Insights into the Variation in Bioactivities of Closely Related Streptomyces Strains from Marine Sediments of the Visayan Sea against ESKAPE and Ovarian Cancer
Abstract
1. Introduction
2. Results and Discussions
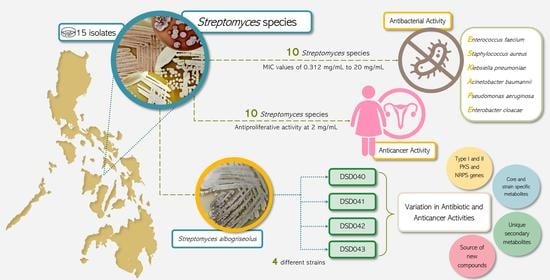
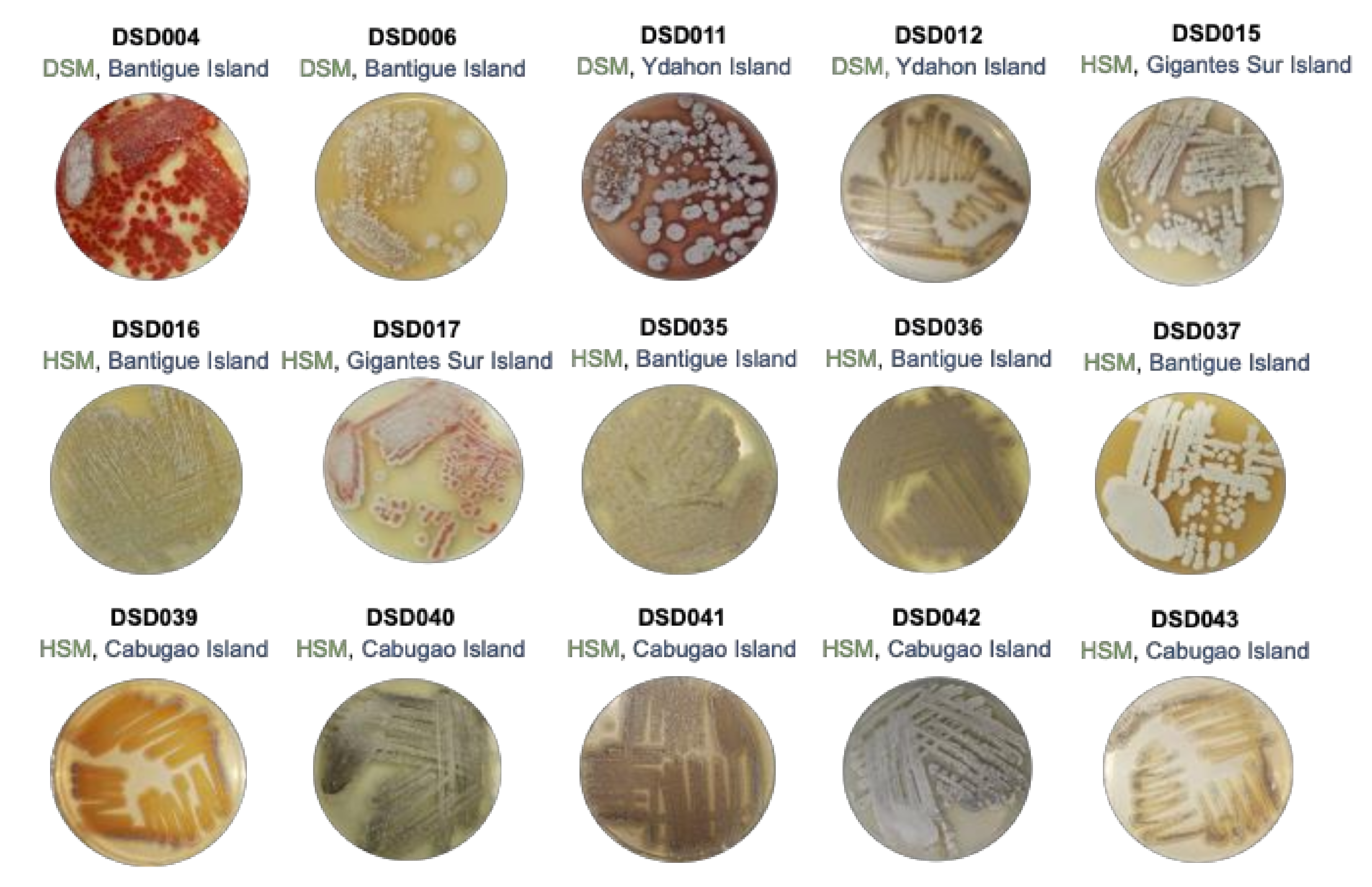
2.1. Cultivable Streptomycete Isolates in Marine Sediments from the Visayan Sea
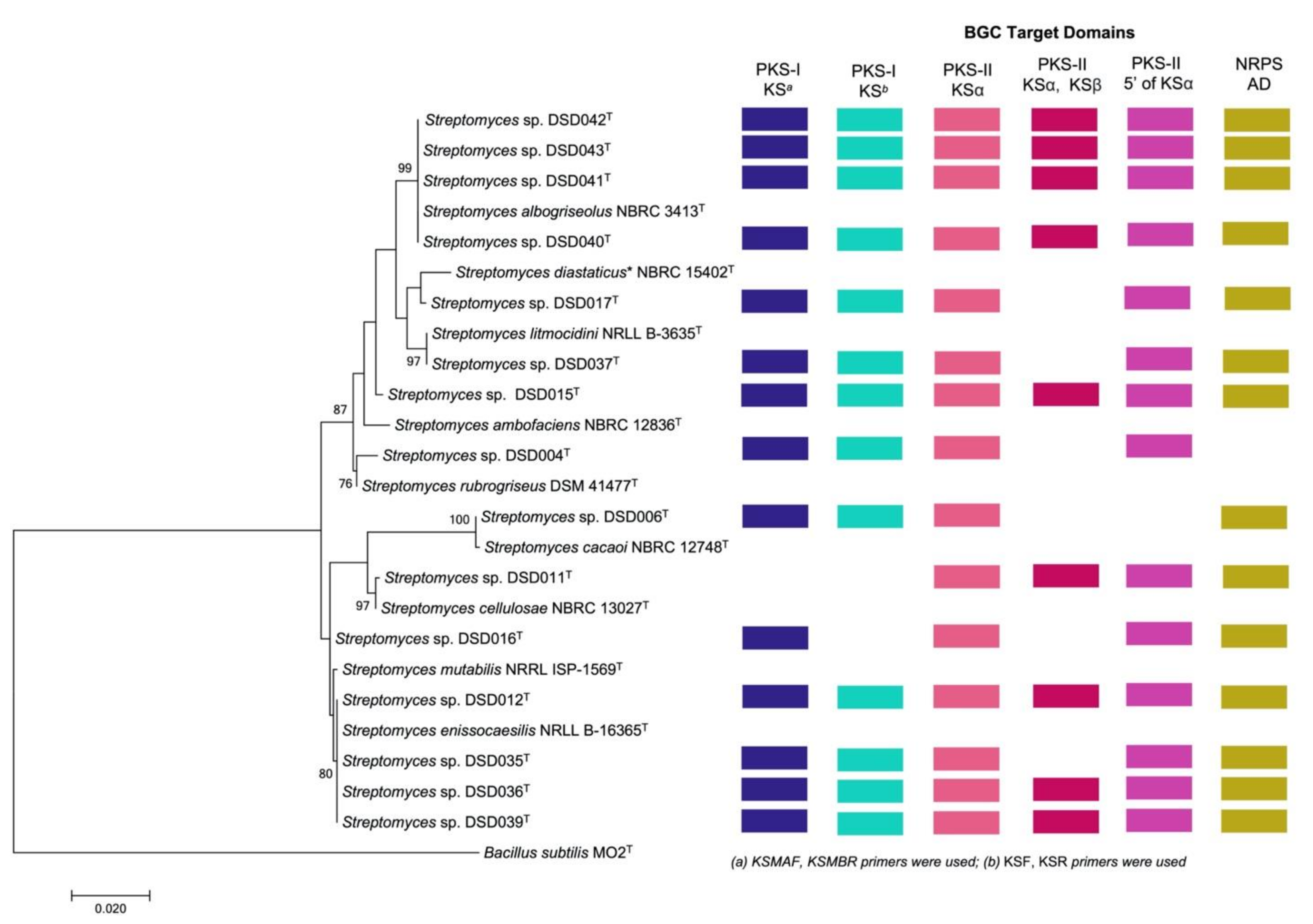
2.2. The Isolates from Marine Sediments of Visayan Sea Belong to Streptomyces Species
2.3. Detection of PKS (Types I and II) and NRPS Domains in Streptomyces Isolates
2.4. Antibiotic Activity against ESKAPE Pathogens
2.5. Antiproliferative Activity against Ovarian Cancer Cells
2.6. Bioactivity Correlation with Biosynthetic Gene Clusters
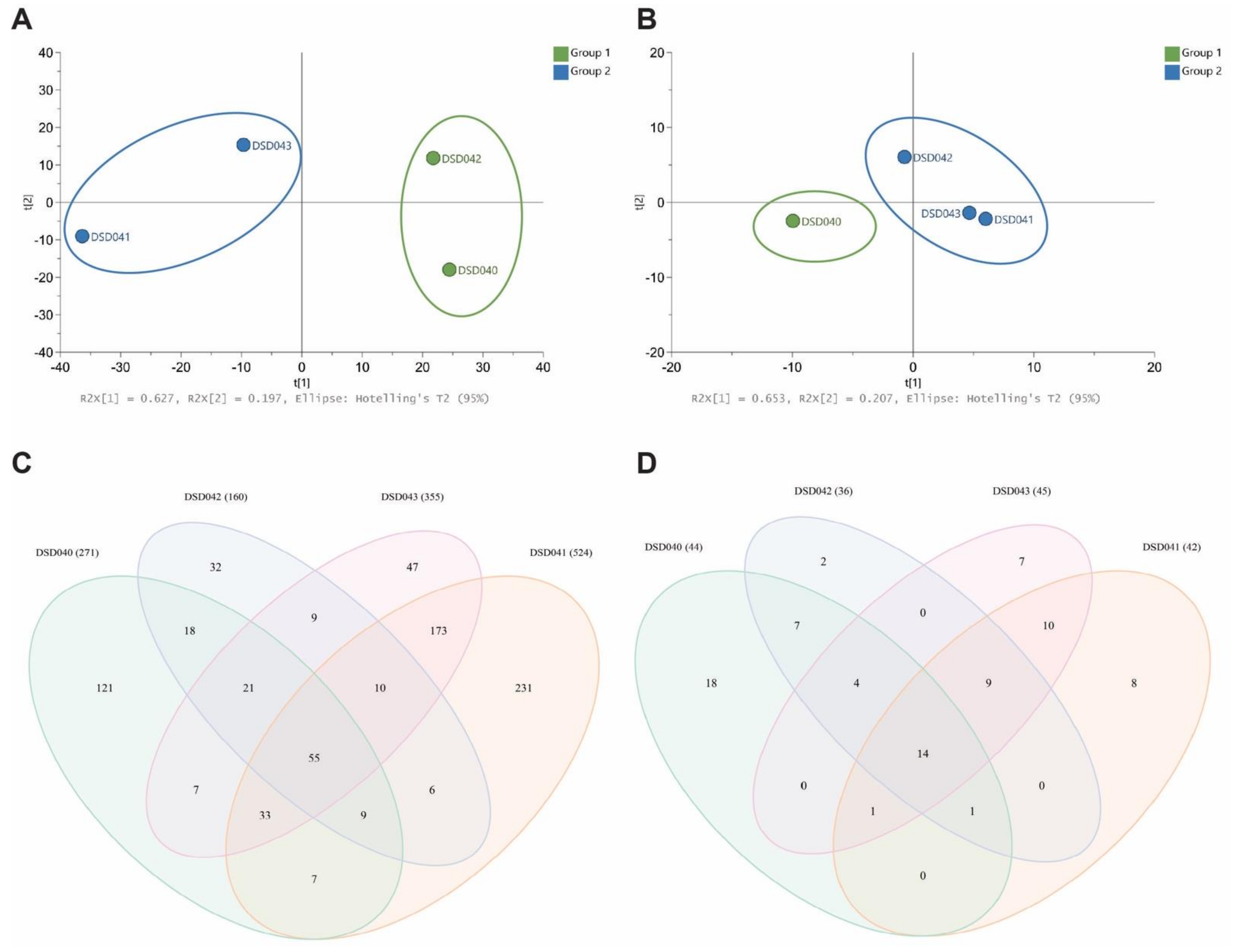
2.7. Metabolomics Analysis and Metabolite Profiling of Closely Related Marine Sediment-Derived Streptomyces albogriseolus Strains
2.8. Antibiotic activity of Streptomyces albogriseolus Strain DSD042T against ESKAPE Pathogens
3. Materials and Methods
3.1. Marine Sediment Selection and Culture Dependent Isolation
3.2. Salt Tolerance
3.3. Phylogenetic Analysis Using Small Subunit 16S rRNA Gene Sequences
3.4. PCR Amplification of Secondary Metabolite Biosynthetic Genes
3.5. Cultivation and Extraction of Biomass
3.6. Determination of Minimum Inhibitory Concentration against ESKAPE Pathogens
3.7. Anticancer Assay against Ovarian Cancer Cells
3.8. Gel Permeation Chromatography (GPC)
3.9. Disk Diffusion Assay of Streptomyces albogriseolus Strain DSD042T GPC Fractions
3.10. High-Pressure Liquid Chromatography Mass Spectrometry (HPLCMS), Multivariate and Venn Analyses of the Crude Extracts of Streptomyces Isolates from the Visayan Sea
3.11. High-Pressure Liquid Chromatography Mass Spectrometry (HPLCMS) and MS/MS Analysis of GPC Fractions and Dereplication
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015, 14, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Forsyth, C. Repairing the Antibiotic Pipeline: Can the GAIN Act Do It? Wash. JL Tech. Arts 2013, 9, 1. [Google Scholar]
- Butler, M.S.; Blaskovich, M.A.; Cooper, M.A. Antibiotics in the clinical pipeline at the end of 2015. J. Antibiot. 2017, 70, 3–24. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Alzate, K.Y.; Bauermeister, A.; Tangerina, M.M.P.; Lotufo, T.M.C.; Ferreira, M.J.P.; Jimenez, P.C.; Padilla, G.; Lopes, N.P.; Costa-Lotufo, L.V. Marine bacteria from Rocas Atoll as a rich source of pharmacologically active compounds. Mar. Drugs 2019, 17, 671. [Google Scholar] [CrossRef]
- Hassan, S.S.U.; Shaikh, A.L. Marine Actinobacteria as a drug treasure house. Biomed. Pharmacother. 2017, 87, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Landwehr, W.; Wolf, C.; Wink, J. Actinobacteria and Myxobacteria-two of the most important bacterial resources for novel antibiotics. Curr. Top Microbiol. Immunol. 2016, 398, 273–302. [Google Scholar] [PubMed]
- Xia, H.; Zhan, X.; Mao, X.M.; Li, Y.Q. The regulatory cascades of antibiotic production in Streptomyces. World J. Microbiol. Biotechnol. 2020, 36, 13. [Google Scholar] [CrossRef] [PubMed]
- Moumbock, A.F.A.; Gao, M.; Qaseem, A.; Li, J.; Kirchner, P.A.; Ndingkokhar, B.; Bekono, B.D.; Simoben, C.V.; Babiaka, S.B.; Malange, Y.I.; et al. StreptomeDB 3.0: An updated compendium of streptomycetes natural products. Nucleic Acids Res. 2021, 49, D600–D604. [Google Scholar] [CrossRef] [PubMed]
- Al-Amoudi, S.; Essack, M.; Simões, M.F.; Bougouffa, S.; Soloviev, I.; Archer, J.A.; Lafi, F.F.; Bajic, V.B. Bioprospecting Red Sea coastal ecosystems for culturable microorganisms and their antimicrobial potential. Mar. Drugs 2016, 14, 165. [Google Scholar] [CrossRef]
- Paderog, M.J.V.; Suarez, A.F.L.; Sabido, E.M.; Low, Z.J.; Saludes, J.P.; Dalisay, D.S. Anthracycline shunt metabolites from Philippine marine sediment-derived Streptomyces destroy cell membrane integrity of multidrug-resistant Staphylococcus aureus. Front. Microbiol. 2020, 11, 743. [Google Scholar] [CrossRef]
- Sabido, E.M.; Tenebro, C.P.; Suarez, A.F.L.; Ong, S.D.C.; Trono, D.J.V.L.; Amago, D.S.; Evangelista, J.E., Jr.; Reynoso, A.M.Q.; Villalobos, I.G.M.; Alit, L.D.D.; et al. Marine sediment-derived Streptomyces strain produces angucycline antibiotics against multidrug-resistant Staphylococcus aureus harboring SCCmec Type 1 gene. J. Mar. Sci. Eng. 2020, 8, 734. [Google Scholar] [CrossRef]
- Dalisay, D.S.; Williams, D.E.; Wang, X.L.; Centko, R.; Chen, J.; Andersen, R.J. Marine sediment-derived Streptomyces bacteria from British Columbia, Canada are a promising microbiota resource for the discovery of antimicrobial natural products. PLoS ONE 2013, 8, e77078. [Google Scholar] [CrossRef]
- Jensen, P.R.; Dwight, R.; Fenical, W. Distribution of actinomycetes in near-shore tropical marine sediments. Appl. Environ. Microbiol. 1991, 57, 1102–1108. [Google Scholar] [CrossRef]
- Hong, K.; Gao, A.H.; Xie, Q.Y.; Gao, H.; Zhuang, L.; Lin, H.P.; Yu, H.P.; Li, J.; Yao, X.S.; Goodfellow, M.; et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 2009, 7, 24–44. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; MacIntyre, L.; Abdelmohsen, U.R.; Horn, H.; Polymenakou, P.N.; Edrada-Ebel, R.; Hentschel, U. Biodiversity, anti-trypanosomal activity screening, and metabolomic profiling of Actinomycetes isolated from mediterranean sponges. PLoS ONE 2015, 10, e0138528. [Google Scholar] [CrossRef] [PubMed]
- Carr, S.A.; Orcutt, B.N.; Mandernack, K.W.; Spear, J.R. Abundant Atribacteria in deep marine sediment from the Adélie Basin, Antarctica. Front. Microbiol. 2015, 6, 872. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, T.; Doi, H.; Uramoto, G.-I.; Wörmer, L.; Adhikari, R.R.; Xiao, N.; Morono, Y.; D’Hondt, S.; Hinrichs, K.-U.; Inagaki, F. Global diversity of microbial communities in marine sediment. Proc. Natl. Acad. Sci. USA 2020, 117, 27587. [Google Scholar] [CrossRef]
- Stincone, P.; Brandelli, A. Marine bacteria as source of antimicrobial compounds. Crit. Rev. Biotechnol. 2020, 40, 306–319. [Google Scholar] [CrossRef]
- Tangerina, M.M.P.; Furtado, L.C.; Leite, V.M.B.; Bauermeister, A.; Velasco-Alzate, K.; Jimenez, P.C.; Garrido, L.M.; Padilla, G.; Lopes, N.P.; Costa-Lotufo, L.V.; et al. Metabolomic study of marine Streptomyces sp.: Secondary metabolites and the production of potential anticancer compounds. PLoS ONE 2020, 15, e0244385. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; De Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhang, L.; Zhou, Z.; Yan, X. Diversity of gene clusters for polyketide and nonribosomal peptide biosynthesis revealed by metagenomic analysis of the Yellow Sea sediment. Front. Microbiol. 2018, 9, 295. [Google Scholar] [CrossRef] [PubMed]
- Ridley, C.P.; Lee, H.Y.; Khosla, C. Evolution of polyketide synthases in bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 4595–4600. [Google Scholar] [CrossRef]
- Rao, H.C.; Rakshith, D.; Gurudatt, D.M.; Satish, S. Implication of PKS type I gene and chromatographic strategy for the biodiscovery of antimicrobial polyketide metabolites from endosymbiotic Nocardiopsis prasina CLA68. Naturwissenschaften 2016, 103, 45. [Google Scholar] [CrossRef]
- Otsuka, M.; Ichinose, K.; Fujii, I.; Ebizuka, Y. Cloning, sequencing, and functional analysis of an iterative type I polyketide synthase gene cluster for biosynthesis of the antitumor chlorinated polyenone neocarzilin in “Streptomyces carzinostaticus”. Antimicrob. Agents Chemother. 2004, 48, 3468–3476. [Google Scholar] [CrossRef]
- Kharel, M.K.; Pahari, P.; Shepherd, M.D.; Tibrewal, N.; Nybo, S.E.; Shaaban, K.A.; Rohr, J. Angucyclines: Biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 2012, 29, 264–325. [Google Scholar] [CrossRef]
- Hertweck, C.; Luzhetskyy, A.; Rebets, Y.; Bechthold, A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007, 24, 162–190. [Google Scholar] [CrossRef]
- Zothanpuia, A.K.P.; Gupta, V.K.; Singh, B.P. Detection of antibiotic-resistant bacteria endowed with antimicrobial activity from a freshwater lake and their phylogenetic affiliation. PeerJ 2016, 4, e2103. [Google Scholar] [CrossRef]
- Seow, K.T.; Meurer, G.; Gerlitz, M.; Wendt-Pienkowski, E.; Hutchinson, C.R.; Davies, J. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: A means to access and use genes from uncultured microorganisms. J. Bacteriol. 1997, 179, 7360–7368. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gontang, E.A.; Gaudêncio, S.P.; Fenical, W.; Jensen, P.R. Sequence-based analysis of secondary-metabolite biosynthesis in marine Actinobacteria. Appl. Environ. Microbiol. 2010, 76, 2487. [Google Scholar] [CrossRef]
- Doroghazi, J.R.; Metcalf, W.W. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genom. 2013, 14, 611. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.V.; Kallifidas, D.; Kim, J.H.; Charlop-Powers, Z.; Feng, Z.; Brady, S.F. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl. Environ. Microbiol. 2012, 78, 3744–3752. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, P.; Raja, A.; LaBonte, J.; Lebbos, J. Daptomycin. Nat. Rev. Drug Discov. 2003, 2, 943–944. [Google Scholar] [PubMed]
- Chau, F.; Lefort, A.; Benadda, S.; Dubée, V.; Fantin, B. Flow cytometry as a tool to determine the effects of cell wall-active antibiotics on vancomycin-susceptible and -resistant Enterococcus faecalis strains. Antimicrob. Agents Chemother. 2011, 55, 395–398. [Google Scholar] [CrossRef]
- Wei, H.-X.; Fang, X.-W.; Xie, X.-S.; Zhang, S.-P.; Jiang, Y.; Wu, S.-H. Secondary metabolites of a soil-derived Streptomyces kunmingensis. Chem. Nat. Compd. 2017, 53, 794–796. [Google Scholar] [CrossRef]
- Singh, R.; Dubey, A.K. Diversity and Applications of endophytic actinobacteria of plants in special and other ecological niches. Front. Microbiol. 2018, 9, 1767. [Google Scholar] [CrossRef]
- Manivasagan, P.; Kang, K.H.; Sivakumar, K.; Li-Chan, E.C.; Oh, H.M.; Kim, S.K. Marine actinobacteria: An important source of bioactive natural products. Environ. Toxicol. Pharmacol. 2014, 38, 172–188. [Google Scholar] [CrossRef]
- Morono, Y.; Ito, M.; Hoshino, T.; Terada, T.; Hori, T.; Ikehara, M.; D’Hondt, S.; Inagaki, F. Aerobic microbial life persists in oxic marine sediment as old as 101.5 million years. Nat. Commun. 2020, 11, 3626. [Google Scholar] [CrossRef] [PubMed]
- Petro, C.; Zäncker, B.; Starnawski, P.; Jochum, L.M.; Ferdelman, T.G.; Jørgensen, B.B.; Røy, H.; Kjeldsen, K.U.; Schramm, A. Marine deep biosphere microbial communities assemble in near-surface sediments in Aarhus Bay. Front. Microbiol. 2019, 10, 758. [Google Scholar] [CrossRef]
- Yang, N.; Song, F. Bioprospecting of novel and bioactive compounds from marine actinomycetes isolated from South China Sea sediments. Curr. Microbiol. 2018, 75, 142–149. [Google Scholar] [CrossRef]
- Undabarrena, A.; Beltrametti, F.; Claverías, F.P.; González, M.; Moore, E.R.; Seeger, M.; Cámara, B. Exploring the diversity and antimicrobial potential of marine actinobacteria from the Comau Fjord in Northern Patagonia, Chile. Front. Microbiol. 2016, 7, 1135. [Google Scholar] [CrossRef]
- Tangerina, M.M.; Correa, H.; Haltli, B.; Vilegas, W.; Kerr, R.G. Bioprospecting from cultivable bacterial communities of marine sediment and invertebrates from the underexplored Ubatuba region of Brazil. Arch. Microbiol. 2017, 199, 155–169. [Google Scholar] [CrossRef]
- Prieto-Davó, A.; Dias, T.; Gomes, S.E.; Rodrigues, S.; Parera-Valadez, Y.; Borralho, P.M.; Pereira, F.; Rodrigues, C.M.; Santos-Sanches, I.; Gaudêncio, S.P. The Madeira Archipelago as a significant source of marine-derived Actinomycete diversity with anticancer and antimicrobial potential. Front. Microbiol. 2016, 7, 1594. [Google Scholar] [CrossRef] [PubMed]
- Parera-Valadez, Y.; Yam-Puc, A.; López-Aguiar, L.K.; Borges-Argáez, R.; Figueroa-Saldivar, M.A.; Cáceres-Farfán, M.; Márquez-Velázquez, N.A.; Prieto-Davó, A. Ecological strategies behind the selection of cultivable Actinomycete strains from the yucatan peninsula for the discovery of secondary metabolites with antibiotic activity. Microb. Ecol. 2019, 77, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Meena, B.; Anburajan, L.; Vinithkumar, N.V.; Kirubagaran, R.; Dharani, G. Biodiversity and antibacterial potential of cultivable halophilic actinobacteria from the deep sea sediments of active volcanic Barren Island. Microb. Pathog. 2019, 132, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.E.; Dalisay, D.S.; Chen, J.; Polishchuck, E.A.; Patrick, B.O.; Narula, G.; Ko, M.; Av-Gay, Y.; Li, H.; Magarvey, N.; et al. Aminorifamycins and Sporalactams produced in culture by a Micromonospora sp. isolated from a northeastern-pacific marine sediment are potent antibiotics. Org. Lett. 2017, 19, 766–769. [Google Scholar] [CrossRef]
- An, J.S.; Shin, B.; Kim, T.H.; Hwang, S.; Shin, Y.H.; Cui, J.; Du, Y.E.; Yi, J.; Nam, S.J.; Hong, S.; et al. Dumulmycin, an antitubercular bicyclic macrolide from a riverine sediment-derived Streptomyces sp. Org. Lett. 2021, 23, 3359–3363. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.E.; Izard, F.; Arnould, S.; Dalisay, D.S.; Tantapakul, C.; Maneerat, W.; Matainaho, T.; Julien, E.; Andersen, R.J. Structures of nahuoic acids B-E produced in culture by a Streptomyces sp. isolated from a marine sediment and evidence for the inhibition of the histone methyl transferase setd8 in human cancer cells by nahuoic acid A. J. Org. Chem. 2016, 81, 1324–1332. [Google Scholar] [CrossRef]
- Williams, D.E.; Dalisay, D.S.; Li, F.; Amphlett, J.; Maneerat, W.; Chavez, M.A.; Wang, Y.A.; Matainaho, T.; Yu, W.; Brown, P.J.; et al. Nahuoic acid A produced by a Streptomyces sp. isolated from a marine sediment is a selective SAM-competitive inhibitor of the histone methyltransferase SETD8. Org. Lett. 2013, 15, 414–417. [Google Scholar] [CrossRef]
- Messaoudi, O.; Sudarman, E.; Bendahou, M.; Jansen, R.; Stadler, M.; Wink, J. Kenalactams A-E, Polyene Macrolactams Isolated from Nocardiopsis CG3. J. Nat. Prod. 2019, 82, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Becerril-Espinosa, A.; Freel, K.C.; Jensen, P.R.; Soria-Mercado, I.E. Marine Actinobacteria from the Gulf of California: Diversity, abundance and secondary metabolite biosynthetic potential. Antonie Van Leeuwenhoek 2013, 103, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, R.N.; Demko, A.M.; Patin, N.V.; Kapono, C.A.; Donia, M.S.; Dorrestein, P.; Jensen, P.R. Detection of natural products and their producers in ocean sediments. Appl. Environ. Microbiol. 2019, 85, e02830-18. [Google Scholar] [CrossRef]
- Coral Triangle Initiative on Coral Reefs, Fisheries and Food Security. Available online: https://coraltriangleinitiative.org/library/statistics-coral-triangle-facts-figures-and-calculations-patterns-biodiversity-and-endemism (accessed on 1 May 2021).
- Carpenter, K.E.; Springer, V.G. The center of the center of marine shore fish biodiversity: The Philippine Islands. Environ. Biol. Fishes 2005, 72, 467–480. [Google Scholar] [CrossRef]
- Goodfellow, M.; Williams, S.T. Ecology of actinomycetes. Annu. Rev. Microbiol. 1983, 37, 189–216. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Zeng, Q.; Huang, H.; Zhu, J.; Fang, Z.; Sun, Q.; Bao, S. A new nematicidal compound produced by Streptomyces albogriseolus HA10002. Antonie Van Leeuwenhoek 2013, 103, 1107–1111. [Google Scholar] [CrossRef]
- Li, X.L.; Xu, M.J.; Zhao, Y.L.; Xu, J. A novel benzo[f][1,7]naphthyridine produced by Streptomyces albogriseolus from mangrove sediments. Molecules 2010, 15, 9298–9307. [Google Scholar] [CrossRef] [PubMed]
- Thirumurugan, D.; Vijayakumar, R.; Vadivalagan, C.; Karthika, P.; Alam Khan, M.K. Isolation, structure elucidation and antibacterial activity of methyl-4,8-dimethylundecanate from the marine actinobacterium Streptomyces albogriseolus ECR64. Microb. Pathog. 2018, 121, 166–172. [Google Scholar] [CrossRef]
- Cui, C.B.; Liu, H.B.; Gu, J.Y.; Gu, Q.Q.; Cai, B.; Zhang, D.Y.; Zhu, T.J. Echinosporins as new cell cycle inhibitors and apoptosis inducers from marine-derived Streptomyces albogriseolus. Fitoterapia 2007, 78, 238–240. [Google Scholar] [CrossRef]
- Hu, L.; Chen, X.; Han, L.; Zhao, L.; Miao, C.; Huang, X.; Chen, Y.; Li, P.; Li, Y. Two new phenazine metabolites with antimicrobial activities from soil-derived Streptomyces species. J. Antibiot. 2019, 72, 574–577. [Google Scholar] [CrossRef]
- Li, Y.; Han, L.; Rong, H.; Li, L.; Zhao, L.; Wu, L.; Xu, L.; Jiang, Y.; Huang, X. Diastaphenazine, a new dimeric phenazine from an endophytic Streptomyces diastaticus subsp. ardesiacus. J. Antibiot. 2015, 68, 210–212. [Google Scholar] [CrossRef]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Igarashi, Y.; Tamura, T. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters among taxonomically close Streptomyces strains. Sci. Rep. 2018, 8, 6888. [Google Scholar] [CrossRef]
- Lanoot, B.; Vancanneyt, M.; Cleenwerck, I.; Wang, L.; Li, W.; Liu, Z.; Swings, J. The search for synonyms among streptomycetes by using SDS-PAGE of whole-cell proteins. Emendation of the species Streptomyces aurantiacus, Streptomyces cacaoi subsp. cacaoi, Streptomyces caeruleus and Streptomyces violaceus. Int. J. Syst. Evol. Microbiol. 2002, 52 Pt 3, 823–829. [Google Scholar]
- Kaweewan, I.; Hemmi, H.; Komaki, H.; Kodani, S. Isolation and structure determination of a new antibacterial peptide pentaminomycin C from Streptomyces cacaoi subsp. cacaoi. J. Antibiot. 2020, 73, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Yılmaz, S.; Aksoy, S.; Uzel, A.; Tosun, Ç.; Kirmizibayrak, P.B.; Bedir, E. Polyethers isolated from the marine actinobacterium Streptomyces cacaoi inhibit autophagy and induce apoptosis in cancer cells. Chem. Biol. Interact. 2019, 307, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Rawlings, B.J.; Harrison, P.H.; Vederas, J.C. Production of new polyene antibiotics by Streptomyces cellulosae after addition of ethyl (Z)-16-phenylhexadec-9-enoate. J. Antibiot. 1989, 42, 577–584. [Google Scholar] [CrossRef]
- Toumatia, O.; Yekkour, A.; Goudjal, Y.; Riba, A.; Coppel, Y.; Mathieu, F.; Sabaou, N.; Zitouni, A. Antifungal properties of an actinomycin D-producing strain, Streptomyces sp. IA1, isolated from a Saharan soil. J. Basic Microbiol. 2015, 55, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Fehr, T.; King, H.D.; Kuhn, M. Mutalomycin, a new polyether antibiotic taxonomy, fermentation, isolation and characterization. J. Antibiot. 1977, 30, 903–907. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hamed, A.; Abdel-Razek, A.S.; Frese, M.; Wibberg, D.; El-Haddad, A.F.; Ibrahim, T.M.A.; Kalinowski, J.; Sewald, N.; Shaaban, M. N-Acetylborrelidin B: A new bioactive metabolite from Streptomyces mutabilis sp. MII. Z Naturforsch. C J. Biosci. 2018, 73, 49–57. [Google Scholar] [CrossRef]
- Yassien, M.A.; Abdallah, H.M.; El-Halawany, A.M.; Jiman-Fatani, A.A. Anti-tuberculous activity of treponemycin produced by a Streptomyces strain MS-6-6 isolated from Saudi Arabia. Molecules 2015, 20, 2576–2590. [Google Scholar] [CrossRef]
- Kumar, P.; Kundu, A.; Kumar, M.; Solanki, R.; Kapur, M.K. Exploitation of potential bioactive compounds from two soil derived actinomycetes, Streptomyces sp. strain 196 and RI.24. Microbiol. Res. 2019, 229, 126312. [Google Scholar] [CrossRef]
- Lewis, R.A.; Devi, J.; Green, K.; Li, J.; Hopkins, A.; Hayles, J.; Nurse, P.; Errington, J.; Allenby, N.E.E. Screening and purification of natural products from actinomycetes that induce a “rounded” morphological phenotype in fission yeast. Nat. Prod. Bioprospect. 2021, 11, 431–445. [Google Scholar] [CrossRef]
- Shaaban, M.; El-Mahdy, A.M. Biosynthesis of Ag, Se, and ZnO nanoparticles with antimicrobial activities against resistant pathogens using waste isolate. IET Nanobiotechnol. 2018, 12, 741–747. [Google Scholar] [CrossRef]
- Hutchinson, E.; Murphy, B.; Dunne, T.; Breen, C.; Rawlings, B.; Caffrey, P. Redesign of polyene macrolide glycosylation: Engineered biosynthesis of 19-(O)-perosaminyl-amphoteronolide B. Chem. Biol. 2010, 17, 174–182. [Google Scholar] [CrossRef]
- Román-Hurtado, F.; Sánchez-Hidalgo, M.; Martín, J.; Ortiz-López, F.J.; Genilloud, O. Biosynthesis and heterologous expression of cacaoidin, the first member of the lanthidin family of RiPPs. Antibiotics 2021, 10, 403. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Progress and challenges in implementing the research on ESKAPE pathogens. Infect. Control. Hosp. Epidemiol. 2010, 31 (Suppl. S1), S7–S10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Willems, R.J.L.; Friedrich, A.W.; Rossen, J.W.A.; Bathoorn, E. Enterococcus faecium: From microbiological insights to practical recommendations for infection control and diagnostics. Antimicrob. Resist Infect. Control 2020, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zheng, W.; Rong, X.; Huang, Y. A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: Use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 2008, 58 Pt 1, 149–159. [Google Scholar] [CrossRef]
- Doroghazi, J.R.; Buckley, D.H. Intraspecies comparison of Streptomyces pratensis genomes reveals high levels of recombination and gene conservation between strains of disparate geographic origin. BMC Genom. 2014, 15, 970. [Google Scholar] [CrossRef]
- Doroghazi, J.R.; Buckley, D.H. Widespread homologous recombination within and between Streptomyces species. ISME J. 2010, 4, 1136–1143. [Google Scholar] [CrossRef]
- Park, C.J.; Andam, C.P. Within-species genomic variation and variable patterns of recombination in the tetracycline producer Streptomyces rimosus. Front. Microbiol. 2019, 10, 552. [Google Scholar] [CrossRef]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020, 10, 2003. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Jha, B. Intertidal marine sediment harbours Actinobacteria with promising bioactive and biosynthetic potential. Sci. Rep. 2017, 7, 10041. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F. Strain-level diversity of secondary metabolism in Streptomyces albus. PLoS ONE 2015, 10, e0116457. [Google Scholar] [CrossRef]
- Gao, D.; Zhou, T.; Da, L.T.; Bruhn, T.; Guo, L.L.; Chen, Y.H.; Xu, J.; Xu, M.J. Characterization and nonenzymatic transformation of three types of alkaloids from Streptomyces albogriseolus MGR072 and discovery of inhibitors of indoleamine 2,3-dioxygenase. Org. Lett. 2019, 21, 8577–8581. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.F.; Chen, M.J.; Liang, D.E.; Shi, L.M.; Ying, Y.M.; Shan, W.G.; Li, G.Q.; Zhan, Z.J. SY67903 produces eunicellin diterpenoids structurally similar to terpenes of the gorgonian. J. Nat. Prod. 2020, 83, 1641–1645. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Ginolhac, A.; Jarrin, C.; Robe, P.; Perrière, G.; Vogel, T.M.; Simonet, P.; Nalin, R. Type I polyketide synthases may have evolved through horizontal gene transfer. J. Mol. Evol. 2005, 60, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Izumikawa, M.; Murata, M.; Tachibana, K.; Ebizuka, Y.; Fujii, I. Cloning of modular type I polyketide synthase genes from salinomycin producing strain of Streptomyces albus. Bioorg. Med. Chem. 2003, 11, 3401–3405. [Google Scholar] [CrossRef]
- Ayuso-Sacido, A.; Genilloud, O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: Detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 2005, 49, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48 (Suppl. S1), 5–16. [Google Scholar] [CrossRef] [PubMed]


| Isolate Code | GenBank Accession Number | Nearest Related Strain | % Identity | Antibacterial Activity * (MIC90) | |||||
|---|---|---|---|---|---|---|---|---|---|
| E. faecium | S. aureus | K. pneumoniae | A. baumannii | P. aeruginosa | E. cloacae | ||||
| DSD004T | MZ323891 | Streptomyces rubrogriseus DSM 41477T | 99.39 | ||||||
| DSD006T | MZ323976 | Streptomyces cacaoi NBRC 12748T | 99.66 | ||||||
| DSD011T | MT820508 | Streptomyces cellulosae NBRC 13027T | 99.93 | ||||||
| DSD012T | MZ323983 | Streptomyces enissocaesilis NRLL B-16365T | 99.93 | ||||||
| DSD015T | MZ323974 | Streptomyces ambofaciens NBRC 12836T | 99.48 | ||||||
| DSD016T | MZ323997 | Streptomyces mutabilis NRRL ISP-1569T | 99.78 | ||||||
| DSD017T | MZ336041 | Streptomyces diastaticus ** NBRC 15402T | 99.11 | ||||||
| DSD035T | MZ323985 | Streptomyces enissocaesilis NRLL B-16365T | 99.86 | ||||||
| DSD036T | MZ323984 | Streptomyces enissocaesilis NRLL B-16365T | 100.00 | ||||||
| DSD037T | MZ323989 | Streptomyces litmocidini NRLL B-3635T | 99.91 | ||||||
| DSD039T | MZ323987 | Streptomyces enissocaesilis NRLL B-16365T | 99.73 | ||||||
| DSD040T | MZ323957 | Streptomyces albogriseolus NBRC 3413T | 99.86 | ||||||
| DSD041T | MZ336042 | Streptomyces albogriseolus NBRC 3413T | 99.93 | ||||||
| DSD042T | MZ323972 | Streptomyces albogriseolus NBRC 3413T | 100.00 | ||||||
| DSD043T | MZ323973 | Streptomyces albogriseolus NBRC 3413T | 99.93 | ||||||
| % Hit rate | 13 | 40 | 33 | 40 | 26 | 26 | |||
| Treatment Code | Disk Content (mg) | Zone of Inhibition (mm) | |||||
|---|---|---|---|---|---|---|---|
| E. faecium ATCC 70021 | S. aureus ATCC BAA-44 | K. pneumoniae ATCC BAA-1705 | A. baumannii ATCC BAA-1605 | P. aeruginosa ATCC BAA-1744 | E. cloacae ATCC BAA-2341 | ||
| G1 | 5.000 | - | - | - | - | - | - |
| G2 | 5.000 | - | - | - | - | - | - |
| G3 | 5.000 | - | 12.6 ± 2.0 | 9.6 ± 0.5 | 11.0 ± 1.7 c | 9.3 ± 0.5 b | 12.6 ± 2.0 c |
| G4 | 5.000 | 11.0 ± 1.0 a | 16.6 ± 1.0 c | 11.3 ± 0.5 c | 10.6 ± 0.5 | - | 13.0 ± 1.7 |
| Cefoxitin | 0.030 | 13.0 ± 1.7 | - | ||||
| Ciprofloxacin | 0.030 | - | - | ||||
| Imipenem | 0.010 | 11.3+ ± 2.0 c | - | 16.0+ ± 1.7 b | - | ||
| Meropenem | 0.010 | 16.0 ± 1.7 | 9.3 ± 0.5 c | 15.0+ ± 1.0 c | |||
| Vancomycin | 0.030 | 22.0 ± 1 a | 16.1 ± 1 c | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabido, E.M.; Tenebro, C.P.; Trono, D.J.V.L.; Vicera, C.V.B.; Leonida, S.F.L.; Maybay, J.J.W.B.; Reyes-Salarda, R.; Amago, D.S.; Aguadera, A.M.V.; Octaviano, M.C.; et al. Insights into the Variation in Bioactivities of Closely Related Streptomyces Strains from Marine Sediments of the Visayan Sea against ESKAPE and Ovarian Cancer. Mar. Drugs 2021, 19, 441. https://doi.org/10.3390/md19080441
Sabido EM, Tenebro CP, Trono DJVL, Vicera CVB, Leonida SFL, Maybay JJWB, Reyes-Salarda R, Amago DS, Aguadera AMV, Octaviano MC, et al. Insights into the Variation in Bioactivities of Closely Related Streptomyces Strains from Marine Sediments of the Visayan Sea against ESKAPE and Ovarian Cancer. Marine Drugs. 2021; 19(8):441. https://doi.org/10.3390/md19080441
Chicago/Turabian StyleSabido, Edna M., Chuckcris P. Tenebro, Dana Joanne Von L. Trono, Carmela Vannette B. Vicera, Sheeny Fane L. Leonida, Jose Jeffrey Wayne B. Maybay, Rikka Reyes-Salarda, Diana S. Amago, Angelica Marie V. Aguadera, May C. Octaviano, and et al. 2021. "Insights into the Variation in Bioactivities of Closely Related Streptomyces Strains from Marine Sediments of the Visayan Sea against ESKAPE and Ovarian Cancer" Marine Drugs 19, no. 8: 441. https://doi.org/10.3390/md19080441
APA StyleSabido, E. M., Tenebro, C. P., Trono, D. J. V. L., Vicera, C. V. B., Leonida, S. F. L., Maybay, J. J. W. B., Reyes-Salarda, R., Amago, D. S., Aguadera, A. M. V., Octaviano, M. C., Saludes, J. P., & Dalisay, D. S. (2021). Insights into the Variation in Bioactivities of Closely Related Streptomyces Strains from Marine Sediments of the Visayan Sea against ESKAPE and Ovarian Cancer. Marine Drugs, 19(8), 441. https://doi.org/10.3390/md19080441