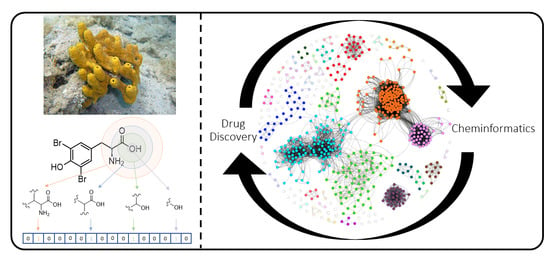
Application of Networking Approaches to Assess the Chemical Diversity, Biogeography, and Pharmaceutical Potential of Verongiida Natural Products
Abstract
:1. Introduction
2. Results and Discussion
2.1. Biosynthesis and Distribution of BTAs
2.2. Biogeography and Hotspots for Verongiida NPs
2.3. Natural Product Diversity across Genera of Verongiida—A Network Analysis Investigation
- To what degree do these genera differ with regard to their secondary metabolites?
- What compound classes contribute to the largest amount of variance between the genera studied?
- Which sponges offer biosynthetic outcomes that are the most exploitable in terms of drug discovery?
2.4. Verongiida NP Drug Score and Drug-Likeness Assessment
3. Methodology
3.1. Collection of Chemical Compound Data
3.2. Network Considerations
3.3. Molecular Fingerprints, Similarity and Scaffolding
3.4. Creation and Visualisation of Networks as Applied to Data for the Verongiida Sponge Order
3.4.1. Bipartite Networks
3.4.2. Scaffold Networks (SNs)
3.4.3. Chemical Similarity Networks (CSNs)
3.5. PCA and Drug Score Assessment
3.6. Limitations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grant, R.E. Animal Kingdom. In The Cyclopaedia of Anatomy and Physiology; Sherwood, Gilber and Piper: London, UK, 1836; pp. 107–118. [Google Scholar]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vries, D.J.; Hall, M.R. Marine biodiversity as a source of chemical diversity. Drug. Dev. Res. 1994, 33, 161–173. [Google Scholar] [CrossRef]
- Wiese, J.; Imhoff, J.F. Marine bacteria and fungi as promising source for new antibiotics. Drug Dev. Res. 2019, 80, 24–27. [Google Scholar] [CrossRef] [Green Version]
- Sigwart, J.D.; Blasiak, R.; Jaspars, M.; Jouffray, J.-B.; Tasdemir, D. Unlocking the potential of marine biodiscovery. Nat. Prod. Rep. 2021, 38, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.C.; Munro, M.H.; Blunt, J.W.; Puga, J.; Jesus, B.; Calado, R.; Rosa, R.; Madeira, C. Biogeography and biodiscovery hotspots of macroalgal marine natural products. Nat. Prod. Rep. 2013, 30, 1380–1390. [Google Scholar] [CrossRef]
- Leal, M.C.; Puga, J.; Serodio, J.; Gomes, N.C.; Calado, R. Trends in the discovery of new marine natural products from invertebrates over the last two decades—Where and what are we bioprospecting? PLoS ONE 2012, 7, e30580. [Google Scholar]
- Bergquist, P.R. Sponges; Hutchinson; University of California Press: Berkeley, CA, USA; London, UK; Los Angeles, CA, USA, 1978; pp. 1–268. [Google Scholar]
- El-Demerdash, A.; Atanasov, A.G.; Horbanczuk, O.K.; Tammam, M.A.; Abdel-Mogib, M.; Hooper, J.N.A.; Sekeroglu, N.; Al-Mourabit, A.; Kijjoa, A. Chemical Diversity and Biological Activities of Marine Sponges of the Genus Suberea: A Systematic Review. Mar. Drugs 2019, 17, 115. [Google Scholar] [CrossRef] [Green Version]
- Lira, N.S.; Montes, R.C.; Tavares, J.F.; da Silva, M.S.; da Cunha, E.V.; de Athayde-Filho, P.F.; Rodrigues, L.C.; da Silva Dias, C.; Barbosa-Filho, J.M. Brominated compounds from marine sponges of the genus Aplysina and a compilation of their 13C NMR spectral data. Mar. Drugs 2011, 9, 2316–2368. [Google Scholar] [CrossRef]
- Niemann, H.; Marmann, A.; Lin, W.; Proksch, P. Sponge derived bromotyrosines: Structural diversity through natural combinatorial chemistry. Nat. Prod. Comm. 2015, 10, 219–231. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Li, J.; Hamann, M.T. The Marine Bromotyrosine Derivatives. Alkaloids Chem. Biol. 2005, 61, 59–262. [Google Scholar]
- Rama Rao, M.; Venkatesham, U.; Sridevi, K.V.; Venkateswarlu, Y. Chemical constituents and their biological activities of the sponge family Aplysinellidae: A review. Ind. J. Chem. 2000, 39B, 723–733. [Google Scholar]
- Erwin, P.M.; Thacker, R.W. Phylogenetic analyses of marine sponges within the order Verongida: A comparison of morphological and molecular data. Invertebr. Biol. 2007, 126, 220–234. [Google Scholar] [CrossRef]
- Hooper, J.N.; Van Soest, R.W. Systema Porifera. A guide to the classification of sponges. In Systema Porifera; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–7. [Google Scholar]
- Thoms, C.; Schupp, P.J. Activated chemical defense in marine sponges—A case study on Aplysinella Rhax. J. Chem. Ecol. 2008, 34, 1242–1252. [Google Scholar] [CrossRef] [PubMed]
- Thoms, C.; Wolff, M.; Padmakumar, K.; Ebel, R.; Proksch, P. Chemical defense of Mediterranean sponges Aplysina cavernicola and Aplysina aerophoba. Z. Naturforsch. 2004, 59c, 113–122. [Google Scholar] [CrossRef]
- Ortlepp, S.; Sjogren, M.; Dahlstrom, M.; Weber, H.; Ebel, R.; Edrada, R.; Thoms, C.; Schupp, P.; Bohlin, L.; Proksch, P. Antifouling activity of bromotyrosine-derived sponge metabolites and synthetic analogues. Mar. Biotechnol. 2007, 9, 776–785. [Google Scholar] [CrossRef] [PubMed]
- Teeyapant, R.; Woerdenbag, H.J.; Kreis, P.; Hacker, J.; Wray, V.; Witte, L.; Proksch, P. Antibiotic and cytotoxic activity of brominated compounds from the marine sponge Verongia aerophoba. Z. Naturforsch. 1993, 48c, 939–945. [Google Scholar] [CrossRef]
- Wang, Q.; Tang, X.L.; Luo, X.C.; de Voog, N.J.; Li, P.L.; Li, G.Q. Aplysinopsin-type and Bromotyrosine-derived Alkaloids from the South China Sea Sponge Fascaplysinopsis reticulata. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuñez, C.V.; de Almeida, E.V.R.; Granato, A.C.; Marques, S.O.; Santos, K.O.; Pereira, F.R.; Macedo, M.L.; Ferreira, A.G.; Hajdu, E.; Pinheiro, U.S.; et al. Chemical variability within the marine sponge Aplysina fulva. Biochem. Syst. Ecol. 2008, 36, 283–296. [Google Scholar] [CrossRef]
- Ciminiello, P.; Fattorusso, E.; Forino, M.; Magno, S. Chemistry of Verongida sponges VIII Bromocompounds from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. Tetrahedron 1997, 18, 6565–6572. [Google Scholar] [CrossRef]
- Ciminiello, P.; Fattorusso, E.; Magno, S.; Pansini, M. Chemistry of Verongiida sponges VI Comparison of the secondary metabolic composition of Aplysina insularis and Aplysina fulva. Biochem. Syst. Ecol. 1996, 24, 105–113. [Google Scholar] [CrossRef]
- Quinoa, E.; Crews, P. Phenolic constituents of psammaplysilla. Tetrahedron Lett. 1987, 28, 3229–3232. [Google Scholar] [CrossRef]
- Kumar, M.S.L.; Ali, K.; Chaturvedi, P.; Meena, S.; Datta, D.; Panda, G. Design, synthesis and biological evaluation of oxime lacking Psammaplin inspired chemical libraries as anti-cancer agents. J. Mol. Struct. 2021, 1225, 129173. [Google Scholar] [CrossRef]
- Jing, Q.; Hu, X.; Ma, Y.; Mu, J.; Liu, W.; Xu, F.; Li, Z.; Bai, J.; Hua, H.; Li, D. Marine-Derived Natural Lead Compound Disulfide-Linked Dimer Psammaplin A: Biological Activity and Structural Modification. Mar. Drugs 2019, 17, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Y.; Xu, Q.; Wang, L.; Wei, Y.; Hu, B.; Wang, J.; Liu, D.; Zhao, L.; Jing, Y. Studying Histone Deacetylase Inhibition and Apoptosis Induction of Psammaplin A Monomers with Modified Thiol Group. ACS Med. Chem. Lett. 2021, 12, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine pharmacology in 2009–2011: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2013, 11, 2510–2573. [Google Scholar]
- Lebouvier, N.; Jullian, V.; Desvignes, I.; Maurel, S.; Parenty, A.; Dorin-Semblat, D.; Doerig, C.; Sauvain, M.; Laurent, D. Antiplasmodial activities of homogentisic acid derivative protein kinase inhibitors isolated from a Vanuatu marine sponge Pseudoceratina sp. Mar. Drugs 2009, 7, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine pharmacology in 2012–2013: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2017, 15, 273. [Google Scholar]
- Mayer, A.; Guerrero, A.J.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Nakamura, F.; Fusetani, N. Marine pharmacology in 2014–2015: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, antiviral, and anthelmintic activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2020, 18, 5. [Google Scholar]
- Xu, M.; Davis, R.A.; Feng, Y.; Sykes, M.L.; Shelper, T.; Avery, V.M.; Camp, D.; Quinn, R.J. Ianthelliformisamines A-C, antibacterial bromotyrosine-derived metabolites from the marine sponge Suberea ianthelliformis. J. Nat. Prod. 2012, 75, 1001–1005. [Google Scholar] [CrossRef]
- Pieri, C.; Borselli, D.; Di Giorgio, C.; De Meo, M.; Bolla, J.-M.; Vidal, N.; Combes, S.; Brunel, J.M. New ianthelliformisamine derivatives as antibiotic enhancers against resistant Gram-negative bacteria. J. Med. Chem. 2014, 57, 4263–4272. [Google Scholar] [CrossRef]
- Mani, L.; Jullian, V.; Mourkazel, B.; Valentin, A.; Dubois, J.; Cresteil, T.; Folcher, E.; Hooper, J.N.A.; Erpenbeck, D.; Aalbersberg, W.; et al. New antiplasmodial bromotyrosine derivatives from Suberea ianthelliformis. Chem. Biodivers. 2012, 9, 1436–1451. [Google Scholar] [CrossRef]
- Buchanan, M.S.; Carroll, A.R.; Wessling, D.; Jobling, M.; Avery, V.M.; Davis, R.A.; Feng, Y.; Hooper, J.N.A.; Quinn, R.J. Clavatadines C-E Guanidine alkaloids from the Australian sponge Suberea clavata. J. Nat. Prod. 2009, 72, 973–975. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, M.S.; Carroll, A.R.; Wessling, D.; Jobling, M.; Avery, V.M.; Davis, R.A.; Feng, Y.; Xue, Y.; Oster, L.; Fex, T.; et al. Clavatadine A, A natural product with selective recognition and irreversible inhibition of factor XIa. J. Med. Chem. 2008, 51, 3583–3587. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Bowden, B.F.; Kapoor, V. Ianthellamide A, a selective kynurenine-3-hydroxylase inhibitor from the Australian marine sponge Ianthella quadrangulata. Bioorg. Med. Chem. Lett. 2012, 22, 3398–3401. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.W.; Feng, Y.; Shimizu, Y.; Pfeifer, T.; Wellington, C.; Hooper, J.N.; Quinn, R.J. Aplysinellamides A-C, bromotyrosine-derived metabolites from an Australian Aplysinella sp. marine sponge. J. Nat. Prod. 2014, 77, 1210–1214. [Google Scholar] [CrossRef]
- Mohanty, I.; Tapadar, S.; Moore, S.G.; Biggs, J.S.; Freeman, C.J.; Gaul, D.A.; Garg, N.; Agarwal, V. Presence of bromotyrosine alkaloids in marine sponges is independent of metabolic and microbiome architectures. Msystems 2021, 6, 1–17. [Google Scholar] [CrossRef]
- Nicacio, K.J.; Ioca, L.P.; Froes, A.M.; Leomil, L.; Appolinario, L.R.; Thompson, C.C.; Thompson, F.L.; Ferreira, A.G.; Williams, D.E.; Andersen, R.J.; et al. Cultures of the Marine Bacterium Pseudovibrio denitrificans Ab134 Produce Bromotyrosine-Derived Alkaloids Previously Only Isolated from Marine Sponges. J. Nat. Prod. 2017, 80, 235–240. [Google Scholar] [CrossRef]
- Thompson, J.E.; Barrow, K.D.; Faulkner, J.D. Localization of two brominated metabolites aerothionin and homoaerothionin in spherulous cells of the marine sponge Aplysina fistularis. Acta. Zool. 1983, 64, 199–210. [Google Scholar] [CrossRef]
- Turon, X.; Becerro, M.A.; Uriz, M.J. Distribution of brominated compounds within the sponge Aplysina aerophoba: Coupling of X-ray microanalysis with cryofixation techniques. Cell Tissue Res. 2000, 301, 311–322. [Google Scholar] [CrossRef]
- Ebel, R.; Brenzinger, M.; Kunze, A.; Gross, H.J.; Proksch, P. Wound activation of protoxins in marine sponge Aplysina aerophoba. J. Chem. Ecol. 1997, 23, 1451–1462. [Google Scholar] [CrossRef]
- Pita, L.; Turon, X.; Lopez-Legentil, S.; Erwin, P.M. Host rules: Spatial stability of bacterial communities associated with marine sponges (Ircinia spp.) in the Western Mediterranean Sea. FEMS Microbiol. Ecol. 2013, 86, 268–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardenas, C.A.; Bell, J.J.; Davy, S.K.; Hoggard, M.; Taylor, M.W. Influence of environmental variation on symbiotic bacterial communities of two temperate sponges. FEMS Microbiol. Ecol. 2014, 88, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Steinert, G.; Taylor, M.W.; Deines, P.; Simister, R.L.; de Voogd, N.J.; Hoggard, M.; Schupp, P.J. In four shallow and mesophotic tropical reef sponges from Guam the microbial community largely depends on host identity. PeerJ 2016, 4, e1936. [Google Scholar] [CrossRef]
- Steinert, G.; Rohde, S.; Janussen, D.; Blaurock, C.; Schupp, P.J. Host-specific assembly of sponge-associated prokaryotes at high taxonomic ranks. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Nardo, G.D. De Spongiis. Isis von Oken 1834, 714–716. [Google Scholar]
- Bergquist, P.R. Dictyoceratida, Dendroceratida and Verongida from the New Caledonia Lagoon (Porifera: Demospongiae). Mem. Qld. Mus. 1995, 38, 1–51. [Google Scholar]
- Salim, A.A.; Khalil, Z.G.; Capon, R.J. Structural and stereochemical investigations into bromotyrosine-derived metabolites from southern Australian marine sponges, Pseudoceratina spp. Tetrahedron 2012, 68, 9802–9807. [Google Scholar] [CrossRef]
- Ragini, K.; Fromont, J.; Piggott, A.M.; Karuso, P. Enantiodivergence in the Biosynthesis of Bromotyrosine Alkaloids from Sponges? J. Nat. Prod. 2017, 80, 215–219. [Google Scholar] [CrossRef]
- Carter, H.J. Descriptions of Sponges from the Neighbourhood of Port Phillip Heads, South Australia. Ann. Mag. Nat. Hist. 1885, 15, 107–117. [Google Scholar] [CrossRef]
- Higgin, T. On a new sponge of the genus Luffaria, from Yucatan, in the Liverpool Free Museum. Ann. Mag. Nat. Hist. 1875, 16, 223–227. [Google Scholar] [CrossRef]
- De Lamarck, J.B. Sur les polypiers empâtés. Ann. Mus. Natl. d’Hist. Nat. 1814, 20, 294–312. [Google Scholar]
- Gray, J.E. Note on Ianthella, a new genus of keratose sponges. Proc. Zool. Soc. Lond. 1869, 1869, 49–51. [Google Scholar] [CrossRef]
- Hyatt, A. Revision of the North American Poriferae; with Remarks upon Foreign Species. Part I. Mem. Boston Soc. Nat. Hist. 1875, 2, 399–408. [Google Scholar]
- Topsent, E. Matériaux pour servir à l’étude de la faune des spongiaires de France. Mémoires Société Zool. France 1896, 9, 113–133. [Google Scholar]
- Bergquist, P.R. A revision of the supraspecific classification of the orders Dictyoceratida, Dendroceratida and Verongida (class Demospongiae). N. Zealand J. Zool. 1980, 7, 443–503. [Google Scholar] [CrossRef] [Green Version]
- De Laubenfels, M.W. The Sponges of the West-Central Pacific; Oregon State Monographs: Studies in Zoology; Oregon State College: Corvallis, OR, USA, 1954; pp. 35–41. [Google Scholar]
- Verrill, A.E. The Bermuda Islands: Part V. An account of the Coral Reefs (Characteristic Life of the Bermuda Coral Reefs). Porifera: Sponges. Trans. Conn. Acad. Arts Sci. 1907, 12, 330–344. [Google Scholar]
- Teeyapant, R.; Proksch, P. Biotransformation of brominated compounds in the marine sponge Verongia aerophoba—Evidence for an induced chemical defense? Sci. Nat. 1993, 80, 369–370. [Google Scholar] [CrossRef]
- Teeyapant, R.; Kreis, P.; Wray, V.; Witte, L.; Proksch, P. Brominated secondary compounds from the marine sponge Verongia aerophoba and the sponge feeding gastropod Tylodina perversa. Z. Naturforsch. 1993, 48, 640–644. [Google Scholar] [CrossRef]
- Kunze, K.; Niemann, H.; Ueberlein, S.; Schulze, R.; Ehrlich, H.; Brunner, E.; Proksch, P.; van Pee, K.H. Brominated skeletal components of the marine demosponges, Aplysina cavernicola and Ianthella basta: Analytical and biochemical investigations. Mar. Drugs 2013, 11, 1271–1287. [Google Scholar] [CrossRef] [Green Version]
- Ocean Biodiversity Information System. 2021. Available online: https://obis.org/ (accessed on 21 March 2021).
- Van Soest, R.W.M.; Boury-Esnault, N.; Hooper, J.N.A.; Rutzler, K.; de Voogd, N.J.; Alvarez, B.; Hajdu, E.; Pisera, A.B.; Manconi, R.; Schonberg, C.; et al. World Porifera Database. 2021. Available online: http://www.marinespecies.org/porifera (accessed on 6 June 2021).
- Yagi, H.; Matsunaga, S.; Fusetani, N. Purpuramines A-I, New bromotyrosine-derived metabolites from the marine sponge Psammaplysilla purpurea. Tetrahedron 1993, 49, 3749–3754. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Kato, H.; Hirota, H.; Fusetani, N. Ceratinamides A and B: New antifouling dibromotyrosine derivatives from the marine sponge Pseudoceratina purpurea. Tetrahedron 1996, 52, 8181–8186. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Kato, H.; Hirota, H.; Fusetani, N. Ceratinamine: An unprecedented antifouling cyanoformamide from the marine sponge Pseudoceratina purpurea. J. Org. Chem. 1996, 61, 2936–2937. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.H.; van Soest, R.W.M.; Fusetani, N.; Matsunaga, S. Pseudoceratins A and B, antifungal bicyclic bromotyrosine-derived metabolites from the marine sponge Pseudoceratina purpurea. J. Org. Chem. 2007, 72, 1211–1217. [Google Scholar] [CrossRef]
- Ma, K.; Yang, Y.; Deng, Z.; de Voogd, N.J.; Proksch, P.; Lin, W. Two new bromotyrosine derivatives from the marine sponge Pseudoceratina sp. Chem. Biodivers. 2008, 5, 1313–1320. [Google Scholar] [CrossRef]
- Huang, X.-P.; Deng, Z.-W.; van Soest, R.W.M.; Lin, W.-H. Brominated derivatives from the Chinese sponge Pseudoceratina sp. J. Asian Nat. Prod. Res. 2008, 10, 239–242. [Google Scholar] [CrossRef]
- Li, H.; Yu, H.; Wu, W.; Sun, P. Chemical constituents of sponge Pseudoceratina sp.; their chemotaxonomic significance. Biochem. Syst. Ecol. 2020, 89, 104002. [Google Scholar] [CrossRef]
- Jimenez, C.; Crews, P. Novel marine sponge derived amino acids 13. additional psammaplin derivatives from Psammplysilla purpurea. Tetrahedron 1991, 47, 2097–2102. [Google Scholar] [CrossRef]
- Kijjoa, A.; Bessa, J.; Wattanadilok, R.; Sawangwong, P.; Nascimento, N.S.J.; Pedro, M.; Silva, A.M.S.; Eaton, G.; van Soest, R.; Herz, W. Dibromotyrosine derivatives, a maleimide, aplysamine-2 and other constituents of the marine sponge Pseudoceratina purpurea. Z. Naturforsch. 2005, 60, 904–908. [Google Scholar] [CrossRef]
- Morris, S.A.; Anderson, R.J. Brominated bis(indole) alkaloids from the marine sponge Hexadella sp. Tetrahedron 1990, 46, 715–720. [Google Scholar] [CrossRef]
- Morris, S.A.; Anderson, R.J. Nitrogenous metabolites from the deep water sponge Hexadella sp. Can. J. Chem. 1989, 67, 677–681. [Google Scholar] [CrossRef]
- Ichiba, T.; Scheuer, P.J. Three Bromotyrosine Derivatives, One Terminating in an Unprecedented Diketocyclopentenylidene Enamine. J. Org. Chem. 1993, 58, 4149–4150. [Google Scholar] [CrossRef]
- Van Soest, R.W.; Boury-Esnault, N.; Vacelet, J.; Dohrmann, M.; Erpenbeck, D.; De Voogd, N.J.; Santodomingo, N.; Vanhoorne, B.; Kelly, M.; Hooper, J.N. Global diversity of sponges (Porifera). PLoS ONE 2012, 7, e35105. [Google Scholar] [CrossRef]
- Carter, H.J. Report on Specimens dredged up from the Gulf of Manaar and presented to the Liverpool Free Museum by Capt.W.H. Cawne Warren. Ann. Mag. Nat. Hist. 1880, 6, 35–61. [Google Scholar] [CrossRef] [Green Version]
- Kernan, M.R.; Cambie, R.C. Chemistry of sponges, VII. 11, 19-Dideoxyfistularin 3 and 11-hydroxyaerothionin, bromotyrosine derivatives from Pseudoceratina durissima. J. Nat. Prod. 1990, 53, 615–622. [Google Scholar] [CrossRef]
- Benharref, A.; Pais, M. Bromotyrosine alkaloids from the sponge Pseudoceratina verrucosa. J. Nat. Prod. 1996, 59, 177–180. [Google Scholar] [CrossRef]
- Tran, T.D.; Pham, N.B.; Fechner, G.; Hooper, J.N.; Quinn, R.J. Bromotyrosine alkaloids from the Australian marine sponge Pseudoceratina verrucosa. J. Nat. Prod. 2013, 76, 516–523. [Google Scholar] [CrossRef]
- Kassuhlke, K.E.; Faulkner, J.D. Two new dibromotyrosine derivatives from the Caribbean sponge pseudoceratina crassa. Tetrahedron 1991, 47, 1809–1814. [Google Scholar] [CrossRef]
- Albrizio, S.; Ciminiello, P.; Fattorusso, E.; Magno, S. Chemistry of Verongida sponges. I. constituents of the caribbean sponge Pseudoceratina crassa. Tetrahedron 1994, 50, 783–788. [Google Scholar] [CrossRef]
- Ciminiello, P.; Fattorusso, E.; Magno, S. Chemistry of Verongida sponges, IV. comparison of the secondary metabolite composition of several specimens of Pseudoceratina crassa. J. Nat. Prod. 1995, 58, 689–696. [Google Scholar] [CrossRef]
- Rahelivao, M.P.; Lubken, T.; Gruner, M.; Kataeva, O.; Ralambondrahety, R.; Andriamanantoanina, H.; Checinski, M.P.; Bauer, I.; Knolker, H.J. Isolation and structure elucidation of natural products of three soft corals and a sponge from the coast of Madagascar. Org. Biomol. Chem. 2017, 15, 2593–2608. [Google Scholar] [CrossRef] [Green Version]
- Keller, C. Die Spongienfauna des rothen Meeres (I. Hälfte). Z. Wiss. Zool. 1889, 48, 311–405. [Google Scholar]
- Badhr, J.M.; Shaala, L.A.; Abou-Shoer, M.I.; Tawfik, M.K.; Abdel-Azim, H.M. Bioactive brominated metabolites from the Red Sea sponge Pseudoceratina arabica. J. Nat. Prod. 2008, 71, 1472–1474. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.; Sulaiman, M.; Behery, F.A.; Foudah, A.I.; Sayed, K.A. Subereamolline A as a potent breast cancer migration, invasion and proliferation inhibitor and bioactive dibrominated alkaloids from the Red Sea sponge Pseudoceratina arabica. Mar. Drugs 2012, 10, 2492–2508. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.A.; Badr, J.M.; Sulaiman, M.; Khedr, A.; El Sayed, K.A. Bioactive alkaloids from the Red Sea marine Verongid sponge Pseudoceratina arabica. Tetrahedron 2015, 71, 7837–7841. [Google Scholar] [CrossRef]
- Bergquist, P.R.; Kelly-Borges, M. Systematics and biogeography of the genus Ianthella (Demospongiae: Verongida: Ianthellidae) in the south-west Pacific. Beagle Rec. Mus. Art Galleries North. Territ. 1995, 12, 151–176. [Google Scholar]
- Balansa, W.; Islam, R.; Gilbert, D.F.; Fontaine, F.; Xiao, X.; Zhang, H.; Piggott, A.M.; Lynch, J.W.; Capon, R.J. Australian marine sponge alkaloids as a new class of glycine-gated chloride channel receptor modulator. Bioorg. Med. Chem. 2013, 21, 4420–4425. [Google Scholar] [CrossRef] [PubMed]
- Motti, C.A.; Freckelton, M.L.; Tapiolas, D.M.; Willis, R.H. FTICR-MS and LC-UV/MS-SPE-NMR Applications for the rapid dereplication of a crude extract from the sponge Ianthella flabelliformis. J. Nat. Prod. 2009, 72, 290–294. [Google Scholar] [CrossRef]
- Carroll, A.R.; Kaiser, S.M.; Davis, R.A.; Moni, R.W.; Hooper, J.N.A.; Quinn, R.J. A Bastadin with Potent and Selective δ-Opioid Receptor Binding Affinity from the Australian Sponge Ianthella flabelliformis. J. Nat. Prod. 2010, 73, 1173–1176. [Google Scholar] [CrossRef]
- Okamoto, Y.; Ojika, M.; Kato, S.; Sakagami, Y. Ianthesines A–D, Four Novel Dibromotyrosine-Derived Metabolites from a Marine Sponge, Ianthella sp. Tetrahedron 2000, 56, 5813–5818. [Google Scholar] [CrossRef]
- Jaspars, M.; Rali, T.; Laney, M.; Schatzman, R.C.; Diaz, M.C.; Schmitz, F.J.; Pordesimo, E.O.; Crews, P. The search for inosine 5′-Phosphate dehydrogenase (IMPDH) inhibitors from marine sponges. Evaluation of the bastadin alkaloids. Tetrahedron 1994, 50, 7367–7374. [Google Scholar] [CrossRef]
- Pallas, P.S. Elenchus zoophytorum sistens generum adumbrationes generaliores et specierum cognitarum succintas descriptiones, cum selectis auctorum synonymis. In Fransiscum Varrentrapp Hagae; Hagae-Comitum: Apud Petrum van Cleef: Hagae, The Netherlands, 1766. [Google Scholar]
- Pordesimo, E.O.; Schmitz, F.J. New bastadins from the sponge Ianthella Basta. J. Org. Chem. 1990, 55, 4704–4709. [Google Scholar] [CrossRef]
- Masuno, M.N.; Hoepker, A.C.; Pessah, I.N.; Molinski, T.F. 1-O-Sulfatobastadins-1 and -2 from Ianthella basta (Pallas). Antagonists of the RyR1-FKBP12 Ca2+ Channel. Mar. Drugs 2004, 2, 176–184. [Google Scholar] [CrossRef] [Green Version]
- Miao, S.; Anderson, R.J. Cytotoxic metabolites from the sponge Ianthella basta collected in Papua New Guinea. J. Nat. Prod. 1990, 53, 1441–1446. [Google Scholar] [CrossRef] [PubMed]
- Pettit, G.R.; Butler, M.S.; Bass, C.G.; Doubek, D.L.; Williams, M.D.; Schmidt, J.M.; Pettit, R.K.; Hooper, J.N.A.; Tackett, L.P.; Filiatrault, M.J. Antineoplastic agents, 326. The stereochemistry of bastadins 8, 10, and 12 from the Bismarck Archipelago marine sponge Ianthella basta. J. Nat. Prod. 1995, 58, 680–688. [Google Scholar] [CrossRef]
- Pettit, G.R.; Butler, M.S.; Williams, M.D.; Filiatrault, M.J.; Pettit, R.K. Isolation and Structure of Hemibastadinols 1−3 from the Papua New Guinea Marine Sponge Ianthella basta. J. Nat. Prod. 1996, 59, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, K.; Kato, H.; Fujiwara, Y.; Losung, F.; Mangindaan, R.E.; de Voogd, N.J.; Takeya, M.; Tsukamoto, S. Bastadins, brominated-tyrosine derivatives, suppress accumulation of cholesterol ester in macrophages. Bioorg. Med. Chem. Lett. 2015, 25, 5389–5392. [Google Scholar] [CrossRef] [PubMed]
- Park, S.K.; Jurek, J.; Carney, J.R.; Scheuer, P.J. Two more bastadins, 16 and 17, from an Indonesian sponge Ianthella basta. J. Nat. Prod. 1994, 57, 407–410. [Google Scholar] [CrossRef]
- Mathieu, V.; Wauthoz, N.; Lefranc, F.; Niemann, H.; Amighi, K.; Kiss, R.; Proksch, P. Cyclic versus hemi-bastadins. pleiotropic anti-cancer effects: From apoptosis to anti-angiogenic and anti-migratory effects. Molecules 2013, 18, 3543–3561. [Google Scholar] [CrossRef]
- Niemann, H.; Lin, W.; Muller, W.E.; Kubbutat, M.; Lai, D.; Proksch, P. Trimeric hemibastadin congener from the marine sponge Ianthella basta. J. Nat. Prod. 2013, 76, 121–125. [Google Scholar] [CrossRef]
- Park, S.K.; Park, H.; Scheuer, P.J. Isolation and structure determination of a new bastadin from an Indonesian sponge Ianthella Basta. Bull. Korean Chem. Soc. 1994, 15, 534–537. [Google Scholar]
- Aoki, S.; Cho, S.H.; Hiramatsu, A.; Kotoku, N.; Kobayashi, M. Bastadins, cyclic tetramers of brominated-tyrosine derivatives, selectively inhibit the proliferation of endothelial cells. J. Nat. Med. 2006, 60, 231–235. [Google Scholar] [CrossRef]
- Mack, M.M.; Molinski, T.F.; Buck, E.D.; Pessah, I.N. Novel modulators of skeletal muscle FKBP12/calcium channel complex from Ianthella basta. J. Biol. Chem. 1994, 269, 23236–23249. [Google Scholar] [CrossRef]
- Kazlauskas, R.; Lidgard, R.O.; Murphy, P.T.; Wells, R.J.; Blount, J.F. Brominated tyrosine-derived metabolites from the sponge Ianthella basta. Aust. J. Chem. 1981, 34, 765–786. [Google Scholar] [CrossRef]
- Butler, M.S.; Lim, T.K.; Capon, R.J.; Hammond, L.S. The Bastadins Revisited: New Chemistry From the Australian Marine Sponge Ianthella basta. Aust. J. Chem. 1991, 44, 287–296. [Google Scholar] [CrossRef]
- Franklin, M.A.; Penn, S.G.; Lebrilla, C.B.; Lam, T.H.; Pessah, I.N.; Molinski, T.F. Bastadin 20 and Bastadin O-Sulfate Esters from Ianthella basta: Novel Modulators of the Ry1R FKBP12 Receptor Complex. J. Nat. Prod. 1996, 59, 1121–1127. [Google Scholar] [CrossRef]
- Gartshore, C.J.; Salib, M.N.; Renshaw, A.A.; Molinski, T.F. Isolation of bastadin-6-O-sulfate and expedient purifications of bastadins-4, -5 and -6 from extracts of Ianthella basta. Fitoterapia 2018, 126, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Greve, H.; Meis, S.; Kassack, M.U.; Kehraus, S.; Krick, A.; Wright, A.D.; Konig, G.M. New Iantherans from the Marine Sponge Ianthella quadrangulata Novel Agonists of the P2Y11 Receptor. J. Med. Chem. 2007, 50, 5600–5607. [Google Scholar] [CrossRef]
- Greve, H.; Kehraus, S.; Krick, A.; Kelter, G.; Maier, A.; Fiebig, H.-H.; Wright, A.D.; Konig, G.M. Cytotoxic Bastadin 24 from the Australian Sponge Ianthella quadrangulata. J. Nat. Prod. 2008, 71, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Coll, J.C.; Kearns, P.S.; Rideout, J.A.; Sankar, V. Bastadin 21, a Novel Isobastarane Metabolite from the Great Barrier Reef Marine Sponge Ianthella quadrangulata. J. Nat. Prod. 2002, 65, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, Y.; Ojika, M.; Suzuki, S.; Murakami, M.; Sakagami, Y. Iantherans A and B, unique dimeric polybrominated benzofurans as Na, K-ATPase inhibitors from a marine sponge, Ianthella sp. Bioorg. Med. Chem. 2001, 9, 179–183. [Google Scholar] [CrossRef]
- Okamoto, Y.; Ojika, M.; Sakagami, Y. Iantheran A, a dimeric polybrominated benzofuran as a Na,K-ATPase inhibitor from a marine sponge, Ianthella sp. Tetrahedron Lett. 1999, 40, 507–510. [Google Scholar] [CrossRef]
- Zhang, H.; Conte, M.M.; Huang, X.C.; Khalil, Z.; Capon, R.J. A search for BACE inhibitors reveals new biosynthetically related pyrrolidones, furanones and pyrroles from a southern Australian marine sponge, Ianthella sp. Org. Biomol. Chem. 2012, 10, 2656–2663. [Google Scholar] [CrossRef]
- Zhang, H.; Conte, M.M.; Khalil, Z.; Huang, X.-C.; Capon, R.J. New dictyodendrins as BACE inhibitors from a southern Australian marine sponge, Ianthella sp. RSC Adv. 2012, 2, 4209–4214. [Google Scholar] [CrossRef]
- Carter, H.J. Notes introductory to the study and classification of the Spongida. Part II. Proposed classification of the Spongida. Ann. Mag. Nat. Hist. 1875, 4, 126–145. [Google Scholar] [CrossRef]
- Nardo, G.D. Auszug aus einem neuen System der Spongiarien, wonach bereits die Aufstellung in der Universitäts-Sammlung zu Padua gemacht ist. Isis, Order Encyclopadische Zeitung Coll (Oken: Jena) 1833, 519–523. Available online: http://ras.biodiversity.aq/aphia.php?p=sourcedetails&id=7979 (accessed on 1 October 2021).
- Vacelet, J. Répartition générale des éponges et systématique des éponges cornées de la région de Marseille et de quelques stations méditerranéennes. Recl. Trav. Stn. Mar. d’Endoume 1959, 16, 39–101. [Google Scholar]
- Putz, A.; Kloeppel, A.; Pfannkuchen, M.; Brummer, F.; Proksch, P. Depth-related alkaloid variation in Mediterranean Aplysina sponges. Z. Naturforsch. 2009, 64c, 279–287. [Google Scholar] [CrossRef]
- Sacristan-Soriano, O.; Banaigs, B.; Becerro, M.A. Relevant spatial scales of chemical variation in Aplysina aerophoba. Mar. Drugs 2011, 9, 2499–2513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacristan-Soriano, O.; Banaigs, B.; Becerro, M.A. Temporal trends in the secondary metabolite production of the sponge Aplysina aerophoba. Mar. Drugs 2012, 10, 677–693. [Google Scholar] [CrossRef] [Green Version]
- Reverter, M.; Perez, T.; Ereskovsky, A.V.; Banaigs, B. Secondary Metabolome Variability and Inducible Chemical Defenses in the Mediterranean Sponge Aplysina cavernicola. J. Chem. Ecol. 2016, 42, 60–70. [Google Scholar] [CrossRef]
- Silva, M.M.; Bergamasco, J.; Lira, S.P.; Lopes, N.P.; Hajdu, E.; Peixinho, S.; Berlinck, R.G.S. Dereplication of bromotyrosine-derived metabolites by LC-PDA-MS and analysis of the chemical profile of 14 Aplysina sponge specimens from the Brazilian coastline. Aust. J. Chem. 2010, 63, 886–894. [Google Scholar] [CrossRef]
- Ciminiello, P.; Costantino, V.; Fattorusso, E.; Magno, S.; Mangoni, A. Chemistry of Verongida sponges, II. Constituents of the Caribbean sponge Aplysina Fistularis forma fulva. J. Nat. Prod. 1994, 57, 705–712. [Google Scholar] [CrossRef]
- Rogers, E.W.; Fernanda de Oliveira, M.; Berlinck, R.G.S.; Konig, G.M.; Molinski, T.F. Stereochemical Heterogeneity in Verongid Sponge Metabolites. Absolute Stereochemistry of (+)-Fistularin-3 and (+)-11-epi-Fistularin-3 by Microscale LCMS-Marfey’s Analysis. J. Nat. Prod. 2005, 68, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Rogers, E.W.; Molinski, T.F. Highly polar spiroisoxazolines from the sponge Aplysina fulva. J. Nat. Prod. 2007, 70, 1191–1194. [Google Scholar] [CrossRef] [Green Version]
- Gunasekera, M.; Gunasekera, S.P. Dihydroxyaerothionin and aerophobin 1. Two brominated tyrosine metabolites from the deep water marine sponge Verongula rigida. J. Nat. Prod. 1989, 52, 753–756. [Google Scholar] [CrossRef]
- Mierzwa, R.; King, A.; Conover, M.A.; Tozzi, S.; Puar, M.S.; Patel, M.; Coval, S.J. Verongamine, a novel bromotyrosine-derived histamine H3-Antagonist from the marine sponge Verongula gigantea. J. Nat. Prod. 1994, 57, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Ciminiello, P.; Fattorusso, E.; Magno, S. Chemistry of Verongida sponges, III. Constituents of a Caribbean Verongula sp. J. Nat. Prod. 1994, 57, 1564–1569. [Google Scholar] [CrossRef]
- Galeano, E.; Thomas, O.P.; Robledo, S.; Munoz, D.; Martinez, A. Antiparasitic bromotyrosine derivatives from the marine sponge Verongula rigida. Mar. Drugs 2011, 9, 1902–1913. [Google Scholar] [CrossRef]
- Ciminiello, P.; Dell’Aversano, C.; Fattorusso, E.; Magno, S.; Pansini, M. Chemistry of Verongida Sponges. 10. Secondary Metabolite Composition of the Caribbean Sponge Verongula gigantea. J. Nat. Prod. 2000, 63, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Kochanowska, A.J.; Rao, K.V.; Childress, S.; El-Alfy, A.; Matsumoto, R.R.; Kelly, M.; Stewart, G.S.; Sufka, K.J.; Hamann, M.T. Secondary Metabolites from Three Florida Sponges with Antidepressant Activity. J. Nat. Prod. 2008, 71, 186–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiso, A.; Kittiwisut, S.; Chantakul, R.; Yuenyongsawad, S.; Putchakarn, S.; Schaberle, T.F.; Temkitthaworn, P.; Ingkaninan, K.; Chaithirayanon, K.; Plubrukarn, A. Quintaquinone, a Merosesquiterpene from the Yellow Sponge Verongula cf rigida Esper. J. Nat. Prod. 2020, 83, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Kochanowska-Karamyan, A.J.; Araujo, H.C.; Zhang, X.; El-Alfy, A.; Carvalho, P.; Avery, M.A.; Holmbo, S.D.; Magolan, J.; Hamann, M.T. Isolation and Synthesis of Veranamine, an Antidepressant Lead from the Marine Sponge Verongula rigida. J. Nat. Prod. 2020, 83, 1092–1098. [Google Scholar] [CrossRef]
- Hwang, I.H.; Oh, J.; Zhou, W.; Park, S.; Kim, J.H.; Chittiboyina, A.G.; Ferreira, D.; Song, G.Y.; Oh, S.; Na, M.; et al. Cytotoxic activity of rearranged drimane meroterpenoids against colon cancer cells via down-regulation of beta-catenin expression. J. Nat. Prod. 2015, 78, 453–461. [Google Scholar] [CrossRef]
- Graham, S.K.; Lambert, L.K.; Pierens, G.K.; Hooper, J.N.A.; Garson, M.J. Psammaplin Metabolites New and Old: An NMR Study Involving Chiral Sulfur Chemistry. Aust. J. Chem. 2010, 63, 867–872. [Google Scholar] [CrossRef]
- Pham, N.B.; Butler, M.S.; Quinn, R.J. Isolation of Psammaplin A 11‘-Sulfate and Bisaprasin 11‘-Sulfate from the Marine Sponge Aplysinella rhax. J. Nat. Prod. 2000, 63, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Tabudravu, J.N.; Eijsink, V.G.H.; Gooday, G.W.; Jaspars, M.; Komander, D.; Legg, M.; Synstad, B.; van Aalten, D.M.F. Psammaplin A, a chitinase inhibitor isolated from the Fijian marine sponge Aplysinella rhax. Bioorg. Med. Chem. 2002, 10, 1123–1128. [Google Scholar] [CrossRef]
- Shin, J.; Lee, H.-S.; Seo, Y.; Rho, J.-R.; Cho, K.W.; Paul, V.J. New Bromotyrosine Metabolites from the Sponge Aplysinella rhax. Tetrahedron 2000, 56, 9071–9077. [Google Scholar] [CrossRef]
- Mudianta, I.W. Bioprospecting of the Balinese marine sponges and nudibranchs. J. Phys. Conf. Ser. 2018, 1040, 1–7. [Google Scholar] [CrossRef]
- Mudianta, I.W.; Skinner-Adams, T.; Andrews, K.T.; Davis, R.A.; Hadi, T.A.; Hayes, P.Y.; Garson, M.J. Psammaplysin derivatives from the Balinese marine sponge Aplysinella strongylata. J. Nat. Prod. 2012, 75, 2132–2143. [Google Scholar] [CrossRef]
- Mandi, A.; Mudianta, I.W.; Kurtan, T.; Garson, M.J. Absolute Configuration and Conformational Study of Psammaplysins A and B from the Balinese Marine Sponge Aplysinella strongylata. J. Nat. Prod. 2015, 78, 2051–2056. [Google Scholar] [CrossRef]
- Ankudey, F.J.; Kiprof, P.; Stromquist, E.R.; Chang, L.C. New bioactive bromotyrosine-derived alkaloid from a marine sponge Aplysinella sp. Planta Med. 2008, 74, 555–559. [Google Scholar] [CrossRef]
- Liu, S.; Schmitz, F.J.; Kelly-Borges, M. Psammaplysin F, a New Bromotyrosine Derivative from a Sponge, Aplysinella sp. J. Nat. Prod. 1997, 60, 614–615. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Schmitz, F.J. 7-Hydroxyceratinamine, a New Cyanoformamide-Containing Metabolite from a Sponge, Aplysinella sp. J. Nat. Prod. 1999, 62, 1072–1073. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.A. Cytotoxic Psammaplysin Analogues from the Verongid Red Sea Sponge Aplysinella Species. Biomolecules 2019, 9, 841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Lendenfeld, R. Descriptive Catalogue of the Sponges in the Australian Museum, Sidney; Taylor & Francis: London, UK, 1888. [Google Scholar]
- El-Demerdash, A.; Moriou, C.; Toullec, J.; Besson, M.; Soulet, S.; Schmitt, N.; Petek, S.; Lecchini, D.; Debitus, C.; Al-Mourabit, A. Bioactive Bromotyrosine-Derived Alkaloids from the Polynesian Sponge Suberea ianthelliformis. Mar. Drugs 2018, 16, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pulitzer-Finali, G. Some new or little-known sponges from the Great Barrier Reef of Australia. Boll. Musei Ist. Biol. Dell’universitá Genova 1982, 48, 87–141. [Google Scholar]
- Spalding, M.D.; Fox, H.E.; Allen, G.R.; Davidson, N.; Ferdana, Z.A.; Finlayson, M.; Halpern, B.S.; Jorge, M.A.; Lombana, A.; Lourie, S.A.; et al. Marine ecoregions of the world: A bioregionalization of coastal and shelf areas. Bioscience 2007, 57, 573–583. [Google Scholar] [CrossRef] [Green Version]
- Kerr, R.; Kelly-Borges, M. Biochemical and morphological heterogeneity in the Caribbean sponge Xestospongia muta (Petrosida: Petrosiidae). In Sponges in Time and Space; van Soest, R.W.M., van Kempen, T.M.G., Braekman, J.C., Eds.; Balkema: Rotterdam, The Netherlands, 1994; pp. 65–73. [Google Scholar]
- Liu, N.; Lai, J.; Lyu, C.; Qiang, B.; Wang, H.; Jin, H.; Zhang, L.; Liu, Z. Chemical Space, Scaffolds, and Halogenated Compounds of CMNPD: A Comprehensive Chemoinformatic Analysis. J. Chem. Inf. Model. 2021, 61, 3323–3336. [Google Scholar] [CrossRef] [PubMed]
- Langdon, S.R.; Brown, N.; Blagg, J. Scaffold diversity of exemplified medicinal chemistry space. J. Chem. Inf. Model. 2011, 51, 2174–2185. [Google Scholar] [CrossRef]
- Wiedenmayer, F. Shallow-Water Sponges of the Western Bahamas; Experientia Supplementum; Springer: Berlin/Heidelberg, Germany, 1977. [Google Scholar]
- Kruger, F.; Stiefl, N.; Landrum, G.A. rdScaffoldNetwork: The Scaffold Network Implementation in RDKit. J. Chem. Inf. Model. 2020, 60, 3331–3335. [Google Scholar] [CrossRef] [PubMed]
- Jeh, G.; Widom, J. Simrank: A measure of structural-context similarity. In Proceedings of the 8th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Edmonton, AB, Canada, 23–26 June 2002; pp. 538–543. [Google Scholar]
- OSIRIS Property Explorer. 2021. Available online: https://www.organic-chemistry.org/prog/peo/ (accessed on 21 March 2021).
- Gonzalez-Medina, M.; Medina-Franco, J.L. Platform for Unified Molecular Analysis: PUMA. J. Chem. Inf. Model. 2017, 57, 1735–1740. [Google Scholar] [CrossRef]
- DIFACQUIM: Computer-Aided drug design at UNAM. 2021. Available online: https://www.difacquim.com/d-tools/ (accessed on 12 March 2021).
- Maggiora, G.M.; Bajorath, J. Chemical space networks: A powerful new paradigm for the description of chemical space. J. Comput. Aided Mol. Des. 2014, 28, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Sun, M.; Bennani, Y.L.; Gopalakrishnan, S.M.; Witte, D.G.; Miller, T.R.; Krueger, K.M.; Browman, K.E.; Thiffault, C.; Wetter, J.; et al. The alkaloid Conessine and Analogues as potent Histamine H3 Reseptor Antagonists. J. Med. Chem. 2008, 51, 5423–5430. [Google Scholar] [CrossRef]
- Nodwell, M.; Zimmerman, C.; Roberge, M.; Andersen, R.J. Synthetic analogues of the microtubule-stabilizing sponge alkaloid ceratamine A are more active than the natural product. J. Med. Chem. 2010, 53, 7843–7851. [Google Scholar] [CrossRef]
- Smith, S.E.; Dello Buono, M.C.; Carper, D.J.; Coleman, R.S.; Day, B.W. Structure elucidation of phase I metabolites of the microtubule perturbagens: Ceratamines A and B. J. Nat. Prod. 2014, 77, 1572–1578. [Google Scholar] [CrossRef]
- Pan, X.; Tao, L.; Ji, M.; Chen, X.; Liu, Z. Synthesis and cytotoxicity of novel imidazo[4,5-d]azepine compounds derived from marine natural product ceratamine A. Bioorg. Med. Chem. Lett. 2018, 28, 866–868. [Google Scholar] [CrossRef]
- Pahwa, S.; Kaur, S.; Jain, R.; Roy, N. Structure based design of novel inhibitors for histidinol dehydrogenase from Geotrichum candidum. Bioorg. Med. Chem. Lett. 2010, 20, 3972–3976. [Google Scholar] [CrossRef]
- Gao, J.; Caballero-George, C.; Wang, B.; Rao, K.V.; Shilabin, A.G.; Hamann, M.T. 5-OHKF and NorKA, Depsipeptides from a Hawaiian Collection of Bryopsis pennata: Binding Properties for NorKA to the Human Neuropeptide Y Y1 Receptor. J. Nat. Prod. 2009, 72, 2172–2176. [Google Scholar] [CrossRef] [Green Version]
- Fdhila, F.; Vazquez, V.; Luis Sanchez, J.; Riguera, R. DD-Diketopiperazines: Antibiotics Active against Vibrio anguillarum Isolated from Marine Bacteria Associated with Cultures of Pecten maximus. J. Nat. Prod. 2003, 66, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, N.; Zhang, H.; Knudson, S.E.; Slayden, R.A.; Tonge, P.J. Synthesis and SAR studies of 1,4-benzoxazine MenB inhibitors: Novel antibacterial agents against Mycobacterium tuberculosis. Bioorg. Med. Chem. Lett. 2010, 20, 6306–6309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campagnola, G.; Gong, P.; Peersen, O.B. High-throughput screening identification of poliovirus RNA-dependent RNA polymerase inhibitors. Antivir. Res. 2011, 91, 241–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de F. Cesario, H.P.S.; Silva, F.C.O.; Ferreira, M.K.A.; de Menezes, J.; Dos Santos, H.S.; Nogueira, C.E.S.; de L. Silva, K.S.B.; Hajdu, E.; Silveira, E.R.; Pessoa, O.D.L. Anxiolytic-like effect of brominated compounds from the marine sponge Aplysina fulva on adult zebrafish (Danio rerio): Involvement of the GABAergic system. Neurochem. Int. 2021, 146, 105021. [Google Scholar] [CrossRef] [PubMed]
- Orfanoudaki, M.; Hartmann, A.; Alilou, M.; Mehic, N.; Kwiatkowski, M.; Johrer, K.; Nguyen Ngoc, H.; Hensel, A.; Greil, R.; Ganzera, M. Cytotoxic Compounds of Two Demosponges (Aplysina aerophoba and Spongia sp.) from the Aegean Sea. Biomolecules 2021, 11, 723. [Google Scholar] [CrossRef]
- Oluwabusola, E.T.; Tabudravu, J.N.; Al Maqbali, K.S.; Annang, F.; Perez-Moreno, G.; Reyes, F.; Jaspars, M. Antiparasitic Activity of Bromotyrosine Alkaloids and New Analogues Isolated from the Fijian Marine Sponge Aplysinella rhax. Chem. Biodivers. 2020, 17, 1–9. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.A. Pseudoceratonic Acid and Moloka’iamine Derivatives from the Red Sea Verongiid Sponge Pseudoceratina arabica. Mar. Drugs 2020, 18, 525. [Google Scholar] [CrossRef]
- Chen, M.; Yan, Y.; Ge, H.; Jiao, W.-H.; Zhang, Z.; Lin, H.-W. Pseudoceroximes A-E and Pseudocerolides A-E—Bromotyrosine Derivatives from a Pseudoceratina sp. Marine Sponge Collected in the South China Sea. Eur. J. Org. Chem. 2020, 2020, 2583–2591. [Google Scholar] [CrossRef]
- Tintillier, F.; Moriou, C.; Petek, S.; Fauchon, M.; Hellio, C.; Saulnier, D.; Ekins, M.; Hooper, J.N.A.; Al-Mourabit, A.; Debitus, C. Quorum Sensing Inhibitory and Antifouling Activities of New Bromotyrosine Metabolites from the Polynesian Sponge Pseudoceratina n. sp. Mar. Drugs 2020, 18, 272. [Google Scholar] [CrossRef]
- Moriou, C.; Lacroix, D.; Petek, S.; El-Demerdash, A.; Trepos, R.; Leu, T.M.; Florean, C.; Diederich, M.; Hellio, C.; Debitus, C.; et al. Bioactive Bromotyrosine Derivatives from the Pacific Marine Sponge Suberea clavata (Pulitzer-Finali, 1982). Mar. Drugs 2021, 19, 143. [Google Scholar] [CrossRef]
- Van Eck, N.J.; Waltman, L. CitNetExplorer: A new software tool for analyzing and visualizing citation networks. J. Informetr. 2014, 8, 802–823. [Google Scholar] [CrossRef] [Green Version]
- Massol, F.; Macke, E.; Callens, M.; Decaestecker, E. A methodological framework to analyse determinants of host-microbiota networks, with an application to the relationships between Daphnia magna’s gut microbiota and bacterioplankton. J. Anim. Ecol. 2021, 90, 102–119. [Google Scholar] [CrossRef]
- Fruchterman, T.M.J.; Reingold, E.M. Graph drawing by Force-directed Placement. Softw. Pract. Exp. 1991, 21, 1129–1164. [Google Scholar] [CrossRef]
- Jacomy, M.; Venturini, T.; Heymann, S.; Bastian, M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE 2014, 9, e98679. [Google Scholar] [CrossRef] [PubMed]
- Wilkens, S.J.; Janes, J.; Su, A.I. HierS: Hierarchical Scaffold Clustering using Topological Chemical Graphs. J. Med. Chem. 2005, 48, 3182–3193. [Google Scholar] [CrossRef] [PubMed]
- Galitz, A.; Nakao, Y.; Schupp, P.J.; Wörheide, G.; Erpenbeck, D. A Soft Spot for Chemistry–Current Taxonomic and Evolutionary Implications of Sponge Secondary Metabolite Distribution. Mar. Drugs 2021, 19, 448. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Nguyen, D.D.; Sresht, V.; Mathiowetz, A.M.; Tu, M.; Wei, G.-W. Are 2D fingerprints still valuable for drug discovery? Phys. Chem. Chem. Phys. 2020, 22, 8373–8390. [Google Scholar] [CrossRef] [Green Version]
- Hert, J.; Willett, P.; Wilton, D.J.; Acklin, P.; Azzaoui, K.; Jacoby, E.; Schuffenhauer, A. Comparison of topological descriptors for similarity-based virtual screening using multiple bioactive reference structures. Org. Biomol. Chem. 2004, 2, 3256–3266. [Google Scholar] [CrossRef] [Green Version]

















| Species | mSIA | bSIA | mSIABT | bSIA BT | BT | BTOx | mSIAGdn | bSIAGdn | BTGdn | BTOxGdn | mSIABTOxGdn |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aiolochroia crassa (Hyatt, 1875) | + | + | + | + | + | ||||||
| Anomoianthella popeae (Bergquist, 1980) | + | ||||||||||
| Aplysina aerophoba (Nardo, 1833) | + | + | + | ||||||||
| Aplysina archeri (Higgin, 1875) | + | + | + | ||||||||
| Aplysina caissara (Pinheiro and Hajdu, 2001) | + | + | |||||||||
| Alplysina cauliformis (Carter, 1882) | + | + | + | + | |||||||
| Aplysina cavernicola (Vacelet, 1959) | + | + | + | ||||||||
| Aplysina fistularis (Pallas, 1766) | + | + | + | + | + | ||||||
| Aplysina fulva (Pallas, 1766) | + | + | + | + | + | ||||||
| Aplysina gerardogreeni (Gomez and Bakus, 1992) | + | ||||||||||
| Aplysina insularis (Duchassaing and Michelotti, 1864) | + | + | + | + | |||||||
| Aplysina lactuca (Pinheiro, Hajdu and Custodio, 2007) | + | + | |||||||||
| Aplysina lacunosa (Lamarck, 1814) | + | + | + | + | + | + | + | + | |||
| Aplysina laevis (=Pseudoceratina durissima Carter, 1885) | |||||||||||
| Aplysina solongeae (Pinheiro, Hajdu and Custodio, 2007) | + | + | |||||||||
| Aplysina sp. (Nardo, 1834) | + | + | + | + | + | + | |||||
| Aplysina thiona (=Aiolochroia thiona Laubenfels, 1930) | + | ||||||||||
| Aplysinella rhax (de Laubenfels, 1954) | |||||||||||
| Aplysinella sp. (Bergquist, 1980) | + | + | + | + | |||||||
| Aplysinella strongylata (Bergquist, 1980) | |||||||||||
| Hexadella dedritifera (Topsent, 1913) | + | ||||||||||
| Hexadella indica (Dendy, 1905) | + | ||||||||||
| Hexadella sp. (Topsent, 1896) | + | + | |||||||||
| Ianthella basta (Pallas, 1766) | + | ||||||||||
| Ianthella flabelliformis (Linnaeus, 1759) | + | ||||||||||
| Ianthella quadrangulata (Bergquist and Kelly-Borges, 1995) | + | ||||||||||
| Ianthella reticulata (Bergquist and Kelly-Borges, 1995) | |||||||||||
| Ianthella sp. (Gray, 1869) | + | + | + | ||||||||
| Pseudoceratina arabica (Keller, 1889) | + | ||||||||||
| Pseudoceratina crassa (=Aiolochroia crassa Hyatt, 1875) | + | + | + | + | + | + | |||||
| Pseudoceratina durissima (Carter, 1885) | + | + | |||||||||
| Pseudoceratina purea (=P. purpurea Carter, 1880) | + | + | + | + | + | + | + | ||||
| Pseudoceratina purpurea (Carter, 1880) | + | + | + | + | + | + | + | + | |||
| Pseudoceratina sp. (Carter, 1885) | + | + | + | + | + | + | + | + | + | + | |
| Pseudoceratina verrucosa (Bergquist, 1995) | + | + | + | + | + | + | + | + | + | ||
| Suberea clavata (Pulitzer-Finali, 1982) | + | + | + | ||||||||
| Suberea creba (Bergquist, 1995) | + | + | + | ||||||||
| Suberea ianthelliformis (Lendenfeld, 1888) | + | + | |||||||||
| Suberea mollis (Row, 1911) | + | + | + | + | |||||||
| Suberea praetensa (Row, 1911) | + | ||||||||||
| Suberea sp. (Bergquist, 1995) | + | + | + | ||||||||
| Verongula gigantea (Hyatt, 1875) | + | + | + | + | + | ||||||
| Verongula rigida (Esper, 1794) | + | + | + | + | + | + | |||||
| Verongula sp. (Verrill, 1907) | + | + | + |
| Genus | Natural Products (M) | Murcko Scaffolds (N) | Singleton Scaffolds (Nsing) | Diversity (N/M) | Novelty (Nsing/M) |
|---|---|---|---|---|---|
| Aiolochroia | 15 | 8 | 0 | 0.533 | 0 |
| Anomoianthella | 1 | 1 | 0 | 1 | 0 |
| Aplysina | 140 | 44 | 20 | 0.314 | 0.143 |
| Aplysinella | 63 | 19 | 8 | 0.301 | 0.127 |
| Hexadella | 12 | 9 | 0 | 0.75 | 0 |
| Ianthella | 95 | 29 | 20 | 0.305 | 0.211 |
| Pseudoceratina | 232 | 67 | 35 | 0.289 | 0.151 |
| Suberea | 115 | 47 | 24 | 0.409 | 0.209 |
| Verongula | 51 | 28 | 13 | 0.549 | 0.255 |
| PC1 | PC2 | PC3 | PC4 | PC5 | |
|---|---|---|---|---|---|
| Eigenvalues | 3.8916 | 1.2130 | 0.5068 | 0.2505 | 0.0841 |
| Proportion of variance | 0.649 | 0.202 | 0.084 | 0.014 | 0.009 |
| Cumulative proportion | 0.649 | 0.851 | 0.935 | 0.991 | 1.000 |
| PC1 | PC2 | PC3 | PC4 | PC5 | |
|---|---|---|---|---|---|
| MW | 0.472 | 0.147 | −0.133 | −0.476 | −0.636 |
| TPSA | 0.458 | −0.338 | 0.020 | 0.049 | 0.538 |
| nHBAcc | 0.471 | −0.199 | 0.122 | −0.453 | 0.277 |
| nHBDon | 0.420 | −0.374 | −0.153 | 0.681 | −0.374 |
| nRotB | 0.307 | 0.522 | 0.748 | 0.270 | −0.002 |
| cLogP | 0.273 | 0.642 | −0.619 | 0.172 | 0.299 |
| Compound | Cluster | Drug Score | Genus | MW | TPSA | nHBAcc | nHBDon | nRotB | cLogP |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 0.81 | Aplysina, Suberea | 323.03 | 23.47 | 2 | 1 | 3 | 2.82 |
| 2 | 3 | 0.84 | Aplysina | 337.05 | 12.47 | 2 | 0 | 4 | 3.09 |
| 3 | 3 | 0.79 | Pseudoceratina | 380.12 | 38.49 | 3 | 1 | 7 | 2.63 |
| 4 | 3 | 0.81 | Pseudoceratina | 396.12 | 58.72 | 4 | 2 | 7 | 1.56 |
| 5 | 3 | 0.79 | Pseudoceratina | 452.19 | 61.8 | 4 | 2 | 9 | 2.42 |
| 6 | 3 | 0.78 | Pseudoceratina | 407.1 | 49.77 | 4 | 1 | 7 | 1.89 |
| 7 | 3 | 0.75 | Aplysina, Suberea, Pseudoceratina | 408.18 | 15.71 | 3 | 0 | 8 | 3.25 |
| 8 | 2 | 0.77 | Aplysina, Pseudoceratina | 460.13 | 99.6 | 6 | 4 | 7 | 2.56 |
| 9 | 2 | 0.88 | Aplysinella, Pseudoceratina, Verongula | 381.23 | 99.6 | 5 | 3 | 7 | 1.84 |
| 10 | 2 | 0.81 | Aplysinella | 323.23 | 118.2 | 5 | 3 | 6 | 0.14 |
| 11 | 2 | 0.8 | Pseudoceratina | 437.13 | 82.95 | 5 | 3 | 8 | 2.39 |
| 12 | 2 | 0.77 | Pseudoceratina | 475.14 | 118.83 | 7 | 5 | 7 | 1.84 |
| 13 | 2 | 0.77 | Aplysinella | 379.23 | 127.43 | 5 | 3 | 7 | 1.19 |
| 14 | 2 | 0.77 | Pseudoceratina | 460.13 | 99.6 | 5 | 3 | 7 | 2.56 |
| 15 | 2 | 0.77 | Pseudoceratina | 434.33 | 96.94 | 5 | 3 | 10 | 3.16 |
| 16 | 2 | 0.75 | Hexadella | 475.14 | 125.62 | 6 | 4 | 7 | 2.25 |
| 17 | 2 | 0.77 | Pseudoceratina | 461.11 | 136.62 | 6 | 5 | 6 | 1.98 |
| 18 | 2 | 0.89 | Pseudoceratina | 367.2 | 110.6 | 5 | 4 | 6 | 1.56 |
| 19 | 2 | 0.75 | Pseudoceratina | 475.14 | 125.62 | 7 | 5 | 7 | 2.25 |
| 20 | 16 | 0.96 | Ianthella | 254.29 | 63.19 | 3 | 2 | 1 | 1.01 |
| 21 | 16 | 0.91 | Verongula | 333.19 | 63.19 | 3 | 2 | 1 | 1.74 |
| 22 | 7 | 0.93 | Suberea | 201.61 | 66.4 | 3 | 2 | 0 | −0.57 |
| 23 | 7 | 0.93 | Aplysina | 201.61 | 66.4 | 3 | 2 | 0 | −0.57 |
| 24 | 21 | 0.79 | Pseudoceratina | 454.12 | 75.08 | 5 | 2 | 3 | 1.94 |
| 25 | 21 | 0.77 | Pseudoceratina | 468.15 | 66.29 | 5 | 1 | 3 | 2.19 |
| 26 | 42 | 0.94 | Pseudoceratina | 126.11 | 61.69 | 3 | 2 | 0 | 0.06 |
| 27 | 50 | 0.95 | Suberea | 154.17 | 97.92 | 4 | 5 | 2 | −1.28 |
| 28 | 39 | 0.98 | Pseudoceratina, Verongula | 137.14 | 46.92 | 2 | 1 | 0 | 0.38 |
| 29 | 19 | 0.81 | Verongula | 346.07 | 19.03 | 1 | 1 | 3 | 3.2 |
| 30 | 43 | 0.79 | Pseudoceratina | 185.23 | 53.6 | 3 | 2 | 2 | 0.02 |
| 31 | 66 | 0.97 | Ianthella | 256.31 | 63.19 | 3 | 2 | 2 | 0.8 |
| 32 | 57 | 0.97 | Suberea | 244.29 | 49.41 | 2 | 1 | 2 | 0.85 |
| Order | Family | Genera (Total NPs) |
|---|---|---|
| Verongiida | Aplysinellidae | Aplysinella (63) Patriciaplysina (0) Suberea (115) |
| Aplysinidae | Aiolochroia (15) Aplysina (140) Verongula (51) | |
| Ernstillidae | Ernstilla (0) | |
| Ianthellidae | Anomoianthella (1) Hexadella (12) Ianthella (95) Vansoestia (0) | |
| Pseudoceratinidae | Pseudoceratina (232) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lever, J.; Brkljača, R.; Rix, C.; Urban, S. Application of Networking Approaches to Assess the Chemical Diversity, Biogeography, and Pharmaceutical Potential of Verongiida Natural Products. Mar. Drugs 2021, 19, 582. https://doi.org/10.3390/md19100582
Lever J, Brkljača R, Rix C, Urban S. Application of Networking Approaches to Assess the Chemical Diversity, Biogeography, and Pharmaceutical Potential of Verongiida Natural Products. Marine Drugs. 2021; 19(10):582. https://doi.org/10.3390/md19100582
Chicago/Turabian StyleLever, James, Robert Brkljača, Colin Rix, and Sylvia Urban. 2021. "Application of Networking Approaches to Assess the Chemical Diversity, Biogeography, and Pharmaceutical Potential of Verongiida Natural Products" Marine Drugs 19, no. 10: 582. https://doi.org/10.3390/md19100582
APA StyleLever, J., Brkljača, R., Rix, C., & Urban, S. (2021). Application of Networking Approaches to Assess the Chemical Diversity, Biogeography, and Pharmaceutical Potential of Verongiida Natural Products. Marine Drugs, 19(10), 582. https://doi.org/10.3390/md19100582