitrant polysaccharides. FEBS J. 2013, 280, 3028–3049. [Google Scholar] [CrossRef]
- Toratani, T.; Kezuka, Y.; Nonaka, T.; Hiragi, Y.; Watanabe, T. Structure of full-length bacterial chitinase containing two fibronectin type III domains revealed by small angle X-ray scattering. Biochem. Biophys. Res. Commun. 2006, 348, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Ishibashi, A.; Ariga, Y.; Hashimoto, M.; Nikaidou, N.; Sugiyama, J.; Matsumoto, T.; Nonaka, T. Trp122 and Trp134 on the surface of the catalytic domain are essential for crystalline chitin hydrolysis by Bacillus circulans chitinase A1. FEBS Lett. 2001, 494, 74–78. [Google Scholar] [CrossRef] [Green Version]
- Synstad, B.; Gåseidnes, S.; Van Aalten, D.M.; Vriend, G.; Nielsen, J.E.; Eijsink, V.G. Mutational and computational analysis of the role of conserved residues in the active site of a family 18 chitinase. Eur. J. Biochem. 2004, 271, 253–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madhuprakash, J.; El Gueddari, N.E.; Moerschbacher, B.M.; Podile, A.R. Catalytic efficiency of chitinase-D on insoluble chitinous substrates was improved by fusing auxiliary domains. PLoS ONE 2015, 10, e0116823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Fong, S.S. Bacterial chitinase: Nature and perspectives for sustainable bioproduction. Bioresour. Bioprocess. 2015, 2, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Hamid, R.; Khan, M.A.; Ahmad, M.; Ahmad, M.M.; Abdin, M.Z.; Musarrat, J.; Javed, S. Chitinases: An update. J. Pharm. BioAllied Sci. 2013, 5, 21. [Google Scholar]
- Shin-Hye, P. Purification and characterization of chitinase from a marine bacterium, Vibrio sp. 98CJ11027. J Microbiol. 2000, 38, 224–229. [Google Scholar]
- Annamalai, N.; Rajeswari, M.V.; Vijayalakshmi, S.; Balasubramanian, T. Purification and characterization of chitinase from Alcaligenes faecalis AU02 by utilizing marine wastes and its antioxidant activity. Ann. Microbiol. 2011, 61, 801–807. [Google Scholar] [CrossRef] [Green Version]
- Mander, P.; Cho, S.S.; Choi, Y.H.; Panthi, S.; Choi, Y.S.; Kim, H.M.; Yoo, J.C. Purification and characterization of chitinase showing antifungal and biodegradation properties obtained from Streptomyces anulatus CS242. Arch. Pharmacal. Res. 2016, 39, 878–886. [Google Scholar] [CrossRef]
- Itoh, T.; Sugimoto, I.; Hibi, T.; Suzuki, F.; Matsuo, K.; Fujii, Y.; Taketo, A.; Kimoto, H. Overexpression, purification, and characterization of Paenibacillus cell surface-expressed chitinase ChiW with two catalytic domains. Biosci. Biotechnol. Biochem. 2014, 78, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Hunt, D.E.; Gevers, D.; Vahora, N.M.; Polz, M.F. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol. 2008, 74, 44–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meibom, K.L.; Li, X.B.; Nielsen, A.T.; Wu, C.-Y.; Roseman, S.; Schoolnik, G.K. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 2004, 101, 2524–2529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Yu, M.; Wang, X.; Zhang, X.-H. Comparative genomic analysis reveals the evolution and environmental adaptation strategies of Vibrios. BMC Genomics 2018, 19, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, A.; Fujii, T.; Yoneyama, T.; REDENBACH, M.; OHNO, T.; WATANABE, T.; MIYASHITA, K. High-multiplicity of chitinase genes in Streptomyces coelicolor A3 (2). Biosci. Biotechnol. Biochem. 1999, 63, 710–718. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; SuGAwARA, N.; Suzuki, M.; Uchiyama, T.; Katouno, F.; Nikaidou, N.; Watanabe, T. Chitinases A, B, and C1 of Serratia marcescens 2170 produced by recombinant Escherichia coli: Enzymatic properties and synergism on chitin degradation. Biosci. Biotechnol. Biochem. 2002, 66, 1075–1083. [Google Scholar] [CrossRef]
- Suzuki, K.; Taiyoji, M.; Sugawara, N.; Nikaidou, N.; Henrissat, B.; Watanabe, T. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 1999, 343, 587–596. [Google Scholar] [CrossRef]
- Svitil, A.L.; Chadhain, S.; Moore, J.A.; Kirchman, D.L. Chitin Degradation Proteins Produced by the Marine Bacterium Vibrio harveyi Growing on Different Forms of Chitin. Appl. Environ. Microbiol. 1997, 63, 408–413. [Google Scholar] [CrossRef] [Green Version]
- Malecki, P.H.; Raczynska, J.E.; Vorgias, C.E.; Rypniewski, W. Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 821–829. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Bendt, A.; Hüller, H.; Kammel, U.; Helmke, E.; Schweder, T. Cloning, expression, and characterization of a chitinase gene from the Antarctic psychrotolerant bacterium Vibrio sp. strain Fi: 7. Extremophiles 2001, 5, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Keyhani, N.O.; Roseman, S. The chitin catabolic cascade in the marine bacterium Vibrio furnissii molecular cloning, isolation, and characterization of a periplasmic chitodextrinase. J. Biol. Chem. 1996, 271, 33414–33424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramli, A.N.; Mahadi, N.M.; Rabu, A.; Murad, A.M.; Bakar, F.D.; Illias, R.M. Molecular cloning, expression and biochemical characterisation of a cold-adapted novel recombinant chitinase from Glaciozyma antarctica PI12. Microb. Cell Fact. 2011, 10, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Shizume, A.; Nogawa, M.; Taguchi, G.; Shimosaka, M. Heterologous expression and functional characterization of a novel chitinase from the chitinolytic bacterium Chitiniphilus shinanonensis. Biosci. Biotechnol. Biochem. 2012, 1203012832. [Google Scholar]
- Ohishi, K.; Yamagishi, M.; Ohta, T.; Suzuki, M.; Izumida, H.; Sano, H.; Nishijima, M.; Miwa, T. Purification and properties of two chitinases from Vibrio alginolyticus H-8. J. Ferment. Bioeng. 1996, 82, 598–600. [Google Scholar] [CrossRef]
- Revathi, M.; Saravanan, R.; Shanmugam, A. Production and characterization of chitinase from Vibrio species, a head waste of shrimp Metapenaeus dobsonii (Miers, 1878) and chitin of Sepiella inermis Orbigny, 1848. Adv. Biosci. Biotechnol. 2012, 3, 392. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Yang, C.; Lu, Y.; Huang, L.; Cai, C.; Lin, Y. Isolation and characterization of chitinase from a marine bacterium Vibrio sp. World J. Microbiol. Biotechnol. 1999, 15, 745–746. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, D.; Kim, I.H.; Lee, C.-E.; Kim, I.-C.; Kim, J.Y.; Kim, S.J.; Lee, H.K.; Yim, J.H. Characteristics of cold-adaptive endochitinase from Antarctic bacterium Sanguibacter antarcticus KOPRI 21702. Enzyme Microb. Technol. 2009, 45, 391–396. [Google Scholar] [CrossRef]
- Suginta, W.; Vongsuwan, A.; Songsiriritthigul, C.; Prinz, H.; Estibeiro, P.; Duncan, R.R.; Svasti, J.; Fothergill-Gilmore, L.A. An endochitinase A from Vibrio carchariae: Cloning, expression, mass and sequence analyses, and chitin hydrolysis. Arch. Biochem. Biophys. 2004, 424, 171–180. [Google Scholar] [CrossRef]
- Kadokura, K.; Rokutani, A.; Yamamoto, M.; Ikegami, T.; Sugita, H.; Itoi, S.; Hakamata, W.; Oku, T.; Nishio, T. Purification and characterization of Vibrio parahaemolyticus extracellular chitinase and chitin oligosaccharide deacetylase involved in the production of heterodisaccharide from chitin. Appl. Microbiol. Biotechnol. 2007, 75, 357. [Google Scholar] [CrossRef]
- Alam, M.; Nikaidou, N.; Tanaka, H.; Watanabe, T. Cloning and sequencing of chiC gene of Bacillus circulans WL-12 and relationship of its product to some other chitinases and chitinase-like proteins. J. Ferment. Bioeng. 1995, 80, 454–461. [Google Scholar] [CrossRef]
- Rush, C.L.; Schüttelkopf, A.W.; Hurtado-Guerrero, R.; Blair, D.E.; Ibrahim, A.F.; Desvergnes, S.; Eggleston, I.M.; van Aalten, D.M. Natural product-guided discovery of a fungal chitinase inhibitor. Chem. Biol. 2010, 17, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.; Hennig, M.; Schlesier, B.; Höhne, W. Structure of jack bean chitinase. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000, 56, 1096–1099. [Google Scholar] [CrossRef] [PubMed]
- Hollis, T.; Monzingo, A.F.; Bortone, K.; Ernst, S.; Cox, R.; Robertus, J.D. The X-ray structure of a chitinase from the pathogenic fungus Coccidioides immitis. Protein Sci. 2000, 9, 544–551. [Google Scholar] [CrossRef] [Green Version]
- Itoi, S.; Kanomata, Y.; Koyama, Y.; Kadokura, K.; Uchida, S.; Nishio, T.; Oku, T.; Sugita, H. Identification of a novel endochitinase from a marine bacterium Vibrio proteolyticus strain No. 442. Biochim. Biophys. Acta. 2007, 1774, 1099–1107. [Google Scholar] [CrossRef] [PubMed]
- Murao, S.; Kawada, T.; Itoh, H.; Oyama, H.; Shin, T. Purification and characterization of a novel type of chitinase from Vibrio alginolyticus TK-22. Biosci. Biotechnol. Biochem. 1992, 56, 368–369. [Google Scholar] [CrossRef]
- Pantoom, S.; Songsiriritthigul, C.; Suginta, W. The effects of the surface-exposed residues on the binding and hydrolytic activities of Vibrio carchariae chitinase A. BMC Biochem. 2008, 9, 2. [Google Scholar] [CrossRef] [Green Version]
- Suginta, W.; Sirimontree, P.; Sritho, N.; Ohnuma, T.; Fukamizo, T. The chitin-binding domain of a GH-18 chitinase from Vibrio harveyi is crucial for chitin-chitinase interactions. Int. J. Biol. Macromol. 2016, 93, 1111–1117. [Google Scholar] [CrossRef]
- Shallom, D.; Golan, G.; Shoham, G.; Shoham, Y. Effect of dimer dissociation on activity and thermostability of the α-glucuronidase from Geobacillus stearothermophilus: Dissecting the different oligomeric forms of family 67 glycoside hydrolases. J. Bacteriol. 2004, 186, 6928–6937. [Google Scholar] [CrossRef] [Green Version]
- Fraser, N.J.; Liu, J.-W.; Mabbitt, P.D.; Correy, G.J.; Coppin, C.W.; Lethier, M.; Perugini, M.A.; Murphy, J.M.; Oakeshott, J.G.; Weik, M. Evolution of protein quaternary structure in response to selective pressure for increased thermostability. J. Mol. Biol. 2016, 428, 2359–2371. [Google Scholar] [CrossRef] [Green Version]
- Schwab, T.; Skegro, D.; Mayans, O.; Sterner, R. A rationally designed monomeric variant of anthranilate phosphoribosyltransferase from Sulfolobus solfataricus is as active as the dimeric wild-type enzyme but less thermostable. J. Mol. Biol. 2008, 376, 506–516. [Google Scholar] [CrossRef]
- Betts, M.J.; Russell, R.B. Amino acid properties and consequences of substitutions. Bioinform. Genet. 2003, 317, 289. [Google Scholar]
- Shortle, D.; Stites, W.E.; Meeker, A.K. Contributions of the large hydrophobic amino acids to the stability of staphylococcal nuclease. Biochemistry 1990, 29, 8033–8041. [Google Scholar] [CrossRef] [PubMed]
- Yutani, K.; Ogasahara, K.; Tsujita, T.; Sugino, Y. Dependence of conformational stability on hydrophobicity of the amino acid residue in a series of variant proteins substituted at a unique position of tryptophan synthase alpha subunit. Proc. Natl. Acad. Sci. USA 1987, 84, 4441–4444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montgomery, M.T.; Kirchman, D.L. Role of chitin-binding proteins in the specific attachment of the marine bacterium Vibrio harveyi to chitin. Appl. Environ. Microbiol. 1993, 59, 373–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardt, M.; Laine, R.A. Mutation of active site residues in the chitin-binding domain ChBDChiA1 from chitinase A1 of Bacillus circulans alters substrate specificity: Use of a green fluorescent protein binding assay. Arch. Biochem. Biophys. 2004, 426, 286–297. [Google Scholar] [CrossRef]
- Liang, J.; Liu, J.; Wang, X.; Lin, H.; Liu, J.; Zhou, S.; Sun, H.; Zhang, X.-H. Spatiotemporal dynamics of free-living and particle-associated Vibrio communities in the northern Chinese marginal seas. Appl. Environ. Microbiol. 2019, 85, e00217–e00219. [Google Scholar] [CrossRef] [Green Version]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2014, 43, D204–D212. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, H. Predicting secretory proteins with SignalP. In Protein Function Prediction; Springer Humana Press: New York, NY, USA, 2017; Volume 1611, pp. 59–73. [Google Scholar]
- Roberts, W.K.; Selitrennikoff, C.P. Plant and bacterial chitinases differ in antifungal activity. Microbiology 1988, 134, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.; Su, Y.; Brackman, G.; Cui, F.; Zhang, Y.; Shi, X.; Coenye, T.; Zhang, X.-H. MomL, a novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 2015, 81, 774–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680. [Google Scholar] [CrossRef] [PubMed]
- Davis, B. Gel for nondenature gel electrophoresis. Part II. Clinical applications. Method and application to human serum proteins. Ann. N. Y. Acad. Sci. 1964, 121, 405–427. [Google Scholar]
- Lee, Y.-S.; Park, I.-H.; Yoo, J.-S.; Chung, S.-Y.; Lee, Y.-C.; Cho, Y.-S.; Ahn, S.-C.; Kim, C.-M.; Choi, Y.-L. Cloning, purification, and characterization of chitinase from Bacillus sp. DAU101. Bioresour. Technol. 2007, 98, 2734–2741. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Raaijmakers, J.G. Statistical analysis of the Michaelis-Menten equation. Biometrics 1987, 793–803. [Google Scholar] [CrossRef]
- Songsiriritthigul, C.; Lapboonrueng, S.; Pechsrichuang, P.; Pesatcha, P.; Yamabhai, M. Expression and characterization of Bacillus licheniformis chitinase (ChiA), suitable for bioconversion of chitin waste. Bioresour. Technol. 2010, 101, 4096–4103. [Google Scholar] [CrossRef]
- Suginta, W.; Sritho, N. Multiple roles of Asp313 in the refined catalytic cycle of chitin degradation by Vibrio harveyi chitinase A. Biosci. Biotechnol. Biochem. 2012, 120559. [Google Scholar]
- Suginta, W.; Vongsuwan, A.; Songsiriritthigul, C.; Svasti, J.; Prinz, H. Enzymatic properties of wild-type and active site mutants of chitinase A from Vibrio carchariae, as revealed by HPLC-MS. FEBS J. 2005, 272, 3376–3386. [Google Scholar] [CrossRef] [PubMed]
- Vaaje-Kolstad, G.; Houston, D.R.; Rao, F.V.; Peter, M.G.; Synstad, B.; van Aalten, D.M.; Eijsink, V.G. Structure of the D142N mutant of the family 18 chitinase ChiB from Serratia marcescens and its complex with allosamidin. Biochim. Biophys. Acta Proteins Proteom. 2004, 1696, 103–111. [Google Scholar] [CrossRef] [PubMed]



| Primer | Sequence (5’-3’) b | Restriction Site and Mutant Amino Acid |
|---|---|---|
| Chi1557-F | 5-CGCGGATCCATGAATGAGATGGTGA-3 | BamH I |
| Chi1557-R | 5-TAGCTCGAGCAACTTATCCCACGCG-3 | Xho I |
| Chi4668-F | 5’-CGCGGATCCATGAATGAAATGGTGA-3’ | BamH I |
| Chi4668-R | 5’-CCCAAGCTTCAACTTATCCCATGCG-3’ | Hind III |
| Y37N-F | 5’-GTAGTCGGTaATTGGCATAACTGGT-3’ | Tyr37 |
| Y37N-R | 5’-GACACCACTATCTGGATTCACCATC-3’ | |
| F71V-F | 5’-AACGTCTCCgTTATGAAGGTGT-3’ | Phe71 |
| F71V-R | 5’-AACCACATTGTACATAGGATCAACT-3’ | |
| D152A-F | 5’-GGTCTGGACATCGcCTTAGAGCA-3’ | Asp152 |
| D152A-R | 5’-ATCAAAGCCGAACTTGTCAGTCAGG-3’ | |
| E154Q-F | 5’-GACATCGACTTAcAGCAATCTGCAG-3’ | Glu154 |
| E154Q-R | 5’-CAGACCATCAAAGCCGAACTTGT-3’ | |
| Q219E-F | 5’-ATCAACCCTgAATTTTACAACCAAG-3’ | Gln219 |
| Q219E-R | 5’-CCAATCGTAGTACCCTTCTAATCCA-3’ | |
| Y221N-F | 5’-CCTCAATTTaACAACCAAGGTGG-3’ | Tyr221 |
| Y221N-R | 5’-GTTGATCCAATCGTAGTACCCTTCT-3’ | |
| W312G-F | 5’-GTAATGACAgGGTCGGTGAACTGGG-3’ | Trp312 |
| W312G-R | 5’-GCCACGAAGTGCCTGCCCTTG-3’ |
| Enzymology Properties | Chi1557-Ni | Chi4668-Ni |
|---|---|---|
| The optimal temperature (°C) | 45–50 | ~50 |
| The optimal pH | 5.0–7.0 | 3.0–6.0 |
| Total enzymatic activity (U) | 2.05 | 3.16 |
| Total protein content (mg mL−1) | 0.13 | 0.18 |
| Specific activity (U mg−1) | 23.42 | 41.14 |
| The kinetic parameters: | ||
| Vmax (mg U−1) | 2.94 | 6.21 |
| Km (mg mL−1) | 7.94 | 2.75 |
| Kcat (s−1) | 3.00 | 5.18 |
| Kcat/Km (s−1M−1) | 0.40 | 1.88 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, X.; Yu, M.; Wu, Y.; Ran, L.; Liu, W.; Zhang, X.-H. Two Highly Similar Chitinases from Marine Vibrio Species have Different Enzymatic Properties. Mar. Drugs 2020, 18, 139. https://doi.org/10.3390/md18030139
He X, Yu M, Wu Y, Ran L, Liu W, Zhang X-H. Two Highly Similar Chitinases from Marine Vibrio Species have Different Enzymatic Properties. Marine Drugs. 2020; 18(3):139. https://doi.org/10.3390/md18030139
Chicago/Turabian StyleHe, Xinxin, Min Yu, Yanhong Wu, Lingman Ran, Weizhi Liu, and Xiao-Hua Zhang. 2020. "Two Highly Similar Chitinases from Marine Vibrio Species have Different Enzymatic Properties" Marine Drugs 18, no. 3: 139. https://doi.org/10.3390/md18030139
APA StyleHe, X., Yu, M., Wu, Y., Ran, L., Liu, W., & Zhang, X.-H. (2020). Two Highly Similar Chitinases from Marine Vibrio Species have Different Enzymatic Properties. Marine Drugs, 18(3), 139. https://doi.org/10.3390/md18030139





