Abstract
Scytonemin is a promising UV-screen and antioxidant small molecule with commercial value in cosmetics and medicine. It is solely biosynthesized in some cyanobacteria. Recently, its biosynthesis mechanism has been elucidated in the model cyanobacterium Nostoc punctiforme PCC 73102. The direct precursors for scytonemin biosynthesis are tryptophan and p-hydroxyphenylpyruvate, which are generated through the shikimate and aromatic amino acid biosynthesis pathway. More upstream substrates are the central carbon metabolism intermediates phosphoenolpyruvate and erythrose-4-phosphate. Thus, it is a long route to synthesize scytonemin from the fixed atmospheric CO2 in cyanobacteria. Metabolic engineering has risen as an important biotechnological means for achieving sustainable high-efficiency and high-yield target metabolites. In this review, we summarized the biochemical properties of this molecule, its biosynthetic gene clusters and transcriptional regulations, the associated carbon flux-driving progresses, and the host selection and biosynthetic strategies, with the aim to expand our understanding on engineering suitable cyanobacteria for cost-effective production of scytonemin in future practices.
1. Introduction
Cyanobacteria are photoautotrophic prokaryotes that can directly convert CO2 into organic compounds using solar energy. Cyanobacteria possess relatively small genomes and can grow with minimal nutrient requirement. Engineering cyanobacteria offers an attractive approach to drive carbon flux to the biosynthesis of fuels, chemicals, medicines, plant secondary metabolites, and other value-added products [1,2,3,4,5].
Cyanobacteria naturally grow in diverse habitats including fresh water, oceans, alkaline waters, cold deserts, polar regions and hot springs. To cope with the extreme and changeable environments, cyanobacteria have developed a wide range of adaptive mechanisms. Some cyanobacteria are directly exposed to strong sunlight in native habitats, such as those dwelling in intertidal mats or on desert soil surfaces [6,7,8]. Intense ultraviolet (UV) radiation can cause cell damage by both direct effects on proteins and nucleic acids and indirect effects via the induced reactive oxygen species [9]. To eliminate the deleterious effects of UV radiation, one of the most important strategies in some cyanobacteria is to generate UV-absorbing compounds, such as mycosporine-like amino acids (MAAs) and scytonemin [10]. Both compounds are located in the exopolysaccharide matrix and constitute the first line of defense against UV penetration and subsequent damage [11,12,13]. Multiple estimates from the fossil record of cyanobacteria and relaxed molecular clock models suggest a minimum age for the evolutionary advent of scytonemin at around 2.1 billion years [14].
Scytonemin is naturally biosynthesized in some cyanobacteria and has a great prospect in cosmetic and biomedical application. However, as far as we know, enhanced production of this compound in cyanobacteria via metabolic engineering has not been reported. To exploit the maximal potential of engineering cyanobacterial hosts for scytonemin biosynthesis is the next important step. In this review, we summarized the research progresses regarding scytonemin, placing emphasis on its biosynthetic route and the metabolic engineering technologies from cyanobacteria and other microorganisms, with the aim to expand our understanding on metabolically engineering of cyanobacterial hosts for cost-effective production of scytonemin in future practices.
2. Functional Roles of Scytonemin
2.1. Molecular Structure and UV Absorption
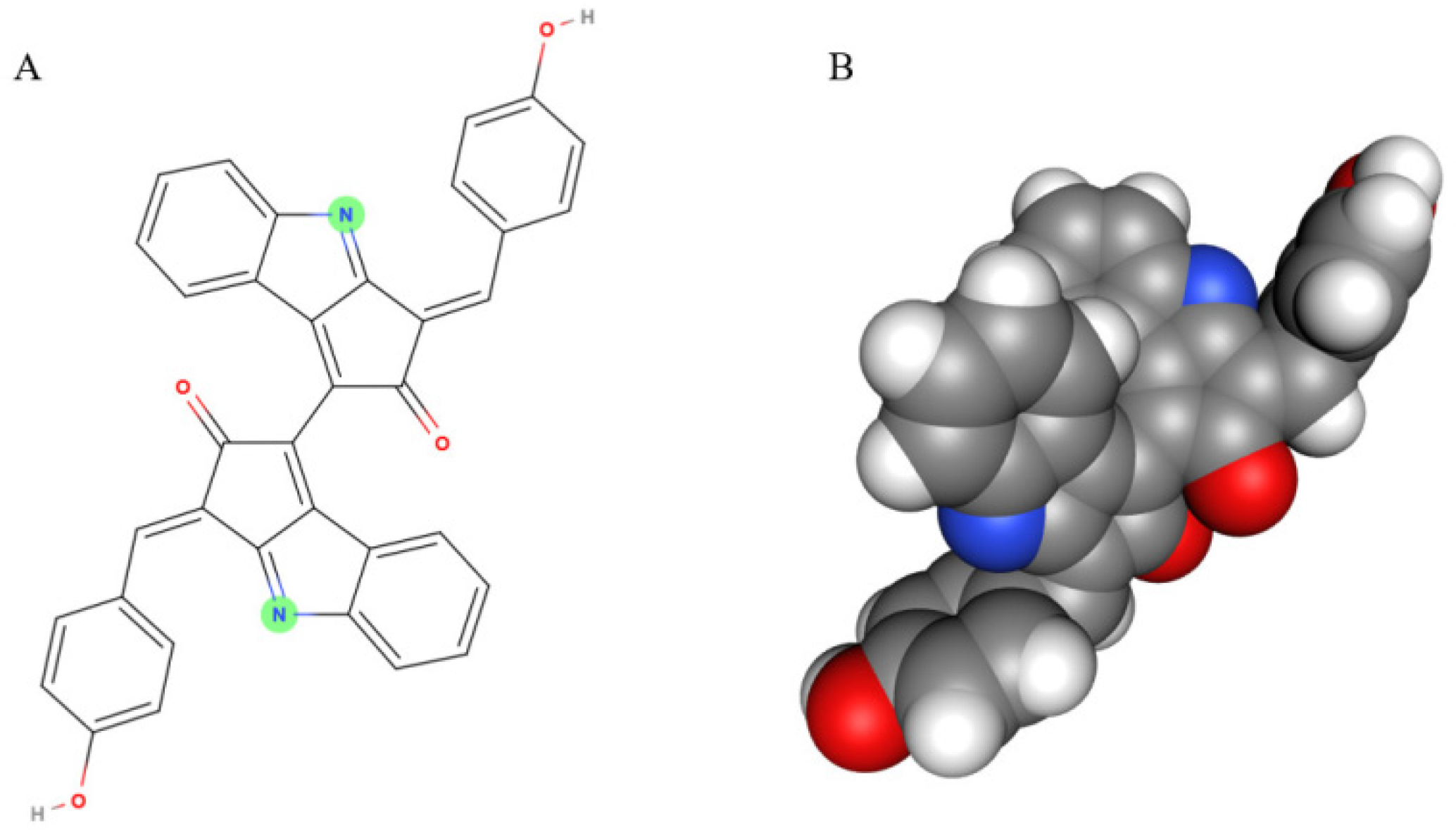
Scytonemin is a lipid soluble and yellow-brown pigment exclusively found in some cyanobacteria [11]. This compound is a dimer composed of indolic and phenolic subunits symmetrically connected through a carbon–carbon bond (Figure 1). Scytonemin has oxidized (yellow; Mw 544 Da) and reduced (red; Mw 546 Da) forms and also three derivatives, dimethoxyscytonemin, tetramethoxyscytonemin, and scytonin [15,16].
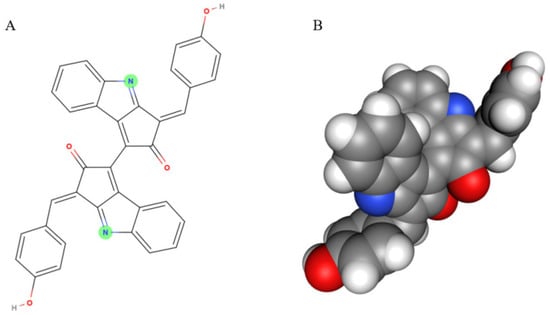
Figure 1.
The scytonemin molecule (A) and its 3D model (B). Two images are generated by the MolView program (https://molview.org/, accessed on 20 August 2020).
The solar UV radiation can be divided into UV-A (315–400 nm), UV-B (280–315 nm), and UV-C (100–280 nm). UV-C does not reach the earth’s surface owing to stratospheric ozone. The remaining UV that reaches the surface of the earth consists of 95% UV-A and 5% UV-B [17]. Scytonemin can absorb the three UV radiations, but mainly the UV-A, and can prevent about 90% of UV-A from entering the cells [11,18,19]. It can also provide substantial protection against UV-B and UV-C damage [20,21]. The absorption in the UV-C range implies that scytonemin is an ancient pigment possibly evolved during the Precambrian [20]. Scytonemin has an in vivo maximum absorption at 370 nm and an in vitro maximum absorption at 384 nm [11,22].
2.2. Solubility and Stability
Scytonemin does not dissolve in water, but dissolves readily in lipids. In liquid suspension culture of Nostoc flagelliforme, yellow-brown scytonemin was observed to be tightly associated with the released exopolysaccharides [23]. Scytonemin was usually extracted from samples with solvents such as 100% acetone or 100% ethyl acetate [11], 100% acetonitrile [24], 80% tetrahydrofuran [13], and methanol:ethyl acetate (1:1, v/v) [9,21]. In addition, we found scytonemin was soluble in N,N-dimethylformamide and the polyvinylpyrrolidone K30 aqueous solution. Its solubility in polyvinylpyrrolidone K30 solution (e.g., 1% aqueous solution) may facilitate its reaction with other water-soluble compounds.
It was reported that scytonemin showed high stability against the abiotic stress conditions of UV-B (0.78 W·m−2), heat (60 °C), and strong oxidizing agent (0.25% H2O2) for 1 h [25]. After continuous exposure to 55 W·m−2 visible light or 5 W·m−2 UV-A radiation for 50 days, the original scytonemin content in dry Nostoc punctiforme samples remained at 93% and 84%, respectively, while chlorophyll α content decreased to an undetectable level by day 10 under both conditions [26]. Its stability was also evidenced by its preservation in the lake sedimentary record [27]. The abundant preservation of scytonemin in deep sea sediments indicates that scytonemin is resistant to degradation during erosion and transport [28]. For this reason, scytonemin is regarded as one of the key biomarkers in both paleoclimatological reconstructions and in terrestrial extreme environments [29,30,31].
2.3. Cellular Distribution and Content
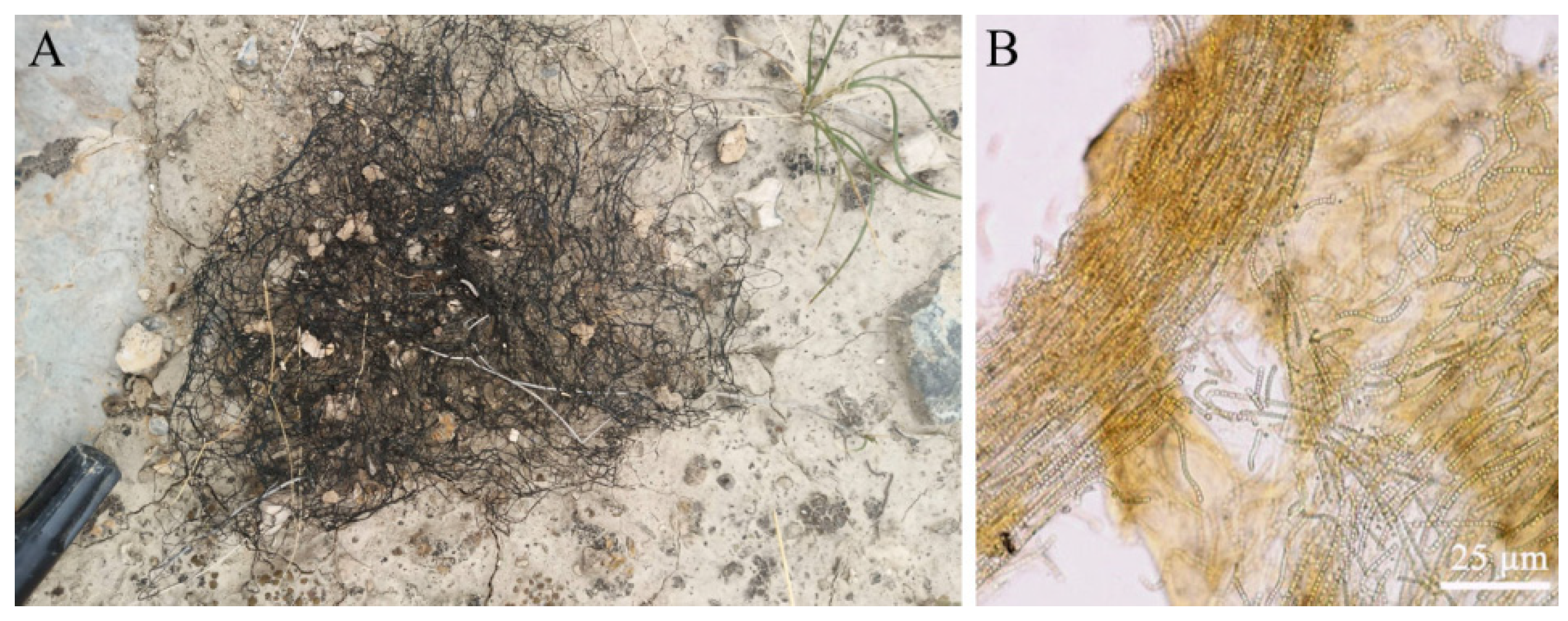
Scytonemin is distributed in the exopolysaccharide sheath of more than 300 cyanobacteria, including Nostoc, Scytonema, Calothrix, Lyngbya, Rivularia, and Chlorogloeopsis [30,32,33]. It is predominantly located in the surface layer of samples. Light microscopy sections of N. flagelliforme clearly showed the distribution of yellow-brown scytonemin in the peripheral region of exopolysaccharide sheath [13]. Figure 2 shows the distribution of scytonemin in N. flagelliforme filaments. CARS microscopy of scytonemin in laboratory-cultivated Nostoc commune further imaged its distribution in the surface layer of the sheath after UV-A radiation [34]. In terrestrial cyanobacterial mats or crusts, cyanobacterial upper layer usually shows high amounts of scytonemin, conferring protection to the cells beneath against UV damage [7,30,35,36].
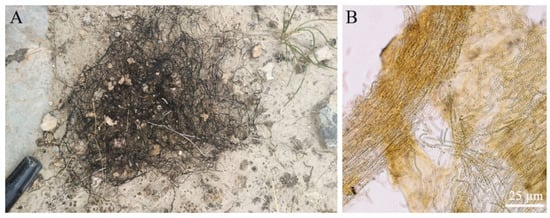
Figure 2.
The filamentous colonies (filaments) of Nostoc flagelliforme in native habitats (A) and the microscopic observation of a crushed filament (B). Yellow-brown scytonemin is distributed throughout a filament as shown in (B).
Scytonemin could constitute up to 5% of cyanobacterial dry weight in the cultured organisms [37]. In some examined cyanobacteria, the contents of scytonemin ranged from 0.08 to 7.98% of dry weight [6]. In N. commune, dry weight percentages were estimated to be in an approximate ratio of 10:1:0.1 for exopolysaccharide, MAAs, and scytonemin, respectively [38].
2.4. Biochemical, Medical and Ecological Values
Scytonemin has received increasing attention for its biochemical and ecological roles owing to UV-absorption and antioxidant functions, and also for its potential application in cosmetic and pharmaceutical industry as an active molecule. It showed strong antioxidant activity and slow radical scavenging activity in the DPPH (2,2-diphenyl-1-picrylhydrazyl) or ABTS (2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid)) assay [24,25,39]. The dose dependent antioxidant activity of scytonemin was 22% and 52% at concentrations of 0.4 and 0.8 mM, respectively, as compared to that of 0.5 mM ascorbic acid used as a positive control [21]. As a natural sunscreen compound, its application for skin protection has attracted great interest among dermatologists and in cosmetics [40,41].
Medically, scytonemin possesses anti-inflammatory and anti-proliferative properties. It inhibits skin inflammation by blocking the expression of inflammatory mediators, partially through down-regulation of NF-κB activity [42]. Moreover, scytonemin inhibits proliferation of human fibroblasts and endothelial cells by selectively inhibiting kinases [43]. The human polo-like kinase 1 that scytonemin inhibits serves as an attractive target of anti-cancerous drugs [44,45]. Scytonemin can also inhibit the activity of other kinases such as Myt1, cyclin-dependent kinase 1 (cyclin B), checkpoint kinase 1, and protein kinase C [46]. In addition, reduced scytonemin can suppress the human T-lymphoid Jurkat cell growth [47], and the LPS/IFNc-stimulated NO production in murine macrophage RAW264 cells [48]. Therefore, scytonemin is a promising small-molecule drug.
Scytonemin possesses important ecological functions and thus shows the great potential in environmental management. It is often found in the upper layers of microbial mats that thrive in areas exposed to intense sunlight. Its photoprotective function is involved in the following mechanisms: UV-absorption and antioxidant functions; reducing the formation of reactive oxygen species and thymine dimers; alleviating the photosynthetic inhibition; heat dissipation from absorbed UV radiation to increase soil surface temperature [22,25,49,50]. In addition, scytonemin has been proposed to confer cyanobacteria with high tolerance to desiccation [26]. One of the mechanisms may be its potential role in stabilizing the exopolysaccharide matrix [8]. Scytonemin may also interact with other extracellular components in the matrix, such as WspA protein, to indirectly function in the desiccation resistance [38]. Scytonemin and iron can form the iron-complexes that possibly facilitate the survival of cyanobacterial colonies on sandstone rocks [51,52].
3. Abiotic Factors Involved in the Induced Biosynthesis of Scytonemin
Although scytonemin is preferentially induced in cyanobacteria by UV radiation, other environmental and nutrient factors have also been reported to incur its induction. In contrast to the strong induction by UV-A radiation, blue, green, or red light has no significant effect on the induction [11]. In Lyngbya aestuarii, the content of scytonemin was lowest at zero salinity level and increased with increasing salinity in the culture medium [53]. In conjunction with UV-A radiation, both high temperature and photooxidative conditions caused an increase of scytonemin production in Chroococcidiopsis, while increased salt concentration inhibited its scytonemin synthesis [54]. The comparison of scytonemin biosynthesis in three desiccation-tolerant cyanobacterial strains, N. punctiforme PCC 73102, Chroococcidiopsis CCMEE 5056, and Chroococcidiopsis CCMEE 246, showed that in presence of UV-A radiation, the former two produced more scytonemin when experiencing periodic desiccation than when continuously hydrated [26]. The production of scytonemin in N. punctiforme PCC 73102 increased 3–7 times in a diazotrophic culture as compared to the non-diazotrophic culture [55]. In nitrogen-deficiency medium, the N. flagelliforme culture treated with 0.2 W·m−2 UV-B for 30 days produced nearly 5 times more scytonemin than the non-treated samples. It was also observed that a light of 60–90 μmol photons·m−2·s−1 induced the production of scytonemin in the exopolysaccharides of N. flagelliforme culture [23].
The exoploysaccharide plays a major role in protecting cells from environmental stresses, and its biosynthesis is similarly induced by salt stress, high temperature, high light, and UV-B radiation [56]. Considering the localization of scytonemin in the polysaccharide-rich extracellular matrix, it is an interesting question whether the biosynthesis of both compounds is simultaneously regulated or tightly coupled. In contrast to scytonemin, the exopolysaccharide is more widely biosynthesized in prokaryotic microorganisms with seemingly more complex mechanisms [56,57]. Without UV induction, scytonemin was rarely biosynthesized in the N. flagelliforme culture [58], but the exopolysaccharide can still be massively biosynthesized [23]. Using a non-scytonemin-producing N. punctiforme mutant, it was found that the exopolysaccharide production was more closely related to oxidative stress than UV-A radiation [59]. Thus, ascertaining the UV-specific cis-regulatory element should be crucial for developing preferential biosynthesis of scytonemin in engineered cyanobacteria.
4. Genes for Scytonemin Biosynthesis and Secretion
4.1. Gene Clusters for the Direct Biosynthesis
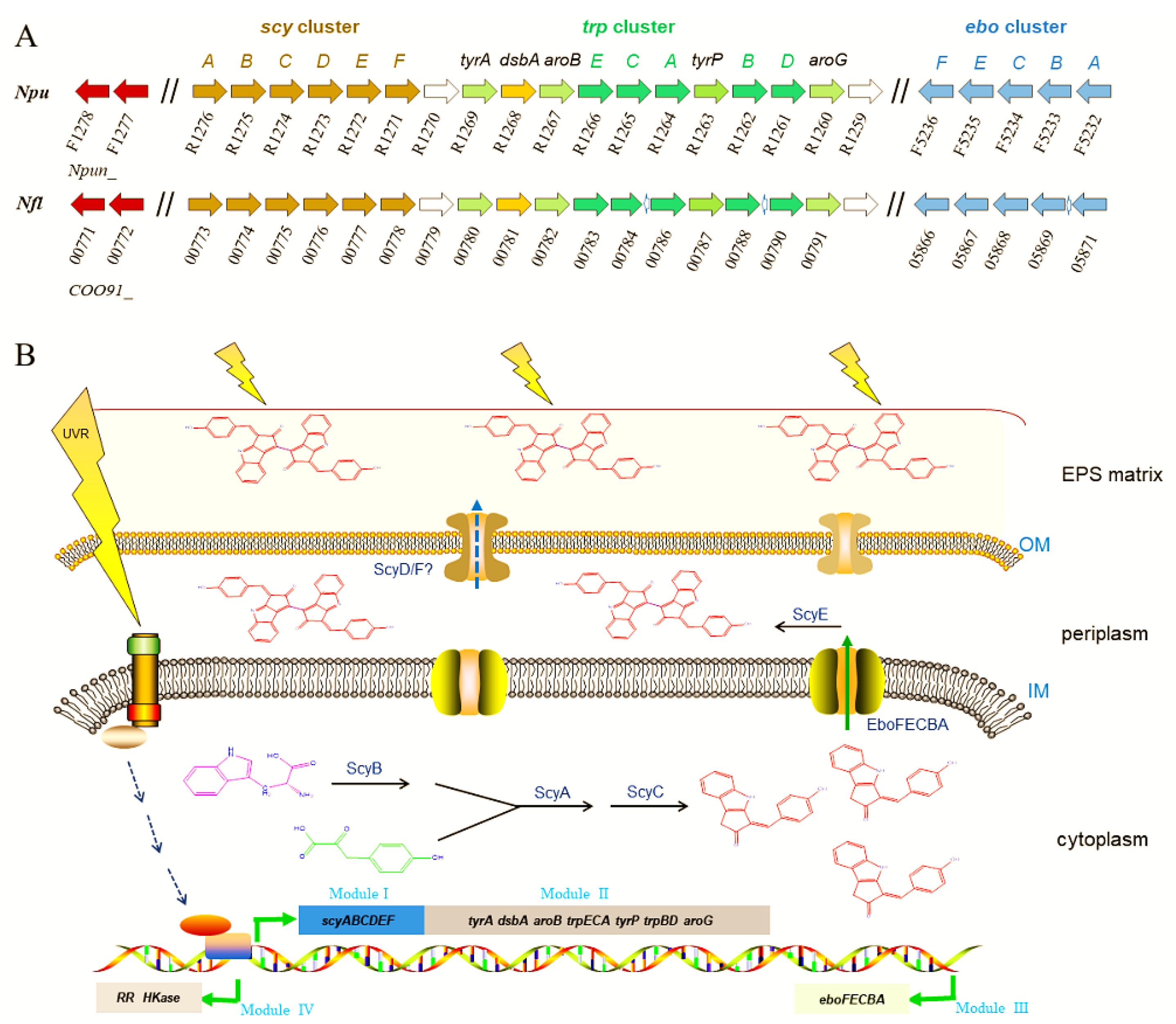
Elucidating the genes for scytonemin biosynthesis is essential for the development of metabolically engineered strains for commercial production of scytonemin. The scytonemin biosynthetic gene cluster (scy cluster) was first identified through the analysis of a transposon mutagenesis-generated non-scytonemin-producing mutant of N. punctiforme ATCC 29133 (PCC 73102) [60]. This mutation was located within a cluster of 18 open reading frames (Npun_R1276–R1259) that were all transcribed in the same direction. Currently, the entire pathway of scytonemin biosynthesis has been basically elucidated, as shown in Figure 3. The genes involved in the biosynthesis, secretion, and regulation processes of scytonemin can be distinguished as four functional sets (or four modules) [61,62,63,64]. Among them, the above-mentioned 18 genes for scytonemin biosynthesis can be divided into two modules: Module I (scyABCDEF, Npun_R1276–R1271) and module II (Npun_R1270–R1259). In module I, ScyABC proteins are involved in catalyzing the formation of the scytonemin monomer within the cells [61], while ScyDEF are thought to be responsible for oxidation and dimerization of the monomer to form the scytonemin dimer in the periplasm [63]. ScyB (Npun_R1275), a leucine dehydrogenase homolog, catalyzes the oxidative deamination of tryptophan (Trp) substrate to yield indole-3-pyruvic acid. ScyA (Npun_R1276), a thiamin diphosphate-dependent enzyme, then mediates the acyloin coupling of indole-3-pyruvic acid and p-hydroxyphenylpyruvate (p-Hpp), to afford a labile β-ketoacid compound. Subsequently, ScyC (Npun_R1274) catalyzes the cyclization and decarboxylation of the β-ketoacid compound to form a ketone, which is one (auto)oxidation state away from what is called the scytonemin monomer [61,65]. ScyE (Npun_R1272) is essential for catalyzing the final oxidative dimerization of the scytonemin monomer to scytonemin in the periplasm, while ScyD (Npun_R1273) and ScyF (Npun_R1271) seem to not be essential for this catalytic process [63]. Since scytonemin is located in the cyanobacterial exopolysaccharide matrix, particularly its peripheral region [13], an unanswered question is how scytonemin is transported or diffused from the periplasm to the exopolysaccharide matrix for final location. Whether ScyD and/or ScyF play a role in this process remains an open topic.
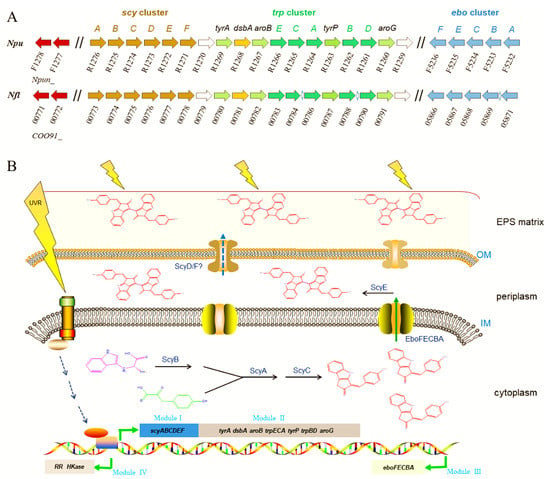
Figure 3.
Homologous comparison of scytonemin biosynthesis-related genes between two genetically close Nostoc species (A) and schematic illustration of UV-induced scytonemin biosynthesis processes in cyanobacteria (B). Npu, Nostoc punctiforme; Nfl, Nostoc flagelliforme. The gene clusters are distinguished as four modules. Module I, scyABCDEF, responsible for scytonemin monomer and dimer biosynthesis. Module II, tyrA~aroG, responsible for biosynthesis of aromatic amino acid substrates. Module III, eboFECBA, responsible for the export of scytonemin monomer. Module IV, responsible for signal transduction in response to UV radiation. Abbreviations: IM, inner membrane; OM, outer membrane; EPS matrix, exopolysaccharide matrix.
Scytonemin synthesis requires the aromatic amino acids as building blocks. The genes in module II are predicted to be involved in the biosynthesis of aromatic amino acid substrates [60,66,67]. Except Npun_R1268 (encoding a DSBA oxidoreductase) and Npun_R1269 (encoding a prephenate dehydrogenase), other genes in Module II have at least two homologous genes. The redundancy of the aromatic amino acid biosynthetic genes should be beneficial for providing more available substrates for scytonemin biosynthesis.
The eboABCEF gene cluster (Npun_F5232–5236), module III, is responsible for translocation of the scytonemin monomer to the periplasm [64]. They function together, and the absence of any one of them prevents this translocation, leading to cytoplasmic accumulation of the scytonemin monomer [64]. The ebo cluster is widespread and conserved in some cyanobacteria, Ceustigmatophytes, Bacteroidetes, and Leptospira [68]. An engineered Escherichia coli strain that harbored the scytonemin biosynthetic genes could only produce the scytonemin monomer, not the final scytonemin dimer [69]. We speculate that the lack of EboABCEF complex might be a potential reason for this defect in E. coli. The ebo cluster was also found in N. flagelliforme (COO91_05871–05866) and non-scytonemin-producing Nostoc PCC 7120 (all0421–0415). In Nostoc PCC 7120, the incompleteness of scytonemin biosynthetic genes, lacking the homologous genes of Npun_R1268 and Npun_R1263, might be one of the key reasons for the incapability in scytonemin biosynthesis in this particular strain.
Two genes that encode a histidine kinase (Npun_F1277) and a response regulator (Npun_F1278), module IV, serve as a two-component regulatory system responsible for multiple biological processes including the biosynthesis of scytonemin [70]. The deletion of Npun_F1278 led to the incapability in scytonemin biosynthesis under either UV-A radiation or white light [70]. The transcription of scyA, the first gene in scy cluster, was measured to be strongly inhibited in the Npun_F1278 mutant strain. Both Npun_F1277 and Npun_F1278 have multiple paralogs in N. punctiforme (e.g., Npun_R3198 and Npun_F4909). Biochemical identification of the specific interaction between the response regulator Npun_F1278 and the transcriptional regulation elements of scy cluster as well as other environmental factors-associated effects need to be further elucidated.
4.2. Transcriptional Regulation under UV-A/B Radiation
Various abiotic stresses are involved in the induction of scytonemin biosynthesis. UV-A radiation is the most effective inducer. Understanding the transcription of scytonemin production-associated genes in response to UV-A is important for strategy design in metabolic engineering. Under 0.64 mW·cm−2 UV-A radiation for 48 h, transcriptional levels of all the genes in module I (Npun_R1276–R1271) and module II (Npun_R1270–R1259) increased 44~82% in N. punctiforme [67]. During the treatment of 0.5 W·cm2 UV-A for 7 days, a dynamic transcription was observed for these genes, mostly reaching a maximum at 48 h, with transcriptional levels elevating 2.8 to 5.2 folds over the control [66]. AroG and AroB (respectively encoded by Npun_R1260 and Npun_R1267) are two major points of regulation in the shikimate pathway [71]. These clues imply that all the six genes of module I for direct scytonemin biosynthesis and the two genes of module II involved in the biosynthesis of aromatic amino acids are critical targets for transcriptional modulation in metabolic engineering. In addition, all the five genes (Npun_F5232–F5236) of module III showed a generally subdued transcriptional induction upon the UV-A radiation, with increases ranging from only 1.9 to 3 folds [66]. Both genes (Npun_F1277 and Npun_F1278) of module IV showed reduced or transient transcriptional induction by UV-A radiation [66,67], emphasizing their regulative or signaling role in the biosynthesis of scytonemin.
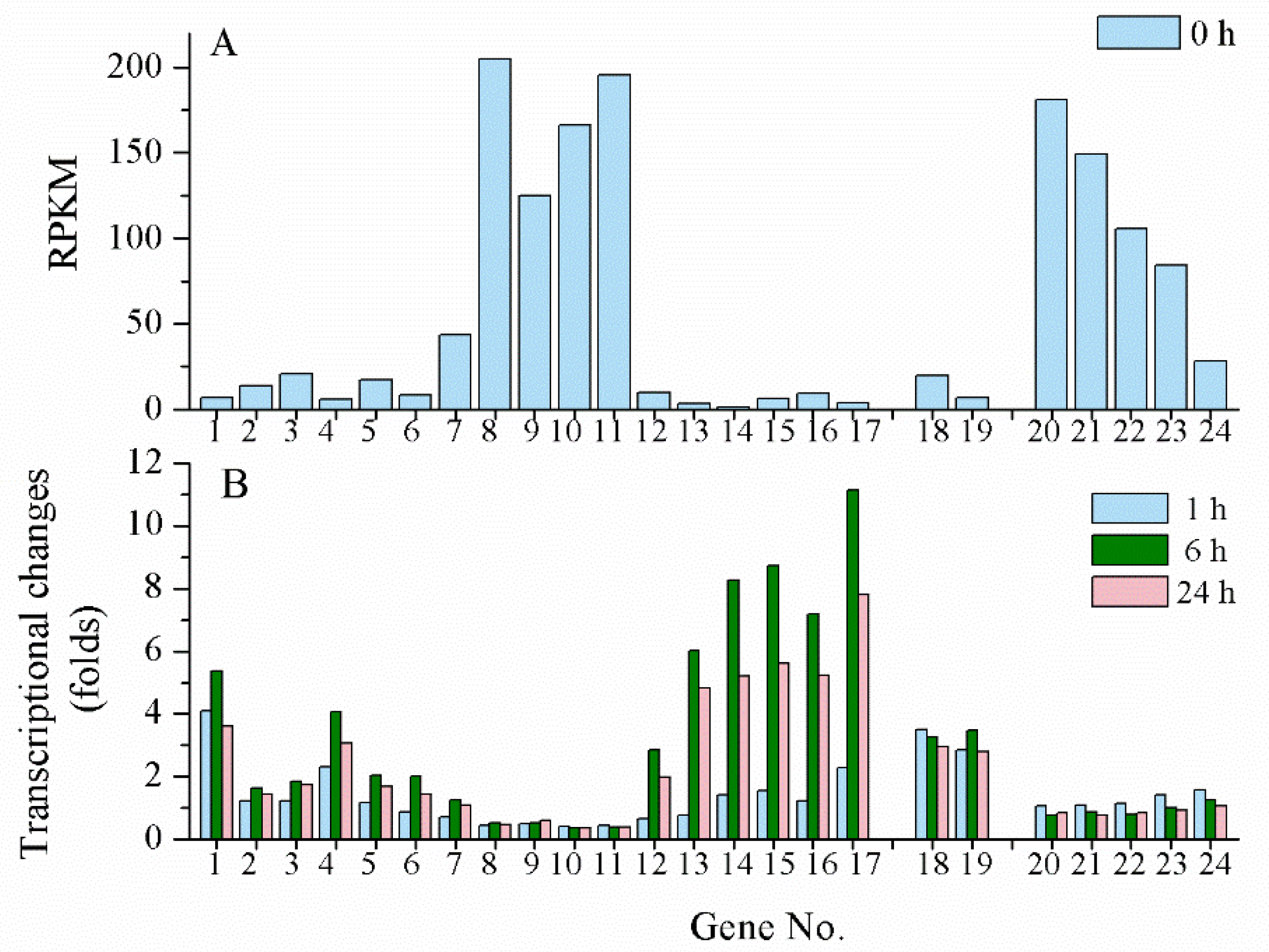
UV-B is another effective inducer for scytonemin biosynthesis. However, its biosynthesis gene clusters in N. flagelliforme exhibited different induction patterns under UV-B radiation according to our transcriptional analysis data (unpublished). When rewetted N. flagelliforme samples were treated by 0.5 W·m2 UV-B for 24 h, the RPKM values of the gene clusters COO91_00783–00779 (similar to Npun_R1266–R1270 in module II) and COO91_05871–05866 (similar to Npun_F5232–F5236 of module III) were much higher than those of other genes before the UV-B treatment (0 h) (Figure 4A), implying their high basal expression levels. However, both clusters were not induced or even inhibited by UV-B (Figure 4B). The gene cluster COO91_00778–00773 (similar to Npun_R1271–R1276 of module I) was highly induced in transcriptional level by UV-B, and the two-component regulatory genes COO91_00772 and COO91_00771 (similar to Npun_F1277 and Npun_F1278 of module IV) and other two genes COO91_00791 and COO91_00787 (similar to Npun_R1260 and Npun_R1263 in module II, respectively) were also significantly induced (Figure 4B). These results indicate a different transcriptional pattern of scytonemin biosynthetic genes in response to UV-B radiation in N. flagelliforme.
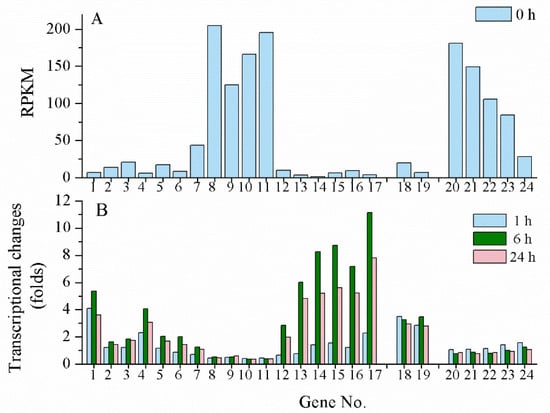
Figure 4.
The transcriptional changes of scytonemin biosynthetic and export-associated genes in response to UV-B radiation. The physiologically recovered Nostoc flagelliforme samples were subjected to 0.5 W·m−2 UV-B radiation for 24 h. (A) The basal RPKM values of the genes according to our transcriptomic analysis. (B) Transcriptional changes of the genes at 1, 6, and 24 h of UV-B radiation. Genes no.1-17, COO91_00791–00773 (excluding COO91_00785 and COO91_00789); genes no. 18-19, COO91_00772–00771; genes no. 20-24, COO91_05871–05866 (excluding COO91_05870).
4.3. Precursors/Substrates Biosynthesis and Key Catalytic Steps
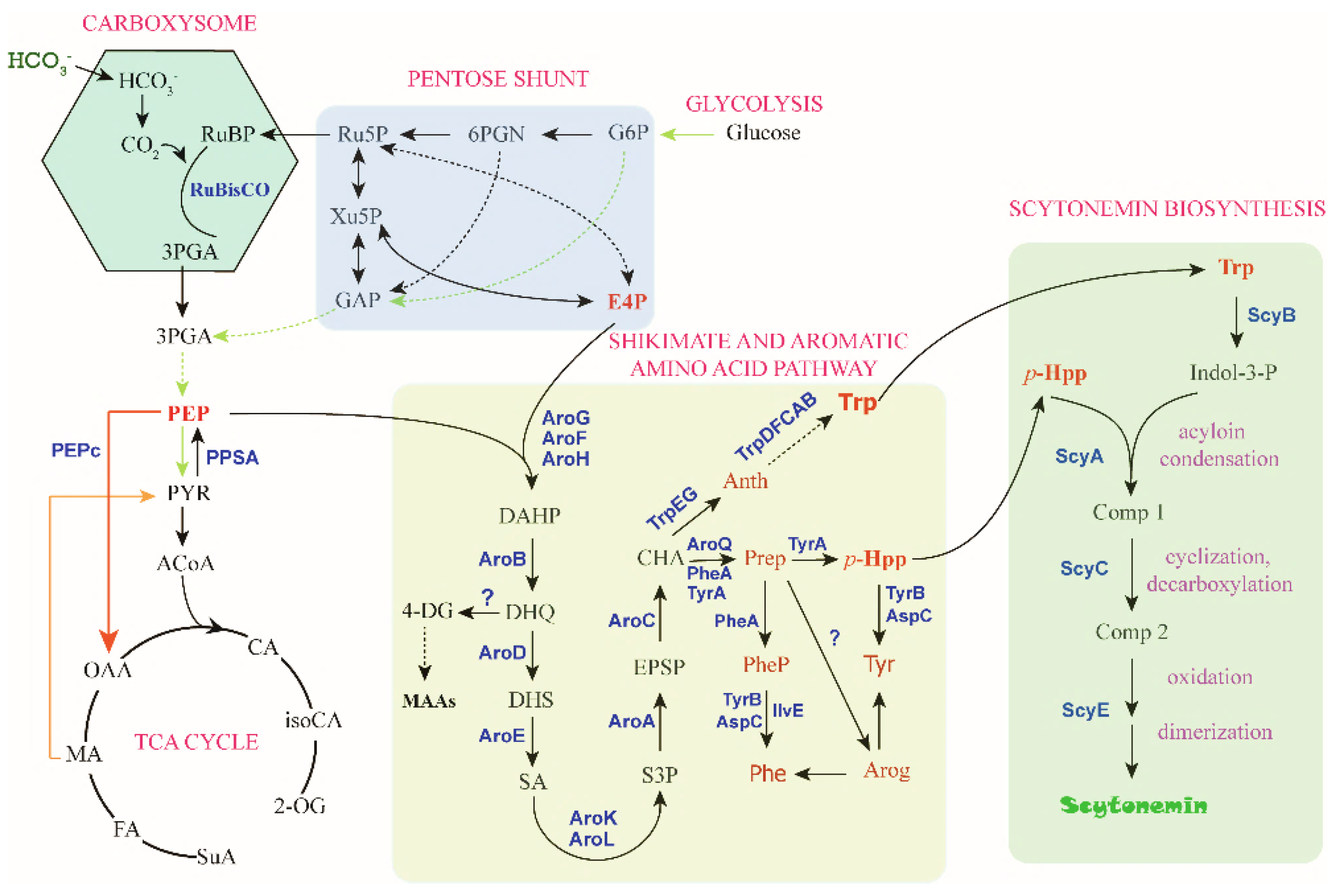
For metabolic engineering of scytonemin production in cyanobacteria, one of the most important steps is to improve the cellular concentrations of the aromatic compound substrates, Trp and p-Hpp. Metabolic engineering for production of aromatic amino acids was recently reported in cyanobacteria [72,73], most of related research is yet from other microorganisms, including E. coli. To a certain extent, cyanobacterial metabolic engineering can take advantage of the rich toolbox or strategy developed for E. coli. Phosphoenolpyruvate (PEP) and erythrose-4-phosphate (E4P) from the central carbon metabolism serve as the substrates for the biosynthesis of Trp and p-Hpp, and the biosynthesis process is realized through the shikimate pathway followed by the branched aromatic amino acid pathway with chorismate (CHA) serving as a major branch point intermediate metabolite (Figure 5). This section summarizes the progress regarding the four critical substrates, PEP, E4P, Trp, and p-Hpp.
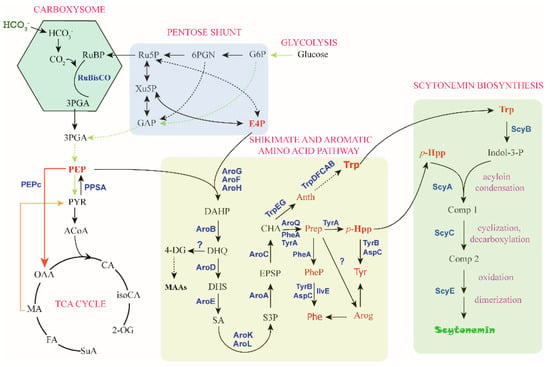
Figure 5.
The “C→C” conversion pathway of scytonemin biosynthesis. The direct precursors for scytonemin synthesis are Trp and p-Hpp, and the substrates for these two metabolites are PEP and E4P. Metabolite abbreviations: RuBP, ribulose-1,5-bisphosphat; Ru5P, ribulose-5-phosphate; Xu5P, xylulose-5-phosphate; GAP, glyceraldehyde-3-phosphate; 6PGN, 6-phosphate gluconate; G6P, glucose-6-phosphate; E4P, erythrose-4-phosphate. 3PGA, 3-phosphoglycerat; PEP, phosphoenolpyruvate; PYR, pyruvate; ACoA, acetyl-CoA; OAA, oxaloacetate; CA, Citrate; iosCA, iso-citrate; 2-OG, alpha-ketoglutarate; MA, malate; FA, fumarate; SuA, succinyl CoA. DAHP, 3-deoxy-D-arabino-heptulosonate-7-phosphate; DHQ, 3-dehydroquinate; DHS, 3-dehydroshikimate; SA, shikimate; S3P, SA-3-phosphate; EPSP, 5-enolpyruvyl-shikimate 3-phosphate; CHA, chorismate; 4-DG, 4-deoxygadusol; MAAs, mycosporine-like amino acids; Anth, anthranilate; Trp, tryptophan; Prep, prephenate; p-HPP, p-hydroxyphenylpyruvate; Phe, phenylalanine; Tyr, tyrosine; PheP, phenylpyruvate; Arog, arogenate. Indol-3-P, indole-3 pyruvate; Comp 1, intermediate compound 1; Comp 2, intermediate compound 2. Enzyme abbreviations: RuBisCO, ribulose-1,5-bisphosphate carboxylase/oxygenase; PEPc, PEP carboxylase.
4.3.1. PEP Biosynthesis
PEP represents one of the important intermediates in the central carbon metabolism of the cells [74]. In cyanobacteria, CO2 is assimilated to generate the building blocks of amino acids and other carbon-containing molecules via the Calvin-Benson-Bassham (CBB) cycle. Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) catalyzes the addition of CO2 to ribulose-1,5-bisphosphate (RuBP), generating two molecules of 3-phosphoglyceric acid (3-PGA) [75]. 3-PGA can diffuse out of the carboxysome and enter into the glycolysis pathway to form PEP. In addition, 3-PGA can also be glycolytically produced from glucose. PEP is then converted to pyruvate by the pyruvate kinase or alternatively converted to oxaloacetate (OAA) by PEP carboxylase (PEPc). PEPc serves as the second major carbon-fixing enzyme in cyanobacteria [76]. Combination of PEPc, malate dehydrogenase and malic enzyme forms a metabolic shunt for the biosynthesis of pyruvate, replenishing the tricarboxylic acid (TCA) cycle [77]. Overexpression of PEPc in Synechocystis (Synechocystis PCC 6803) resulted in improved succinate production via the reductive TCA cycle and improved 2-oxoglutarate (2-OG) production via the oxidative TCA cycle [78,79].
In order to increase the PEP pool, several useful strategies can be employed, including overexpression of PEP-forming enzymes (i.e., PEP synthase and PEP carboxykinase) and inactivation of PEP-degrading enzymes (i.e., pyruvate kinase and PEPc) [78,79,80]. Inhibition of PEPc might be also targeted for maintaining the PEP level, since PEPc can be allosterically inhibited by metabolic effectors, such as malate in several cyanobacteria or after being mutated [81]. A gluconeogenic enzyme phosphoenolpyruvate synthase (PPSA) catalyzes the reverse conversion from pyruvate to PEP [82]. Therefore, enhancement of PPSA should increase the PEP pool. In E. coli, the phosphoenolpyruvate-carbohydrate phosphotransferase system (PTS) is responsible for the transport and phosphorylation of sugars, such as glucose, at the expense of PEP [83]. Inactivation of the PTS reduced the PEP consumption [71]. However, this is not necessary for cyanobacterial engineering, since it should be more cost-effective to use the photosynthetically fixed carbon than to use the sugars as carbon source. The excess fixed carbon of cyanobacteria can be stored as glycogen and can be degraded during the dark/light transition [84], which will contribute to buffer the available PEP.
4.3.2. Erythrose-4-Phosphate (E4P) Biosynthesis
E4P is an intermediate in the pentose phosphate pathway (PPP) and the CBB cycle. The PPP is an alternative pathway of glucose metabolism. The function of the PPP includes: Generating NADPH for reductive metabolic reactions within cells, generating ribose-5-phosphate (R5P) as a precursor for nucleotide biosynthesis, and providing E4P as a precursor for aromatic amino acid biosynthesis [85,86]. PPP consists of an oxidative and a non-oxidative phase. In the oxidative phase, NADPH is generated during the oxidative decarboxylation of glucose-6-phosphate (G6P) to form ribulose-5-phosphate (Ru5P). In the non-oxidative phase, Ru5P is converted into R5P or xylulose-5-phosphate (Xu5P). These two five-carbon compounds are then converted to glyceraldehyde 3-phosphate (GAP) and sedoheptulose-7-phosphate (S7P) by transketolase. GAP and S7P are further converted into E4P and fructose 6-phosphate (F6P) by transaldolase. Overexpression of transketolase and/or transaldolase resulted in an increase of the E4P pool [71,87]. E4P can also be directly produced from the catabolism of sedoheptulose-1,7-bisphosphate by the universally conserved glycolytic enzymes ATP-dependent phosphofructokinase and aldolase, when the intracellular level of S7P is high [88].
4.3.3. Trp and p-Hpp Biosynthesis
The aromatic amino acids are not only the building blocks of proteins, but also precursors for a wide range of secondary metabolites in plants and microorganisms. Plants have evolved with interactive plastidial and cytosolic pathways for the biosynthesis of aromatic amino acids [89]. Unlike plants, cyanobacteria exhibit endo-oriented control of the multi-branched aromatic biosynthesis pathway and biosynthesize moderate amounts of aromatic amino acids under natural conditions [90].
Regulation in the Shikimate Pathway
The direct substrates for scytonemin biosynthesis, catalyzed by ScyABCDEF in cyanobacteria, are Trp and p-Hpp [61]. Both substrates are biosynthesized from PEP and E4P via the shikimate and aromatic amino acid pathway. The shikimate pathway comprises seven enzymatic steps that convert PEP and E4P into CHA [91]. The serial metabolic intermediates are 3-deoxy-d-arabino-heptulosonate-7-phosphate (DAHP), 3-dehydroquinate (DHQ), 3-dehydroshikimate (DHS), shikimate (SA), shikimate-3-phosphate (S3P), 5-enolpyruvylshikimate 3-phosphate (EPSP), and CHA. The most studied enzymes in this pathway are the first and sixth enzyme, respectively, DAHP synthase and EPSP synthase. DAHP synthase is a major point of regulation in the initiation of the shikimate pathway. E. coli has three isozymes of DAHP synthase, AroF (catalyzing ~20% of enzymatic activity), AroG (catalyzing ~80% of enzymatic activity), and AroH (catalyzing ~1% of enzymatic activity), each of which is allosterically feedback-regulated by one of the aromatic amino acids, tyrosine (Tyr), phenylalanine (Phe), and Trp [92,93]. EPSP synthase catalyzes the formation of EPSP from PEP and S3P, which is an inhibitory target by the herbicide glyphosate [94]. Other enzymes, DHQ synthase, SA kinase, and CHA synthase, are also rate-limiting enzymes in the shikimate pathway [95,96,97].
E. coli strains, harboring insensitive mutations of AroG and AroF, showed increased carbon flux toward to SA and aromatic amino acid production through the shikimate pathway [71,87]. In Synechocystis, overexpression of the feedback-inhibition-resistant AroG and TyrA enzymes from E. coli led to the conversion of an estimated 56% of the total fixed carbon to Phe and Tyr [72]. In addition, the intracellular levels of the three DAHP synthase(s) in E. coli are also controlled by transcriptional repression through the repressors TyrR and TrpR, and deletion of tyrR and trpR alleviated the transcriptional control [98,99,100]. Enhancing the expression of DHQ synthase (encoded by aroB) and SA kinase isoenzymes I and II (encoded by aroK and aroL, respectively) via various genetic engineering approaches resulted in increased aromatic amino acid production [87].
Intermediates in the shikimate pathway are also precursors for biosynthesis of diverse secondary metabolites. The shikimate pathway is the predominate route for UV-induced MAA biosynthesis [101]. DHQ is also a possible precursor for biosynthesis of another group of sunscreens, mycosporines and MAAs, via 4-deoxygadusol [102]. In cyanobacteria, MAAs and scytonemin are both effectively induced by UV radiation [13,38], and disruption of the bypass in MAA biosynthesis should be potentially beneficial for scytonemin production.
Regulation in the Aromatic Amino Acid Pathway
CHA is a common precursor for biosynthesis of the three aromatic amino acids, Phe, Tyr, and Trp. In total, five successive enzymatic reactions catalyzed by TrpABCDEFG in E. coli lead to the formation of Trp, in which anthranilate synthase (AS) catalyzes the first committed step and is allosterically regulated by Trp [87,103,104]. Transgenic expression of a feedback-resistant AS from E. coli improved the Trp production in Synechocystis [73]. In E. coli, tryptophanase (TnaA) converts Trp to indole, and deletion of TnaA gene led to reduced Trp degradation [99,105]. Trp production was also affected by the modification of transport systems. Overexpression of an exporter protein YddG and inactivation of a permease AroP led to increased Trp pool [106,107].
The first committed enzyme of Phe and Tyr biosynthesis is CHA mutase (CM or AroQ), which converts CHA to prephenate. Prephenate is subsequently converted to Phe via phenylpyruvate or converted to Tyr via p-Hpp in most microorganisms [91,108]. In plant, the major route of Phe and Tyr biosynthesis occurs via an intermediate metabolite arogenate [89]. CM was only found to be feedback-inhibited by Phe, Tyr, or Trp in a few cyanobacterial strains [109]. However, CM-prephenate dehydrogenase (TyrA) is inhibited by Tyr and CM-prephenate dehydratase (PheA) is inhibited by Phe in E. coli [110,111,112]. Blocking completive Phe and Tyr biosynthetic pathways by deletion of pheA and tyrA could lead to the enhanced production of Trp [99]. However, this strategy may affect the biosynthesis of p-Hpp.
5. Suggested Strategies for Engineering Scytonemin Production
5.1. The Host for Scytonemin Production
The host selection is the first and most important consideration for metabolic engineering. Scytonemin is solely produced in some cyanobacterial species, particularly those exposed to high solar radiation. They are natural ideal hosts for metabolic engineering for scytonemin production. The exploitation of these photosynthetic microorganisms is required to overcome the obstacles of mass cultivation and genetic manipulation. The complete gene clusters or genes for scytonemin biosynthesis were first elucidated in the model cyanobacterium N. punctiforme PCC 73102. Other model cyanobacteria, such as Synechocystis PCC 6803 and Nostoc PCC 7120, do not synthesize scytonemin due to the lack of complete scytonemin-biosynthetic genes [36,67]. For example, the homologous genes of Npun_R1268 and Npun_R1263 are absent in the Nostoc PCC 7120 genome. The supplementation of absent genes is a potential strategy to exploit them as metabolic engineering hosts. Non-photosynthetic microorganisms such as E. coli are also proper selection to engineer the production of scytonemin, but only the scytonemin monomer could be obtained so far [69]. A further processing for oxidative dimerization is necessary. In cyanobacteria, this step is realized after the export of the scytonemin monomer to the periplasm with the help of Ebo proteins [64]. Therefore, the complete biosynthesis of the scytonemin dimer in E. coli is still a challenge.
N. flagelliforme, a close relative of N. punctiforme, was recently sequenced [113]. It is a promising scytonemin producer [8,13]. It is easy to be cultivated in liquid suspension and can produce lots of exopolysaccharides. The dense exopolysaccharide matrix provides an ideal environment for accommodating scytonemin. In addition to COO91_00781, COO91_00780, and COO91_00775 (the homologous genes of Npun_R1268, Npun_R 1269, and Npun_R1274, respectively), other scytonemin-biosynthetic genes all have paralogous genes in the N. flagelliforme genome. Its genetic manipulation with CRISPA system was recently established in our laboratory, which will benefit its exploitation in metabolic engineering.
5.2. Carbon Flux Modification
High productivity of a target metabolite requires optimizing the redirection of carbon flux through primary and secondary metabolisms. Useful strategies for scytonemin production may include: (1) Engineering the CBB cycle or high-density cultivation to enhance photosynthetic carbon fixation and biomass increase [75,114]; (2) re-directing the metabolic flux towards the shikimate and aromatic amino acid pathway, such as improving the availability of the precursors PEP and E4P, relieving the rate-limiting enzymatic reactions, removing the transcriptional and allosteric regulations, and disruption of consumptive bypasses [72,115]; and (3) enhancing the transcription of scy and ebo gene clusters or the critical genes for the direct scytonemin biosynthesis [63,64], especially rewiring or optimizing the above-mentioned four “Modules”. Random mutagenesis is also an important technology for relieving product inhibition or ameliorating metabolic suitability in hosts [73,116].
5.3. Cultivation and Harvest Technology
Cultivation and harvest technologies are also important for final target metabolite yield. Useful strategies may include: (1) Decoupling growth and production or so-called two-stage cultivation strategy. This strategy was usually adopted in algal metabolite production [117,118,119]. (2) Enhancing the product export, accumulation, and collection. It is particularly beneficial in industrial production. Like developing the fatty acid secreting system in cyanobacteria [120], realizing the release of scytonemin from the exopolysaccharide matrix to the surface of liquid culture is also an attractive technology. (3) Adopting stressful conditions during the whole culture process. The biosynthesis of metabolites can be triggered by a number of abiotic stresses [121]. A drawback is that abiotic stresses often slow down or restrain the biomass increase and thus an elaborate balance between the stress factor and cell growth should be established. (4) Supplementing external substrates or elicitor substances from concomitant bacteria [122].
6. Perspective
Scytonemin is a cyanobacterial secondary metabolite with a high potential market value. Metabolic engineering techniques will provide one of the key solutions for achieving cost-effective production of scytonemin in the near future. Previous engineering in the E. coli strain can only produce the scytonemin monomer. Along with the recent elucidation of its biosynthesis mechanism and related carbon flux-rewiring technologies, highly efficient biosynthesis of scytonemin in suitable cyanobacterial hosts, including N. flagelliforme and N. punctiforme, is greatly expected as an important starting point.
Author Contributions
Conceptualization, X.G. and P.L.; writing—original draft preparation, X.G.; writing—re-view and editing, X.G., X.J., X.L. and P.L.; supervision, P.L.; funding acquisition, X.G. All authors read and approved the final version of the manuscript.
Funding
We acknowledge the support from the China Scholarship Council and the Key Project of Natural Science of Shaanxi Province, China (No. 2020JZ-51).
Conflicts of Interest
All authors declare no conflict of interests.
References
- Nunnery, J.K.; Mevers, E.; Gerwick, W.H. Biologically active secondary metabolites from marine cyanobacteria. Curr. Opin. Biotechnol. 2010, 21, 787–793. [Google Scholar] [CrossRef]
- Xue, Y.; He, Q. Cyanobacteria as cell factories to produce plant secondary metabolites. Front. Bioeng. Biotechnol. 2015, 3, 57. [Google Scholar] [CrossRef]
- Knoot, C.J.; Ungerer, J.; Wangikar, P.P.; Pakrasi, H.B. Cyanobacteria: Promising biocatalysts for sustainable chemical production. J. Biol. Chem. 2018, 293, 5044–5052. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Xie, H.; Liu, X.; Lindberg, P.; Lindblad, P. Current processes and future challenges of photoautotrophic production of acetyl-CoA-derived solar fuels and chemicals in cyanobacteria. Curr. Opin. Chem. Biol. 2020, 59, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Zahra, Z.; Choo, D.H.; Lee, H.; Parveen, A. Cyanobacteria: Review of current potentials and applications. Environments 2020, 7, 13. [Google Scholar] [CrossRef]
- Büdel, B.; Karsten, U.; Garcia-Pichel, F. Ultraviolet-absorbing scytonemin and mycosporine-like amino acid derivatives in exposed, rock-inhabiting cyanobacterial lichens. Oecologia 1997, 112, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Balskus, E.P.; Case, R.J.; Walsh, C.T. The biosynthesis of cyanobacterial sunscreen scytonemin in intertidal microbial mat communities. FEMS Microbiol. Ecol. 2011, 77, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Gao, X. Scytonemin plays a potential role in stabilizing the exopolysaccharidic matrix in terrestrial cyanobacteria. Microb. Ecol. 2017, 73, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, R.P.; Sonani, R.R.; Madamwar, D. The high-energy radiation protectant extracellular sheath pigment scytonemin and its reduced counterpart in the cyanobacterium Scytonema sp. R77DM. Bioresour. Technol. 2014, 171, 396–400. [Google Scholar] [CrossRef]
- Gao, Q.; Garcia-Pichel, F. Microbial ultraviolet sunscreens. Nat. Rev. Microbiol. 2011, 9, 791–802. [Google Scholar] [CrossRef]
- Garcia-Pichel, F.; Castenholz, R.W. Characterization and biological implications of scytonemin, a cyanobacterial sheath pigment1. J. Phycol. 1991, 27, 395–409. [Google Scholar] [CrossRef]
- Sinha, R.P.; Singh, S.P.; Häder, D.-P. Database on mycosporines and mycosporine-like amino acids (MAAs) in fungi, cyanobacteria, macroalgae, phytoplankton and animals. J. Photochem. Photobiol. B 2007, 89, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Ferroni, L.; Klisch, M.; Pancaldi, S.; Häder, D.-P. Complementary UV-absorption of mycosporine-like amino acids and scytonemin is responsible for the UV-insensitivity of photosynthesis in Nostoc flagelliforme. Mar. Drugs 2010, 8, 106–121. [Google Scholar] [CrossRef]
- Garcia-Pichel, F.; Lombard, J.; Soule, T.; Dunaj, S.; Wu, S.H.; Wojciechowski, M.F. Timing the evolutionary advent of cyanobacteria and the Later Great Oxidation Event using gene phylogenies of a sunscreen. mBio 2019, 10, e00561-19. [Google Scholar] [CrossRef]
- Bultel-Poncé, V.; Felix-Theodose, F.; Sarthou, C.; Ponge, J.-F.; Bodo, B. New pigments from the terrestrial cyanobacterium Scytonema sp. collected on the Mitaraka Inselberg, French Guyana. J. Nat. Prod. 2004, 67, 678–681. [Google Scholar] [CrossRef] [PubMed]
- Varnali, T.; Edwards, H.G.M. Raman spectroscopic identification of scytonemin and its derivatives as key biomarkers in stressed environments. Philos. Trans. A Math. Phys. Eng. Sci. 2014, 372, 20140197. [Google Scholar] [CrossRef]
- Gies, P.; Roy, C.; Udelhofen, P. Solar and ultraviolet radiation (chapter). In Prevention of Skin Cancer; Springer: Berlin/Heidelberg, Germany, 2004; Volume 3, pp. 21–54. ISBN 978-90-481-6346-5. [Google Scholar]
- Garcia-Pichel, F.; Sherry, N.D.; Castenholz, R.W. Evidence for an ultraviolet sunscreen role of the extracellular pigment scytonemin in the terrestrial cyanobacterium Chlorogloeopsis sp. Photochem. Photobiol. 1992, 56, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Proteau, P.J.; Gerwick, W.H.; Garcia-Pichel, F.; Castenholz, R. The structure of scytonemin, an ultraviolet sunscreen pigment from the sheaths of cyanobacteria. Experientia 1993, 49, 825–829. [Google Scholar] [CrossRef]
- Dillon, J.; Castenholz, R. Scytonemin, a cyanobacterial sheath pigment, protects against UVC radiation: Implications for early photosynthetic life. J. Phycol. 2002, 35, 673–681. [Google Scholar] [CrossRef]
- Pandey, A.; Pathak, J.; Singh, D.K.; Ahmed, H.; Singh, V.; Kumar, D.; Sinha, R.P. Photoprotective role of UV-screening pigment scytonemin against UV-B-induced damages in the heterocyst-forming cyanobacterium Nostoc sp. strain HKAR-2. Braz. J. Bot. 2020, 43, 67–80. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Sinha, R.P.; Incharoensakdi, A. Partial characterization, UV-induction and photoprotective function of sunscreen pigment, scytonemin from Rivularia sp. HKAR-4. Chemosphere 2013, 93, 1874–1878. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Xu, H.; Zhu, Z.; Gao, X. The effects of the exopolysaccharide and growth rate on the morphogenesis of the terrestrial filamentous cyanobacterium Nostoc flagelliforme. Biol. Open 2017, 6, 1329–1335. [Google Scholar] [CrossRef]
- Matsui, K.; Nazifi, E.; Hirai, Y.; Wada, N.; Matsugo, S.; Sakamoto, T. The cyanobacterial UV-absorbing pigment scytonemin displays radical-scavenging activity. J. Gen. Appl. Microbiol. 2012, 58, 137–144. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Incharoensakdi, A. Characterization of UV-screening compounds, mycosporine-like amino acids, and scytonemin in the cyanobacterium Lyngbya sp. CU2555. FEMS Microbiol. Ecol. 2014, 87, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Fleming, E.D.; Castenholz, R.W. Effects of periodic desiccation on the synthesis of the UV-screening compound, scytonemin, in cyanobacteria. Environ. Microbiol. 2007, 9, 1448–1455. [Google Scholar] [CrossRef]
- Squier, A.H.; Hodgson, D.A.; Keely, B.J. A critical assessment of the analysis and distributions of scytonemin and related UV screening pigments in sediments. Org. Geochem. 2004, 35, 1221–1228. [Google Scholar] [CrossRef]
- Fulton, J.M.; Arthur, M.A.; Freeman, K.H. Subboreal aridity and scytonemin in the Holocene Black Sea. Org. Geochem. 2012, 49, 47–55. [Google Scholar] [CrossRef]
- Varnali, T.; Edwards, H.G.M. Theoretical study of novel complexed structures for methoxy derivatives of scytonemin: Potential biomarkers in iron-rich stressed environments. Astrobiology 2013, 13, 861–869. [Google Scholar] [CrossRef]
- Vítek, P.; Jehlička, J.; Ascaso, C.; Mašek, V.; Gómez-Silva, B.; Olivares, H.; Wierzchos, J. Distribution of scytonemin in endolithic microbial communities from halite crusts in the hyperarid zone of the Atacama Desert, Chile. FEMS Microbiol. Ecol. 2014, 90, 351–366. [Google Scholar] [CrossRef]
- Lalić, D.; Meriluoto, J.; Zorić, M.; Dulić, T.; Mirosavljević, M.; Župunski, M.; Svirčev, Z. Potential of cyanobacterial secondary metabolites as biomarkers for paleoclimate reconstruction. Catena 2020, 185, 104283. [Google Scholar] [CrossRef]
- Sinha, R.P.; Klisch, M.; Gröniger, A.; Häder, D.-P. Ultraviolet-absorbing/screening substances in cyanobacteria, phytoplankton and macroalgae. J. Photochem. Photobiol. B 1998, 47, 83–94. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Sonani, R.R.; Madamwar, D. Cyanobacterial sunscreen scytonemin: Role in photoprotection and biomedical research. Appl. Biochem. Biotechnol. 2015, 176, 1551–1563. [Google Scholar] [CrossRef]
- Venckus, P.; Paliulis, S.; Kostkevičiene, J.; Dementjev, A. CARS microscopy of scytonemin in cyanobacteria Nostoc commune. J. Raman Spectrosc. 2018, 49, 1333–1338. [Google Scholar] [CrossRef]
- Pathak, J.; Sonker, A.S.; Richa, R.; Kannaujiya, V.K.; Singh, V.; Ahmed, H.; Sinha, R.P. Screening and partial purification of photoprotective pigment scytonemin from cyanobacterial crusts dwelling on the historical monuments in and around Varanasi, India. Microbiol. Res. 2017, 8, 6559. [Google Scholar] [CrossRef]
- D’Agostino, P.M.; Woodhouse, J.N.; Liew, H.T.; Sehnal, L.; Pickford, R.; Wong, H.L.; Burns, B.P.; Neilan, B.A. Bioinformatic, phylogenetic and chemical analysis of the UV-absorbing compounds scytonemin and mycosporine-like amino acids from the microbial mat communities of Shark Bay, Australia. Environ. Microbiol. 2019, 21, 702–715. [Google Scholar] [CrossRef]
- Karsten, U.; Maier, J.; Garcia-Pichel, F. Seasonality in UV-absorbing compounds of cyanobacterial mat communities from an intertidal mangrove flat. Aquat. Microb. Ecol. 1998, 16, 37–44. [Google Scholar] [CrossRef]
- Wright, D.J.; Smith, S.C.; Joardar, V.; Scherer, S.; Jervis, J.; Warren, A.; Helm, R.F.; Potts, M. UV irradiation and desiccation modulate the three-dimensional extracellular matrix of Nostoc commune (Cyanobacteria). J. Biol. Chem. 2005, 280, 40271–40281. [Google Scholar] [CrossRef]
- Takamatsu, S.; Hodges, T.W.; Rajbhandari, I.; Gerwick, W.H.; Hamann, M.T.; Nagle, D.G. Marine natural products as novel antioxidant prototypes. J. Nat. Prod. 2003, 66, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J. Microbial sunscreens. Microb. Biotechnol. 2011, 4, 1–7. [Google Scholar] [CrossRef]
- Derikvand, P.; Llewellyn, C.A.; Purton, S. Cyanobacterial metabolites as a source of sunscreens and moisturizers: A comparison with current synthetic compounds. Eur. J. Phycol. 2017, 52, 43–56. [Google Scholar] [CrossRef]
- Kang, M.R.; Jo, S.A.; Lee, H.; Yoon, Y.D.; Kwon, J.-H.; Yang, J.-W.; Choi, B.J.; Park, K.H.; Lee, M.Y.; Lee, C.W.; et al. Inhibition of skin inflammation by scytonemin, an ultraviolet sunscreen pigment. Mar. Drugs 2020, 18, 300. [Google Scholar] [CrossRef]
- Stevenson, C.S.; Capper, E.A.; Roshak, A.K.; Marquez, B.; Grace, K.; Gerwick, W.H.; Jacobs, R.S.; Marshall, L.A. Scytonemin-a marine natural product inhibitor of kinases key in hyperproliferative inflammatory diseases. Inflamm. Res. 2002, 51, 112–114. [Google Scholar] [CrossRef]
- Strebhardt, K.; Ullrich, A. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer 2006, 6, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, Z.; Liu, Z. Scytonemin inhibits cell proliferation and arrests cell cycle through downregulating Plk1 activity in multiple myeloma cells. Tumor. Biol. 2013, 34, 2241–2247. [Google Scholar] [CrossRef]
- Pathak, J.; Mondal, S.; Ahmed, H. In silico study on interaction between human polo-like kinase 1 and cyanobacterial sheath pigment scytonemin by molecular docking approach. Biointerface Res. Appl. Chem. 2019, 9, 4374–4378. [Google Scholar]
- Itoh, T.; Tsuzuki, R.; Tanaka, T.; Ninomiya, M.; Yamaguchi, Y.; Takenaka, H.; Ando, M.; Tsukamasa, Y.; Koketsu, M. Reduced scytonemin isolated from Nostoc commune induces autophagic cell death in human T-lymphoid cell line Jurkat cells. Food Chem. Toxicol. 2013, 60, 76–82. [Google Scholar] [CrossRef]
- Itoh, T.; Koketsu, M.; Yokota, N.; Touho, S.; Ando, M.; Tsukamasa, Y. Reduced scytonemin isolated from Nostoc commune suppresses LPS/IFNγ-induced NO production in murine macrophage RAW264 cells by inducing hemeoxygenase-1 expression via the Nrf2/ARE pathway. Food Chem. Toxicol. 2014, 69, 330–338. [Google Scholar] [CrossRef]
- Dillon, J.G.; Castenholz, R.W. The synthesis of the UV-screening pigment, scytonemin, and photosynthetic performance in isolates from closely related natural populations of cyanobacteria (Calothrix sp.). Environ. Microbiol. 2003, 5, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Couradeau, E.; Karaoz, U.; Lim, H.C.; Nunes da Rocha, U.; Northen, T.; Brodie, E.; Garcia-Pichel, F. Bacteria increase arid-land soil surface temperature through the production of sunscreens. Nat. Commun. 2016, 7, 10373. [Google Scholar] [CrossRef]
- Varnali, T.; Edwards, H.G.M. Iron-scytonemin complexes: DFT calculations on new UV protectants for terrestrial cyanobacteria and astrobiological implications. Astrobiology 2010, 10, 711–716. [Google Scholar] [CrossRef]
- Varnali, T.; Gören, B. Two distinct structures of the sandwich complex of scytonemin with iron and their relevance to astrobiology. Struct. Chem. 2018, 29, 1565–1572. [Google Scholar] [CrossRef]
- Rath, J.; Mandal, S.; Adhikary, S.P. Salinity induced synthesis of UV-screening compound scytonemin in the cyanobacterium Lyngbya aestuarii. J. Photochem. Photobiol. B 2012, 115, 5–8. [Google Scholar] [CrossRef]
- Dillon, J.G.; Tatsumi, C.M.; Tandingan, P.G.; Castenholz, R.W. Effect of environmental factors on the synthesis of scytonemin, a UV-screening pigment, in a cyanobacterium (Chroococcidiopsis sp.). Arch. Microbiol. 2002, 177, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Fleming, E.D.; Castenholz, R.W. Effects of nitrogen source on the synthesis of the UV-screening compound, scytonemin, in the cyanobacterium Nostoc punctiforme PCC 73102. FEMS Microbiol. Ecol. 2008, 63, 301–308. [Google Scholar] [CrossRef]
- Pereira, S.; Zille, A.; Micheletti, E.; Moradas-Ferreira, P.; De Philippis, R.; Tamagnini, P. Complexity of cyanobacterial exopolysaccharides: Composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 2009, 33, 917–941. [Google Scholar] [CrossRef]
- Pereira, S.; Mota, R.; Vieira, C.; Vieira, J.; Tamagnini, P. Phylum-wide analysis of genes/proteins related to the last steps of assembly and export of extracellular polymeric substances (EPS) in cyanobacteria. Sci. Rep. 2015, 5, 14835. [Google Scholar] [CrossRef]
- Yu, H.; Liu, R. Effect of UV-B radiation on the synthesis of UV-absorbing compounds in a terrestrial cyanobacterium, Nostoc flagelliforme. J. Appl. Phycol. 2013, 25, 1441–1446. [Google Scholar] [CrossRef]
- Soule, T.; Shipe, D.; Lothamer, J. Extracellular polysaccharide production in a scytonemin-deficient mutant of Nostoc punctiforme under UVA and oxidative stress. Curr. Microbiol. 2016, 73, 455–462. [Google Scholar] [CrossRef]
- Soule, T.; Stout, V.; Swingley, W.D.; Meeks, J.C.; Garcia-Pichel, F. Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. J. Bacteriol. 2007, 189, 4465–4472. [Google Scholar] [CrossRef]
- Balskus, E.P.; Walsh, C.T. Investigating the initial steps in the biosynthesis of cyanobacterial sunscreen scytonemin. J. Am. Chem. Soc. 2008, 130, 15260–15261. [Google Scholar] [CrossRef]
- Soule, T.; Palmer, K.; Gao, Q.; Potrafka, R.M.; Stout, V.; Garcia-Pichel, F. A comparative genomics approach to understanding the biosynthesis of the sunscreen scytonemin in cyanobacteria. BMC Genom. 2009, 10, 336. [Google Scholar] [CrossRef]
- Ferreira, D.; Garcia-Pichel, F. Mutational studies of putative biosynthetic genes for the cyanobacterial sunscreen scytonemin in Nostoc punctiforme ATCC 29133. Front. Microbiol. 2016, 7, 735. [Google Scholar] [CrossRef] [PubMed]
- Klicki, K.; Ferreira, D.; Hamill, D.; Dirks, B.; Mitchell, N.; Garcia-Pichel, F. The widely conserved ebo Cluster is involved in precursor transport to the periplasm during scytonemin synthesis in Nostoc punctiforme. mBio 2018, 9, e02266-18. [Google Scholar] [CrossRef] [PubMed]
- Balskus, E.P.; Walsh, C.T. An enzymatic cyclopentyl[b]indole formation involved in scytonemin biosynthesis. J. Am. Chem. Soc. 2009, 131, 14648–14649. [Google Scholar] [CrossRef]
- Soule, T.; Garcia-Pichel, F.; Stout, V. Gene expression patterns associated with the biosynthesis of the sunscreen scytonemin in Nostoc punctiforme ATCC 29133 in response to UVA radiation. J. Bacteriol. 2009, 191, 4639–4646. [Google Scholar] [CrossRef]
- Sorrels, C.M.; Proteau, P.J.; Gerwick, W.H. Organization, evolution, and expression analysis of the biosynthetic gene cluster for scytonemin, a cyanobacterial UV-absorbing pigment. Appl. Environ. Microbiol. 2009, 75, 4861–4869. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, T.; Ševčíková, T.; Strnad, H.; Butenko, A.; Eliáš, M. The plastid genome of some eustigmatophyte algae harbours a bacteria-derived six-gene cluster for biosynthesis of a novel secondary metabolite. Open Biol. 2016, 6, 160249. [Google Scholar] [CrossRef] [PubMed]
- Malla, S.; Sommer, M.O.A. A sustainable route to produce the scytonemin precursor using Escherichia coli. Green Chem. 2014, 16, 3255–3265. [Google Scholar] [CrossRef]
- Naurin, S.; Bennett, J.; Videau, P.; Philmus, B.; Soule, T. The response regulator Npun_F1278 is essential for scytonemin biosynthesis in the cyanobacterium Nostoc punctiforme ATCC 29133. J. Phycol. 2016, 52, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.A.; Bolívar, F.; Escalante, A. Shikimic acid production in Escherichia coli: From classical metabolic engineering strategies to omics applied to improve its production. Front. Bioeng. Biotechnol. 2015, 3, 145. [Google Scholar] [CrossRef]
- Brey, L.F.; Włodarczyk, A.J.; Bang Thøfner, J.F.; Burow, M.; Crocoll, C.; Nielsen, I.; Zygadlo Nielsen, A.J.; Jensen, P.E. Metabolic engineering of Synechocystis sp. PCC 6803 for the production of aromatic amino acids and derived phenylpropanoids. Metab. Eng. 2020, 57, 129–139. [Google Scholar] [CrossRef]
- Deshpande, A.; Vue, J.; Morgan, J. Combining random mutagenesis and metabolic engineering for enhanced tryptophan production in Synechocystis sp. Strain PCC 6803. Appl. Environ. Microbiol. 2020, 86, e02816-19. [Google Scholar] [CrossRef]
- Papagianni, M. Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb. Cell Fact. 2012, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Lindberg, P.; Lindblad, P. Engineering photoautotrophic carbon fixation for enhanced growth and productivity. Sustain. Energ. Fuels 2018, 2, 2583–2600. [Google Scholar] [CrossRef]
- Xiong, W.; Morgan, J.A.; Ungerer, J.; Wang, B.; Maness, P.-C.; Yu, J. The plasticity of cyanobacterial metabolism supports direct CO2 conversion to ethylene. Nat. Plants 2015, 1, 1–7. [Google Scholar]
- Scholl, J.; Dengler, L.; Bader, L.; Forchhammer, K. Phosphoenolpyruvate carboxylase from the cyanobacterium Synechocystis sp. PCC 6803 is under global metabolic control by PII signaling. Mol. Microbiol. 2020, 114, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Matsuda, M.; Kato, Y.; Vavricka, C.J.; Kondo, A. Temperature enhanced succinate production concurrent with increased central metabolism turnover in the cyanobacterium Synechocystis sp. PCC 6803. Metab. Eng. 2018, 48, 109–120. [Google Scholar] [CrossRef]
- Durall, C.; Lindberg, P.; Yu, J.; Lindblad, P. Increased ethylene production by overexpressing phosphoenolpyruvate carboxylase in the cyanobacterium Synechocystis PCC 6803. Biotechnol. Biofuels 2020, 13, 16. [Google Scholar] [CrossRef]
- Bongaerts, J.; Krämer, M.; Müller, U.; Raeven, L.; Wubbolts, M. Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab. Eng. 2001, 3, 289–300. [Google Scholar] [CrossRef]
- Takeya, M.; Hirai, M.Y.; Osanai, T. Allosteric inhibition of phosphoenolpyruvate carboxylases is determined by a single amino acid residue in cyanobacteria. Sci. Rep. 2017, 7, 41080. [Google Scholar] [CrossRef]
- Long, C.P.; Au, J.; Sandoval, N.R.; Gebreselassie, N.A.; Antoniewicz, M.R. Enzyme I facilitates reverse flux from pyruvate to phosphoenolpyruvate in Escherichia coli. Nat. Commun. 2017, 8, 14316. [Google Scholar] [CrossRef] [PubMed]
- Escalante, A.; Cervantes, A.S.; Gosset, G.; Bolívar, F. Current knowledge of the Escherichia coli phosphoenolpyruvate-carbohydrate phosphotransferase system: Peculiarities of regulation and impact on growth and product formation. Appl. Microbiol. Biotechnol. 2012, 94, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Shinde, S.; Zhang, X.; Singapuri, S.P.; Kalra, I.; Liu, X.; Morgan-Kiss, R.M.; Wang, X. Glycogen metabolism supports photosynthesis start through the oxidative pentose phosphate pathway in cyanobacteria. Plant Physiol. 2020, 182, 507–517. [Google Scholar] [CrossRef]
- Soderberg, T. Biosynthesis of ribose-5-phosphate and erythrose-4-phosphate in archaea: A phylogenetic analysis of archaeal genomes. Archaea 2005, 1, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Patra, K.C.; Hay, N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef]
- Rodriguez, A.; Martnez, J.A.; Flores, N.; Escalante, A.; Gosset, G.; Bolivar, F. Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb. Cell Fact. 2014, 13, 126. [Google Scholar] [CrossRef]
- Nakahigashi, K.; Toya, Y.; Ishii, N.; Soga, T.; Hasegawa, M.; Watanabe, H.; Takai, Y.; Honma, M.; Mori, H.; Tomita, M. Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Mol. Syst. Biol. 2009, 5, 306. [Google Scholar] [CrossRef]
- Lynch, J.H.; Dudareva, N. Aromatic amino acids: A complex network ripe for future exploration. Trends Plant Sci. 2020, 25, 670–681. [Google Scholar] [CrossRef]
- Żyszka-Haberecht, B.; Niemczyk, E.; Lipok, J. Metabolic relation of cyanobacteria to aromatic compounds. Appl. Microbiol. Biotechnol. 2019, 103, 1167–1178. [Google Scholar] [CrossRef]
- Tzin, V.; Galili, G.; Aharoni, A. Shikimate Pathway and Aromatic Amino Acid Biosynthesis. In eLS; John Wiley & Sons, Ltd.: Chichester, UK, 2012. [Google Scholar] [CrossRef]
- Jossek, R.; Bongaerts, J.; Sprenger, G.A. Characterization of a new feedback-resistant 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase AroF of Escherichia coli. FEMS Microb. Lett. 2001, 202, 145–148. [Google Scholar] [CrossRef]
- Cui, D.; Deng, A.; Bai, H.; Yang, Z.; Liang, Y.; Liu, Z.; Qiu, Q.; Wang, L.; Liu, S.; Zhang, Y.; et al. Molecular basis for feedback inhibition of tyrosine-regulated 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Escherichia coli. J. Struct. Biol. 2019, 206, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Schönbrunn, E.; Eschenburg, S.; Shuttleworth, W.A.; Schloss, J.V.; Amrhein, N.; Evans, J.N.; Kabsch, W. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. USA 2001, 98, 1376–1380. [Google Scholar] [CrossRef]
- Dell, K.A.; Frost, J.W. Identification and removal of impediments to biocatalytic synthesis of aromatics from D-glucose: Rate-limiting enzymes in the common pathway of aromatic amino acid biosynthesis. J. Am. Chem. Soc. 1993, 115, 11581–11589. [Google Scholar] [CrossRef]
- Krämer, M.; Bongaerts, J.; Bovenberg, R.; Kremer, S.; Müller, U.; Orf, S.; Wubbolts, M.; Raeven, L. Metabolic engineering for microbial production of shikimic acid. Metab. Eng. 2003, 5, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Oldiges, M.; Kunze, M.; Degenring, D.; Sprenger, G.A.; Takors, R. Stimulation, monitoring, and analysis of pathway dynamics by metabolic profiling in the aromatic amino acid pathway. Biotechnol. Prog. 2004, 20, 1623–1633. [Google Scholar] [CrossRef][Green Version]
- Sprenger, G.A. From scratch to value: Engineering Escherichia coli wild type cells to the production of L-phenylalanine and other fine chemicals derived from chorismate. Appl. Microbiol. Biotechnol. 2007, 75, 739–749. [Google Scholar] [CrossRef]
- Zhao, Z.-J.; Zou, C.; Zhu, Y.-X.; Dai, J.; Chen, S.; Wu, D.; Wu, J.; Chen, J. Development of L-tryptophan production strains by defined genetic modification in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2011, 38, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Yang, F.; Kang, J.; Wang, Q.; Qi, Q. One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli. Microb. Cell Fact. 2012, 11, 30. [Google Scholar] [CrossRef]
- Pathak, J.; Ahmed, H.; Singh, R.; Singh, S.P.; Häder, D.-P.; Sinha, R.P. Genetic regulation of scytonemin and mycosporine-like amino acids (MAAs) biosynthesis in cyanobacteria. Plant Gene 2019, 17, 100172. [Google Scholar] [CrossRef]
- Fuentes-Tristan, S.; Parra-Saldivar, R.; Iqbal, H.M.N.; Carrillo-Nieves, D. Bioinspired biomolecules: Mycosporine-like amino acids and scytonemin from Lyngbya sp. with UV-protection potentialities. J. Photochem. Photobiol. B 2019, 201, 111684. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Ann. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Fordjour, E.; Adipah, F.K.; Zhou, S.; Du, G.; Zhou, J. Metabolic engineering of Escherichia coli BL21 (DE3) for de novo production of l-DOPA from d-glucose. Microb. Cell Fact. 2019, 18, 74. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Young, K.D. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 2013, 159, 402–410. [Google Scholar] [CrossRef]
- Liu, Q.; Cheng, Y.; Xie, X.; Xu, Q.; Chen, N. Modification of tryptophan transport system and its impact on production of l-tryptophan in Escherichia coli. Bioresour. Technol. 2012, 114, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cheng, L.-K.; Wang, J.; Liu, Q.; Shen, T.; Chen, N. Genetic engineering of Escherichia coli to enhance production of L-tryptophan. Appl. Microbiol. Biotechnol. 2013, 97, 7587–7596. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.; Widhalm, J.R.; Qian, Y.; Maeda, H.; Cooper, B.R.; Jannasch, A.S.; Gonda, I.; Lewinsohn, E.; Rhodes, D.; Dudareva, N. An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine: Phenylpyruvate aminotransferase. Nat. Commun. 2013, 4, 2833. [Google Scholar] [CrossRef] [PubMed]
- Hall, G.C.; Flick, M.B.; Gherna, R.L.; Jensen, R.A. Biochemical diversity for biosynthesis of aromatic amino acids among the cyanobacteria. J. Bacteriol. 1982, 149, 65–78. [Google Scholar] [CrossRef]
- Zhang, S.; Wilson, D.B.; Ganem, B. Probing the catalytic mechanism of prephenate dehydratase by site-directed mutagenesis of the Escherichia coli P-protein dehydratase domain. Biochemistry 2000, 39, 4722–4728. [Google Scholar] [CrossRef]
- Báez-Viveros, J.L.; Osuna, J.; Hernández-Chávez, G.; Soberón, X.; Bolívar, F.; Gosset, G. Metabolic engineering and protein directed evolution increase the yield of L-phenylalanine synthesized from glucose in Escherichia coli. Biotechnol. Bioeng. 2004, 87, 516–524. [Google Scholar] [CrossRef]
- Lütke-Eversloh, T.; Stephanopoulos, G. Feedback inhibition of chorismate mutase/prephenate dehydrogenase (TyrA) of Escherichia coli: Generation and characterization of tyrosine-insensitive mutants. Appl. Environ. Microbiol. 2005, 71, 7224–7228. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.-L.; Chen, M.; Hou, S.; Li, T.; Yang, Y.-W.; Li, Q.; Jiang, H.-B.; Dai, G.-Z.; Zhang, Z.-C.; Hess, W.R.; et al. Genomic and transcriptomic insights into the survival of the subaerial cyanobacterium Nostoc flagelliforme in arid and exposed habitats. Environ. Microbiol. 2019, 21, 845–863. [Google Scholar] [CrossRef] [PubMed]
- Dienst, D.; Wichmann, J.; Mantovani, O.; Rodrigues, J.S.; Lindberg, P. High density cultivation for efficient sesquiterpenoid biosynthesis in Synechocystis sp. PCC 6803. Sci. Rep. 2020, 10, 5932. [Google Scholar] [CrossRef]
- Huccetogullari, D.; Luo, Z.W.; Lee, S.Y. Metabolic engineering of microorganisms for production of aromatic compounds. Microb. Cell Fact. 2019, 18, 41. [Google Scholar] [CrossRef]
- Gérando, H.M.; Fayolle-Guichard, F.; Rudant, L.; Millah, S.K.; Monot, F.; Ferreira, N.L.; López-Contreras, A.M. Improving isopropanol tolerance and production of Clostridium beijerinckii DSM 6423 by random mutagenesis and genome shuffling. Appl. Microbiol. Biotechnol. 2016, 100, 5427–5436. [Google Scholar] [CrossRef]
- Nagappan, S.; Devendran, S.; Tsai, P.-C.; Dahms, H.-U.; Ponnusamy, V.K. Potential of two-stage cultivation in microalgae biofuel production. Fuel 2019, 252, 339–349. [Google Scholar] [CrossRef]
- Kim, U.; Cho, D.-H.; Heo, J.; Yun, J.-H.; Choi, D.-Y.; Cho, K.; Kim, H.-S. Two-stage cultivation strategy for the improvement of pigment productivity from high-density heterotrophic algal cultures. Bioresour. Technol. 2020, 302, 122840. [Google Scholar] [CrossRef]
- Xie, Z.; Pei, H.; Zhang, L.; Yang, Z.; Nie, C.; Hou, Q.; Yu, Z. Accelerating lipid production in freshwater alga Chlorella sorokiniana SDEC-18 by seawater and ultrasound during the stationary phase. Renew. Energy 2020, 161, 448–456. [Google Scholar] [CrossRef]
- Liu, X.; Sheng, J.; Iii, R.C. Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 6899–6904. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, C.; Mitra, M.; Bhayani, K.; Bharadwaj, S.V.V.; Ghosh, T.; Dubey, S.; Mishra, S. Abiotic stresses as tools for metabolites in microalgae. Bioresour. Technol. 2017, 244 Pt 2, 1216–1226. [Google Scholar] [CrossRef]
- Cho, K.; Heo, J.; Cho, D.-H.; Tran, Q.-G.; Yun, J.-H.; Lee, S.-M.; Lee, Y.J.; Kim, H.-S. Enhancing algal biomass and lipid production by phycospheric bacterial volatiles and possible growth enhancing factor. Alg. Res. 2019, 37, 186–194. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).