Ubiquitousness of Haloferax and Carotenoid Producing Genes in Arabian Sea Coastal Biosystems of India
Abstract
:1. Introduction
2. Results
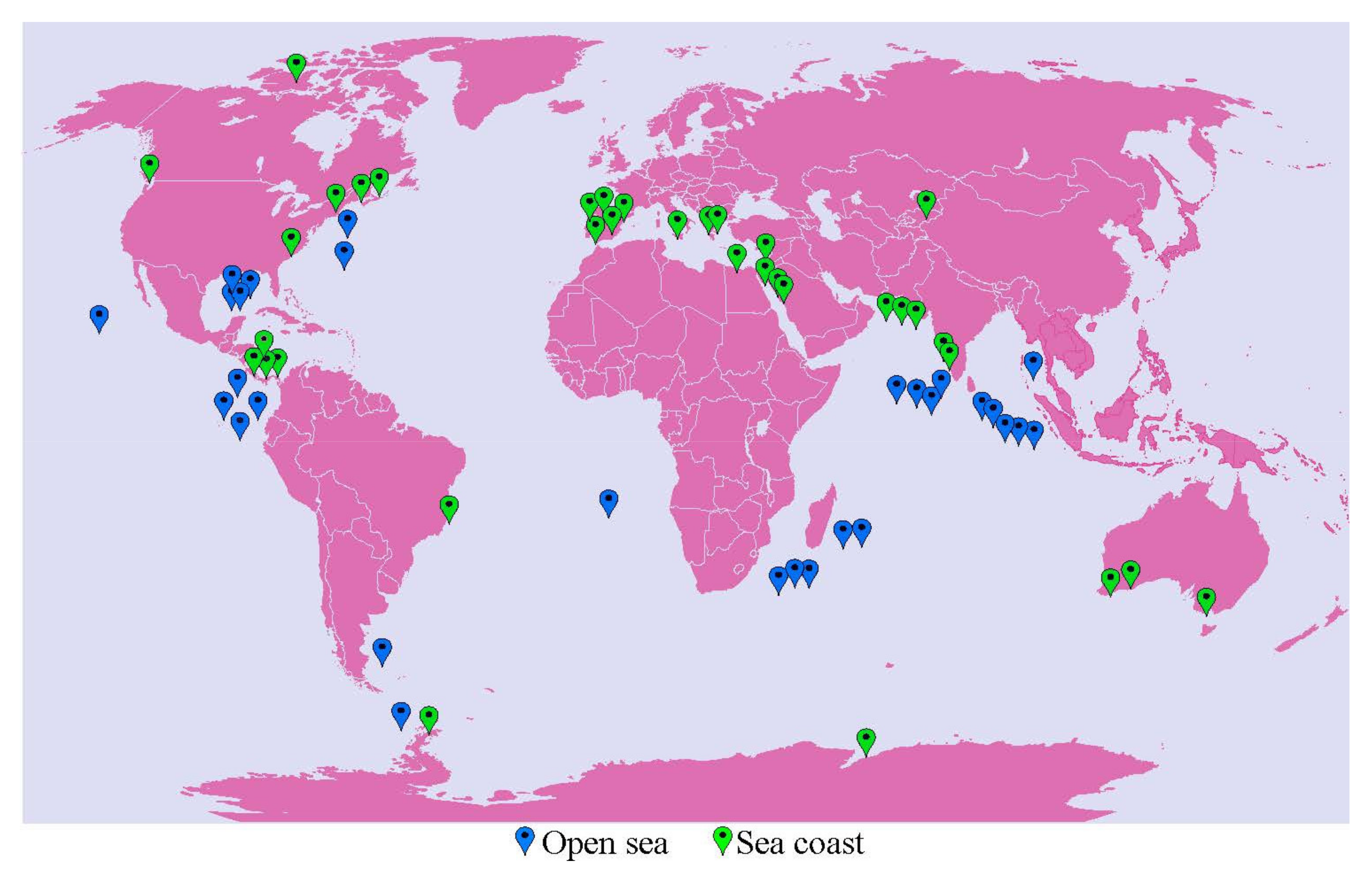
2.1. Microbiome Composition in Open Sea and Seacoast
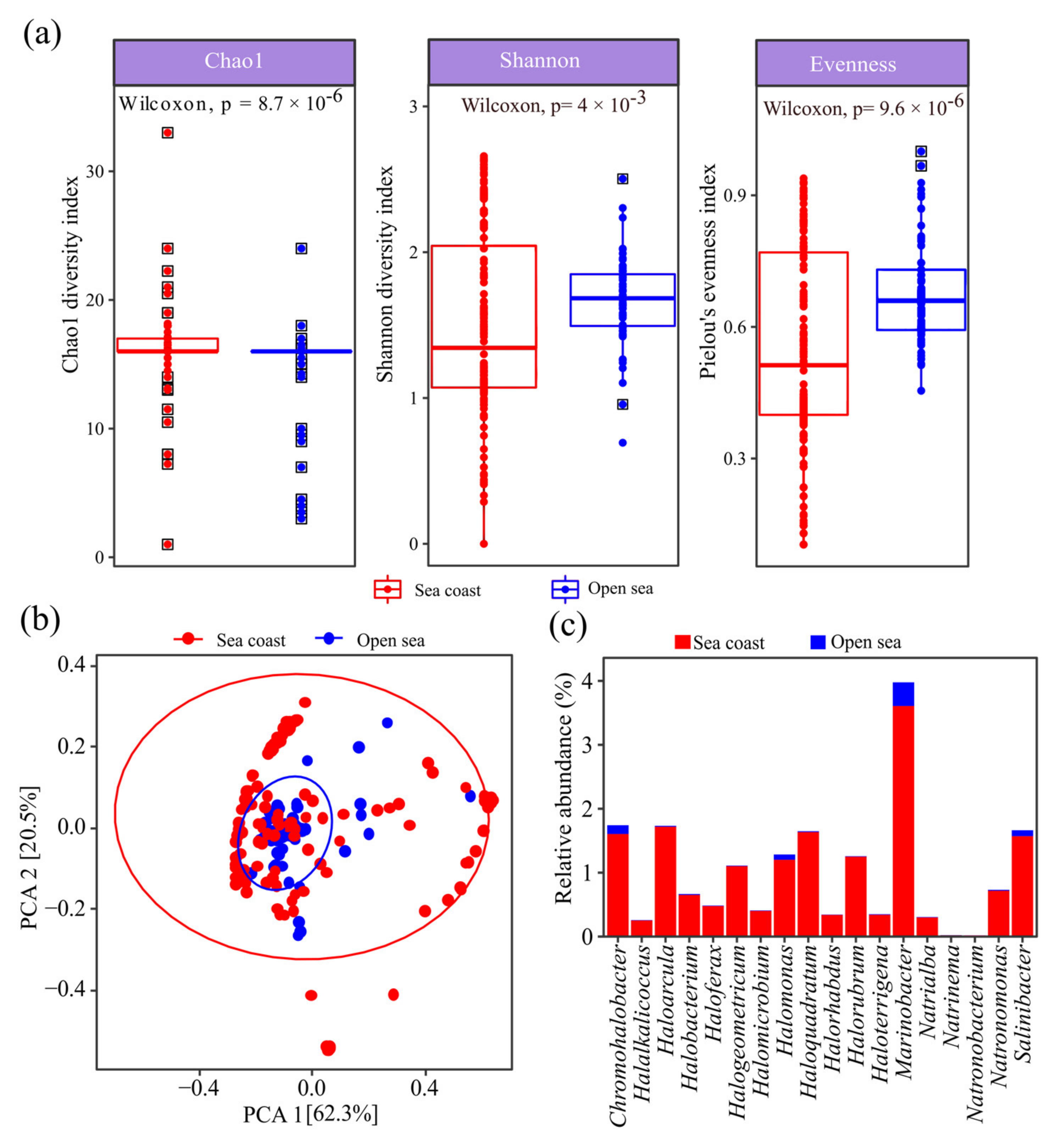
2.2. Diversity of Halophilic Microbiome in Open Sea and Seacoast
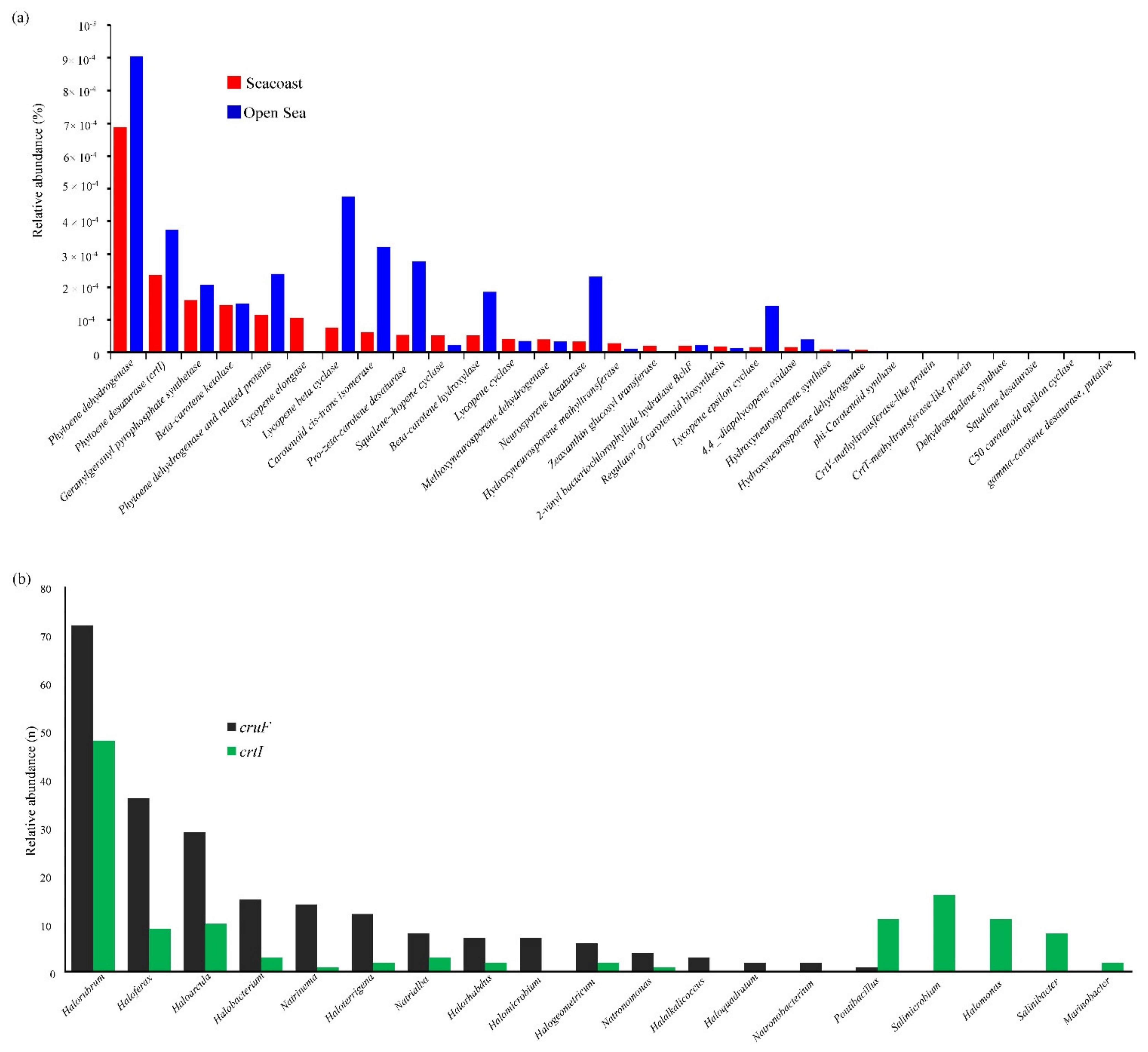
2.3. Diversity of Carotenoid Gene in the Halophiles
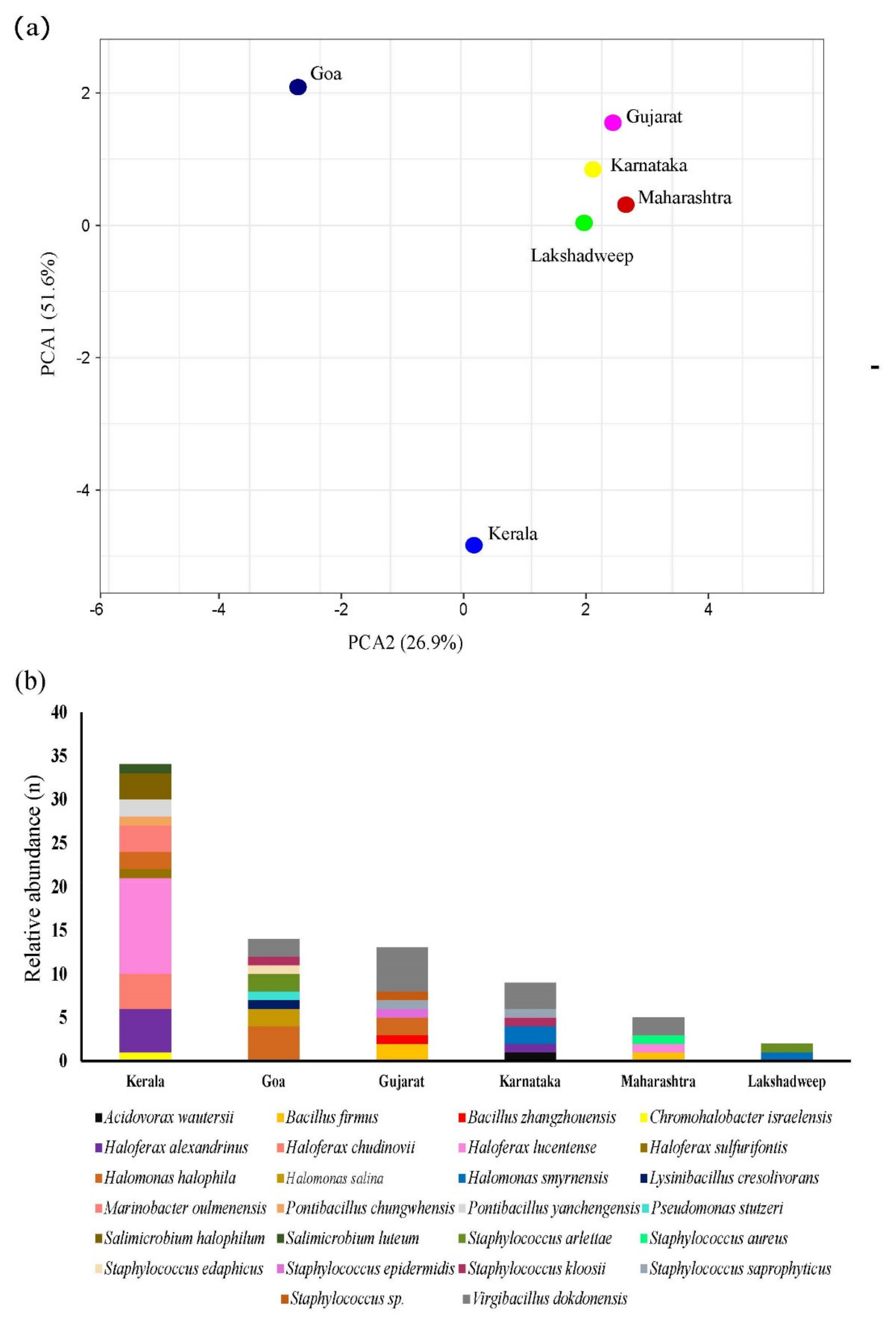
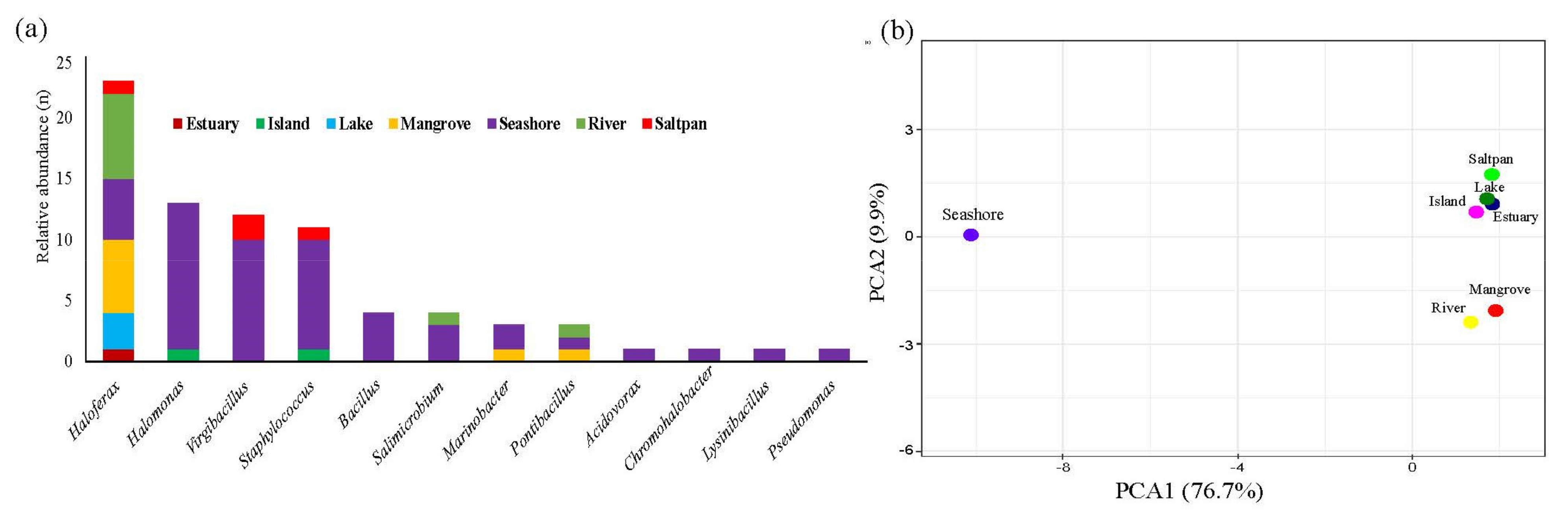
2.4. Isolation of Pigmented Halophiles across the Arabian Sea Coastline
2.5. Morphological Diversity across the Halotolerant Organism
2.6. Biochemical Diversity across the Halotolerant Organisms
2.7. Antibiotic Susceptibility Profiling
2.8. Influence of NaCl Concentration on Pigment Production
2.9. Identification of the Isolates through 16S rDNA Phylogenetic Analysis
2.10. G + C Diversity across the Halotolerant Organisms
3. Discussion
3.1. Extreme Halophiles Are Enriched in Seacoast Biosystems
3.2. Archaeal Extreme Halophiles Are Enriched with crtI and cruF
3.3. Archaeal Halophiles Exhibit Diverse Pigmentation
3.4. Biogeography of Extreme Halophiles in Arabia Sea Coast of India
3.5. Morphological, Biochemical, and Antibiotic Resistance Profiles of Halotolerant Species
4. Materials and Methods
4.1. Metagenomic Analysis
4.2. Sediment Sampling
4.3. Enrichment and Isolation of Pigmented Halophiles
4.4. 16S rRNA Gene Identification and Phylogenetic Analysis
4.5. Optimization of NaCl Concentration on Growth and Pigment Production
4.6. Morphological and Biochemical Analysis
4.7. G + C Content Estimation
4.8. Antibiotics Resistance Profiling
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calegari-Santos, R.; Diogo, R.A.; Fontana, J.D.; Bonfim, T.M.B. Carotenoid Production by Halophilic Archaea Under Different Culture Conditions. Curr. Microbiol. 2016, 72, 641–651. [Google Scholar] [CrossRef]
- Moopantakath, J.; Imchen, M.; Siddhardha, B.; Kumavath, R. 16s rRNA metagenomic analysis reveals predominance of Crtl and CruF genes in Arabian Sea coast of India. Sci. Total Environ. 2020, 743, 140699. [Google Scholar] [CrossRef] [PubMed]
- Kirti, K.; Amita, S.; Priti, S.; Kumar, A.M.; Jyoti, S. Colorful World of Microbes: Carotenoids and Their Applications. Adv. Biol. 2014, 2014, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Rodrigo-Baños, M.; Garbayo, I.; Vílchez, C.; Bonete, M.-J.; Martínez-Espinosa, R.M. Carotenoids from Haloarchaea and Their Potential in Biotechnology. Mar. Drugs 2015, 13, 5508–5532. [Google Scholar] [CrossRef] [Green Version]
- Giani, M.; Garbayo, I.; Vílchez, C.; Martínez-Espinosa, R.M. Haloarchaeal Carotenoids: Healthy Novel Compounds from Extreme Environments. Mar. Drugs 2019, 17, 524. [Google Scholar] [CrossRef] [Green Version]
- Hegazy, G.; Abu-Serie, M.M.; Abo-Elela, G.M.; Ghozlan, H.; Sabry, S.A.; Soliman, N.A.; Abdel-Fattah, Y.R. In vitro dual (anticancer and antiviral) activity of the carotenoids produced by haloalkaliphilic archaeon Natrialba sp. M6. Sci. Rep. 2020, 10, 5986-14. [Google Scholar] [CrossRef]
- Oren, A. Halophilic microbial communities and their environments. Curr. Opin. Biotechnol. 2015, 33, 119–124. [Google Scholar] [CrossRef]
- Edgcomb, V.P.; Biddle, J.F.; Edgcomb, V.P.A.; Altenbach, A.V.; Bernhard, J.M.; Seckbach, J. (Eds.) Cellular Origin, Life in Extreme Habitats and Astrobiology; Springer: Dordrecht, The Netherlands, 2012; Volume 21, ISBN 978-94-007-1895-1. [Google Scholar]
- Jensen, M.W.; Matlock, S.A.; Reinheimer, C.H.; Lawlor, C.J.; Reinheimer, T.A.; Gorrell, A. Potassium stress growth characteristics and energetics in the haloarchaeon Haloarcula marismortui. Extremophiles 2015, 19, 315–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallsworth, J.E. Microbial unknowns at the saline limits for life. Nat. Ecol. Evol. 2019, 3, 1503–1504. [Google Scholar] [CrossRef]
- Steinmuller, H.E.; Foster, T.E.; Boudreau, P.; Hinkle, C.R.; Chambers, L.G. Characterization of herbaceous encroachment on soil biogeochemical cycling within a coastal marsh. Sci. Total Environ. 2020, 738, 139532. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Lovell, C.R. Microbial Surface Colonization and Biofilm Development in Marine Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toulza, E.; Tagliabue, A.; Blain, S.; Piganeau, G. Analysis of the Global Ocean Sampling (GOS) Project for Trends in Iron Uptake by Surface Ocean Microbes. PLoS ONE 2012, 7, e30931. [Google Scholar] [CrossRef] [PubMed]
- De la Calle, F. Marine microbiome as source of natural products. Microb. Biotechnol. 2017, 10, 1293–1296. [Google Scholar] [CrossRef] [PubMed]
- Mena, C.; Reglero, P.; Balbín, R.; Martín, M.; Santiago, R.; Sintes, E. Seasonal Niche Partitioning of Surface Temperate Open Ocean Prokaryotic Communities. Front. Microbiol. 2020, 11, 1749. [Google Scholar] [CrossRef] [PubMed]
- Moran, M.A. The global ocean microbiome. Science 2015, 350, aac8455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Carvalho, C.C.C.R.; Caramujo, M.J. Carotenoids in Aquatic Ecosystems and Aquaculture: A Colorful Business with Implications for Human Health. Front. Mar. Sci. 2017, 4, 93. [Google Scholar] [CrossRef] [Green Version]
- Giani, M.; Martínez-Espinosa, R. Carotenoids as a Protection Mechanism against Oxidative Stress. Haloferax Mediterr. Antioxid. 2020, 9, 1060. [Google Scholar] [CrossRef] [PubMed]
- Giani, M.; Montero-Lobato, Z.; Garbayo, I.; Vílchez, C.; Vega, J.; Martínez-Espinosa, R. Haloferax mediterranei Cells as C50 Carotenoid Factories. Mar. Drugs 2021, 19, 100. [Google Scholar] [CrossRef]
- Paul, S.; Bag, S.K.; Das, S.; Harvill, E.; Dutta, C. Molecular signature of hypersaline adaptation: Insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 2008, 9, R70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartman, A.L.; Norais, C.; Badger, J.H.; Delmas, S.; Haldenby, S.; Madupu, R.; Robinson, J.; Khouri, H.; Ren, Q.; Lowe, T.M.; et al. The Complete Genome Sequence of Haloferax volcanii DS2, a Model Archaeon. PLoS ONE 2010, 5, e9605. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.P.; Nicora, C.D.; Carini, P.; Lipton, M.S.; Norbeck, A.D.; Smith, R.D.; Giovannoni, S.J. Proteome Remodeling in Response to Sulfur Limitation in “Candidatus Pelagibacter ubique. ” mSystems 2016, 1, e00068-16. [Google Scholar] [CrossRef] [Green Version]
- Kasai, Y.; Kishira, H.; Sasaki, T.; Syutsubo, K.; Watanabe, K.; Harayama, S. Predominant growth of Alcanivorax strains in oil-contaminated and nutrient-supplemented sea water. Environ. Microbiol. 2002, 4, 141–147. [Google Scholar] [CrossRef]
- Chernikova, T.N.; Bargiela, R.; Toshchakov, S.V.; Shivaraman, V.; Lunev, E.A.; Yakimov, M.M.; Thomas, D.N.; Golyshin, P.N. Hydrocarbon-Degrading Bacteria Alcanivorax and Marinobacter Associated with Microalgae Pavlova lutheri and Nannochloropsis oculata. Front. Microbiol. 2020, 11, 572931. [Google Scholar] [CrossRef]
- Astriani, M.; Zubaidah, S.; Abadi, A.L.; Suarsini, E. Pseudomonas plecoglossicida as a novel bacterium for phosphate solubilizing and indole-3-acetic acid-producing from soybean rhizospheric soils of East Java, Indonesia. Biodiversitas J. Biol. Divers. 2020, 21, 578–586. [Google Scholar] [CrossRef]
- Thombre, R.S.; Shinde, V.D.; Oke, R.S.; Dhar, S.K.; Shouche, Y.S. Biology and survival of extremely halophilic archaeon Haloarcula marismortui RR12 isolated from Mumbai salterns, India in response to salinity stress. Sci. Rep. 2016, 6, 25642. [Google Scholar] [CrossRef]
- Bonete, M.-J.; Martínez-Espinosa, R.M.; Pire, C.; Zafrilla, B.; Richardson, D.J. Nitrogen metabolism in haloarchaea. Saline Syst. 2008, 4, 9–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolhuis, H.; Martín-Cuadrado, A.B.; Rosselli, R.; Pasic, L.; Rodriguez-Valera, F. Transcriptome analysis of Haloquadratum walsbyi: Vanity is but the surface. BMC Genom. 2017, 18, 1–14. [Google Scholar] [CrossRef]
- Imchen, M.; Vennapu, R.K.; Ghosh, P.; Kumavath, R. Insights into Antagonistic Interactions of Multidrug Resistant Bacteria in Mangrove Sediments from the South Indian State of Kerala. Microorgasims 2019, 7, 678. [Google Scholar] [CrossRef] [Green Version]
- Ask, J.; Rowe, O.; Brugel, S.; Strömgren, M.; Byström, P.; Andersson, A. Importance of coastal primary production in the northern Baltic Sea. Ambio 2016, 45, 635–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldwin, A.H.A.; Mendelssohn, I. Effects of salinity and water level on coastal marshes: An experimental test of disturbance as a catalyst for vegetation change. Aquat. Bot. 1998, 61, 255–268. [Google Scholar] [CrossRef]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum Hydrocarbon-Degrading Bacteria for the Remediation of Oil Pollution Under Aerobic Conditions: A Perspective Analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.T. Enrichment of Potential Halophilic Marinobacter Consortium for Mineralization of Petroleum Hydrocarbons and Also as Oil Reservoir Indicator in Red Sea, Saudi Arabia. Polycycl. Aromat. Compd. 2020, 1–12. [Google Scholar] [CrossRef]
- Lu, Y.; Yuan, J.; Lu, X.; Su, C.; Zhang, Y.; Wang, C.; Cao, X.; Li, Q.; Su, J.; Ittekkot, V.; et al. Major threats of pollution and climate change to global coastal ecosystems and enhanced management for sustainability. Environ. Pollut. 2018, 239, 670–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fathepure, B.Z. Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments. Front. Microbiol. 2014, 5, 173. [Google Scholar] [CrossRef] [Green Version]
- Vargas, C.; Argandona, M.; Reina-Bueno, M.; Rodriguez-Moya, J.; Fernandez-Aunion, C.; Nieto, J.J. Unravelling the adaptation responses to osmotic and temperature stress in Chromohalobacter salexigens, a bacterium with broad salinity tolerance. Saline Syst. 2008, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Giani, M.; Miralles-Robledillo, J.M.; Peiró, G.; Pire, C.; Martínez-Espinosa, R.M. Deciphering Pathways for Carotenogenesis in Haloarchaea. Molecules 2020, 25, 1197. [Google Scholar] [CrossRef] [Green Version]
- Fong, N.J.C.; Burgess, M.L.; Barrow, K.D.; Glenn, D.R. Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl. Microbiol. Biotechnol. 2001, 56, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Dubey, A.P.; Kumar, S.; Dutta, D.; Mishra, M.N.; Singh, B.N.; Tripathi, A.K. Carotenoid Biosynthetic Pathways Are Regulated by a Network of Multiple Cascades of Alternative Sigma Factors in Azospirillum brasilense Sp7. J. Bacteriol. 2016, 198, 2955–2964. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.; Chanotiya, C.S.; Shanker, K.; Tripathi, A.K. Characterization of carotenoids and genes encoding their biosynthetic pathways in Azospirillum brasilense. FEMS Microbiol. Lett. 2021, 368. [Google Scholar] [CrossRef]
- Naziri, D.; Hamidi, M.; Hassanzadeh, S.; Tarhriz, V.; Zanjani, B.M.; Nazemyieh, H.; Hejazi, M.A.; Hejazi, M.S. Analysis of Carotenoid Production by Halorubrum sp. TBZ126; an Extremely Halophilic Archeon from Urmia Lake. Adv. Pharm. Bull. 2013, 4, 61–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalazar, L.; Pagola, P.; Miró, M.V.; Churio, M.S.; Cerletti, M.; Martínez, C.; Iniesta-Cuerda, M.; Soler, A.; Cesari, A.; De Castro, R.; et al. Bacterioruberin extracts from a genetically modified hyperpigmented Haloferax volcanii strain: Antioxidant activity and bioactive properties on sperm cells. J. Appl. Microbiol. 2018, 126, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Ambati, R.R.; Phang, S.-M.; Ravi, S.; Aswathanarayana, R.G. Astaxanthin: Sources, Extraction, Stability, Biological Activities and Its Commercial Applications—A Review. Mar. Drugs 2014, 12, 128–152. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Cui, H.-L. In Vitro Antioxidant, Antihemolytic, and Anticancer Activity of the Carotenoids from Halophilic Archaea. Curr. Microbiol. 2017, 75, 266–271. [Google Scholar] [CrossRef]
- Bernstein, P.S.; Li, B.; Vachali, P.P.; Gorusupudi, A.; Shyam, R.; Henriksen, B.S.; Nolan, J.M. Lutein, zeaxanthin, and meso-zeaxanthin: The basic and clinical science underlying carotenoid-based nutritional interventions against ocular disease. Prog. Retin. Eye Res. 2016, 50, 34–66. [Google Scholar] [CrossRef] [Green Version]
- Sibero, M.T.; Igarashi, Y.; Radjasa, O.K.; Sabdono, A.; Trianto, A.; Zilda, D.S.; Wijaya, Y.J. Sponge-associated fungi from a mangrove habitat in Indonesia: Species composition, antimicrobial activity, enzyme screening and bioactive profiling. Int. Aquat. Res. 2019, 11, 173–186. [Google Scholar] [CrossRef] [Green Version]
- Estrada, M.; Henriksen, P.; Gasol, J.M.O.; Casamayor, E.; Pedrãs-Aliã, C. Diversity of planktonic photoautotrophic microorganisms along a salinity gradient as depicted by microscopy, flow cytometry, pigment analysis and DNA-based methods. FEMS Microbiol. Ecol. 2004, 49, 281–293. [Google Scholar] [CrossRef]
- Amoozegar, M.A.; Makhdoumi, A.; Mehrshad, M.; Fazeli, S.A.S.; Ventosa, A. Halopenitus malekzadehii sp. nov., an extremely halophilic archaeon isolated from a salt lake. Int. J. Syst. Evol. Microbiol. 2013, 63, 3232–3236. [Google Scholar] [CrossRef] [Green Version]
- Repeta, D.J.; Gagosian, R.B. Carotenoid transformations in coastal marine waters. Nat. Cell Biol. 1982, 295, 51–54. [Google Scholar] [CrossRef]
- Leiva, S.; Alvarado, P.; Huang, Y.; Wang, J.; Garrido, I. Diversity of pigmented Gram-positive bacteria associated with marine macroalgae from Antarctica. FEMS Microbiol. Lett. 2015, 362, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-G.; Gwak, J.-H.; Jung, M.-Y.; An, S.-U.; Hyun, J.-H.; Kang, S.; Rhee, S.-K. Distinct temporal dynamics of planktonic archaeal and bacterial assemblages in the bays of the Yellow Sea. PLoS ONE 2019, 14, e0221408. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Vahed, S.Z.; Forouhandeh, H.; Tarhriz, V.; Chaparzadeh, N.; Hejazi, M.A.; Jeon, C.O.; Hejazi, M.S. Halomonas urmiana sp. nov., a moderately halophilic bacterium isolated from Urmia Lake in Iran. Int. J. Syst. Evol. Microbiol. 2020, 70, 2254–2260. [Google Scholar] [CrossRef] [PubMed]
- Whitman, R.L.; Harwood, V.J.; Edge, T.A.; Nevers, M.B.; Byappanahalli, M.N.; Vijayavel, K.; Brandão, J.; Sadowsky, M.J.; Alm, E.W.; Crowe, A.; et al. Microbes in beach sands: Integrating environment, ecology and public health. Rev. Environ. Sci. Bio/Technology 2014, 13, 329–368. [Google Scholar] [CrossRef] [Green Version]
- D’Souza, S.E.; Altekar, W. Adaptive response of Haloferax mediterranei to low concentrations of NaCl (<20%) in the growth medium. Arch. Microbiol. 1997, 168, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Šmarda, P.; Bureš, P.; Horová, L.; Leitch, I.J.; Mucina, L.; Pacini, E.; Tichý, L.; Grulich, V.; Rotreklová, O. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proc. Natl. Acad. Sci. USA 2014, 111, E4096–E4102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yakovchuk, P.; Protozanova, E.; Frank-Kamenetskii, M.D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006, 34, 564–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.L.; Baxter, B.K. Bipyrimidine Signatures as a Photoprotective Genome Strategy in G + C-rich Halophilic archaea. Life 2016, 6, 37. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Kimura, H. Temperature-dependent expression of different guanine-plus-cytosine content 16S rRNA genes in Haloarcula strains of the class Halobacteria. Antonie Leeuwenhoek 2019, 112, 187–201. [Google Scholar] [CrossRef]
- Palidwor, G.A.; Perkins, T.J.; Xia, X. A General Model of Codon Bias Due to GC Mutational Bias. PLoS ONE 2010, 5, e13431. [Google Scholar] [CrossRef]
- Du, M.-Z.; Liu, S.; Zeng, Z.; Alemayehu, L.A.; Wei, W.; Guo, F.-B. Amino acid compositions contribute to the proteins’ evolution under the influence of their abundances and genomic GC content. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Eoren, A. Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front. Microbiol. 2013, 4, 315. [Google Scholar] [CrossRef] [Green Version]
- Bogacz-Radomska, L.; Harasym, J.; Piwowar, A. Commercialization aspects of carotenoids. In Carotenoids: Properties, Processing and Applications; Elsevier BV: Amsterdam, The Netherlands, 2020; pp. 327–357. [Google Scholar]
- Liu, L.; Chen, J. (Eds.) Systems and Synthetic Biotechnology for Production of Nutraceuticals; Springer Singapore: Singapore, 2019; ISBN 978-981-15-0445-7. [Google Scholar]
- Chen, C.W.; Hsu, S.-H.; Lin, M.-T.; Hsu, Y.-H. Mass production of C50 carotenoids by Haloferax mediterranei in using extruded rice bran and starch under optimal conductivity of brined medium. Bioprocess Biosyst. Eng. 2015, 38, 2361–2367. [Google Scholar] [CrossRef]
- Kumar, S.; Grewal, J.; Sadaf, A.; Hemamalini, R.; Khare, S.K. Halophiles as a source of polyextremophilic α-amylase for industrial applications. AIMS Microbiol. 2016, 2, 1–26. [Google Scholar] [CrossRef]
- Haque, R.U.; Paradisi, F.; Allers, T. Haloferax volcanii as immobilised whole cell biocatalyst: New applications for halophilic systems. Appl. Microbiol. Biotechnol. 2019, 103, 3807–3817. [Google Scholar] [CrossRef] [Green Version]
- Pais, J.; Serafim, L.; Freitas, F.; Reis, M.A. Conversion of cheese whey into poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by Haloferax mediterranei. New Biotechnol. 2016, 33, 224–230. [Google Scholar] [CrossRef]
- Alsafadi, D.; Al-Mashaqbeh, O. A one-stage cultivation process for the production of poly-3-(hydroxybutyrate-co-hydroxyvalerate) from olive mill wastewater by Haloferax mediterranei. New Biotechnol. 2017, 34, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Simó-Cabrera, L.; García-Chumillas, S.; Hagagy, N.; Saddiq, A.; Tag, H.; Selim, S.; AbdElgawad, H.; Agüero, A.A.; Sánchez, F.M.; Cánovas, V.; et al. Haloarchaea as Cell Factories to Produce Bioplastics. Mar. Drugs 2021, 19, 159. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, H.-Y. Purification and characterization of novel organic-solvent-tolerant β-amylase and serine protease from a newly isolated Salimicrobium halophilum strain LY20. FEMS Microbiol. Lett. 2012, 329, 204–211. [Google Scholar] [CrossRef] [Green Version]
- Sagar, S.; Esau, L.; Holtermann, K.; Hikmawan, T.; Zhang, G.; Stingl, U.; Bajic, V.B.; Kaur, M. Induction of apoptosis in cancer cell lines by the Red Sea brine pool bacterial extracts. BMC Complement. Altern. Med. 2013, 13, 344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, E.J.; Nam, S.-J.; Paul, L.; Beatty, D.; Kauffman, C.; Jensen, P.R.; Fenical, W. Previously Uncultured Marine Bacteria Linked to Novel Alkaloid Production. Chem. Biol. 2015, 22, 1270–1279. [Google Scholar] [CrossRef] [Green Version]
- Esau, L.; Zhang, G.; Sagar, S.; Stingl, U.; Bajic, V.B.; Kaur, M. Mining the deep Red-Sea brine pool microbial community for anticancer therapeutics. BMC Complement. Altern. Med. 2019, 19, 142. [Google Scholar] [CrossRef]
- Fang, W.; Xue, S.; Deng, P.; Zhang, X.; Wang, X.; Xiao, Y.; Fang, Z. AmyZ1: A novel α-amylase from marine bacterium Pontibacillus sp. ZY with high activity toward raw starches. Biotechnol. Biofuels 2019, 12, 95. [Google Scholar] [CrossRef]
- Rodríguez-Moya, J.; Argandoña, M.; Guerra, F.I.; Nieto, J.J.; Vargas, C. Temperature- and Salinity-Decoupled Overproduction of Hydroxyectoine by Chromohalobacter salexigens. Appl. Environ. Microbiol. 2013, 79, 1018–1023. [Google Scholar] [CrossRef] [Green Version]
- Erdogmus, S.F.; Korcan, S.E.; Konuk, M.; Guven, K.M.B.M. Aromatic Hydrocarbon Utilization Ability of Chromohalobacter sp. Ekoloji 2015, 16, 10–16. [Google Scholar] [CrossRef]
- Llamas, I.; Béjar, V.; Martínez-Checa, F.; Martínez-Cánovas, M.J.; Molina, I.; Quesada, E. Halomonas stenophila sp. nov., a halophilic bacterium that produces sulphate exopolysaccharides with biological activity. Int. J. Syst. Evol. Microbiol. 2011, 61, 2508–2514. [Google Scholar] [CrossRef] [PubMed]
- Haque, R.U.; Paradisi, F.; Allers, T. Haloferax volcanii for biotechnology applications: Challenges, current state and perspectives. Appl. Microbiol. Biotechnol. 2019, 104, 1371–1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, M.-R.; Zhang, D.-F.; Yang, X.-Y.; Zhang, X.-M.; Zhang, Y.-G.; Zhang, Y.-M.; Zhu, H.; Li, W.-J. Halomonas nanhaiensis sp. nov., a halophilic bacterium isolated from a sediment sample from the South China Sea. Antonie Leeuwenhoek 2013, 103, 997–1005. [Google Scholar] [CrossRef]
- Gutierrez, T.; Morris, G.; Ellis, D.; Mulloy, B.; Aitken, M.D. Production and characterisation of a marine Halomonas surface-active exopolymer. Appl. Microbiol. Biotechnol. 2019, 104, 1063–1076. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, M.; Garlapati, V.K. Production and Characterization of a Halo-, Solvent-, Thermo-tolerant Alkaline Lipase by Staphylococcus arlettae JPBW-1, Isolated from Rock Salt Mine. Appl. Biochem. Biotechnol. 2013, 171, 1429–1443. [Google Scholar] [CrossRef]
- Egusa, E.A.; Edwards, D.J.; Thao, M.L.; Kirk, L.L.; Hanne, L.F. Isolation and Characterization of Bacteria that Produce Polyhydroxybutyrate Depolymerases†. J. Microbiol. Biol. Educ. 2018, 19, 10–12. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.P.; Prasad, T.G. Formation and spreading of Arabian Sea high-salinity water mass. J. Geophys. Res. Space Phys. 1999, 104, 1455–1464. [Google Scholar] [CrossRef]
- Arahal, D.R.; García, M.T.; Ludwig, W.; Schleifer, K.H.; Ventosa, A. Transfer of Halomonas canadensis and Halomonas israelensis to the genus Chromohalobacter as Chromohalobacter canadensis comb. nov. and Chromohalobacter israelensis comb. nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 1443–1448. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, C.M.; Kamekura, M.; Holmes, M.L.; Dyall-Smith, M.L.; Ventosa, A. Taxonomic characterization of Haloferax sp. (“H. alicantei“) strain Aa 2.2: Description of Haloferax lucentensis sp. nov. Extremophiles 2002, 6, 479–483. [Google Scholar] [CrossRef]
- Lim, J.-M.; Jeon, C.O.; Song, S.M.; Kim, C.-J. Pontibacillus chungwhensis gen. nov., sp. nov., a moderately halophilic Gram-positive bacterium from a solar saltern in Korea. Int. J. Syst. Evol. Microbiol. 2005, 55, 165–170. [Google Scholar] [CrossRef] [Green Version]
- Saralov, A.I.; Baslerov, R.V.; Kuznetsov, B.B. Haloferax chudinovii sp. nov., a halophilic archaeon from Permian potassium salt deposits. Extremophiles 2013, 17, 499–504. [Google Scholar] [CrossRef]
- DasSarma, S.; DasSarma, P.; Laye, V.J.; Schwieterman, E.W. Extremophilic models for astrobiology: Haloarchaeal survival strategies and pigments for remote sensing. Extremophiles 2020, 24, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Pratap, R.; Geetanjali, S.; Indresh, M.; Maurya, K.; Wei, Y. Microbial Versatility in Varied Environments; Singh, R.P., Manchanda, G., Maurya, I.K., Wei, Y., Eds.; Springer: Singapore, 2020; ISBN 978-981-15-3027-2. [Google Scholar]
- Xie, X.; Pu, L.; Wang, Q.; Zhu, M.; Xu, Y.; Zhang, M. Response of soil physicochemical properties and enzyme activities to long-term reclamation of coastal saline soil, Eastern China. Sci. Total Environ. 2017, 607-608, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Indira, D.; Das, B.; Balasubramanian, P.; Jayabalan, R. Sea Water as a Reaction Medium for Bioethanol Production. In Microbial Biotechnology; Springer Singapore: Singapore, 2018; Volume 2, pp. 171–192. [Google Scholar]
- Pan, C.; Liu, C.; Zhao, H.; Wang, Y. Changes of soil physico-chemical properties and enzyme activities in relation to grassland salinization. Eur. J. Soil Biol. 2013, 55, 13–19. [Google Scholar] [CrossRef]
- Mouhamad, R.S.; Razaq, I.B.; Fadhel, A.S.; Yousir, S.A.; Taha, D.I.; Iqbal, M. Urease Activity under Salinity Stress in Calcareous Soils of Semi-Arid Regions of Iraq. Int. J. Chem. Biochem. Sci. 2014, 6, 68–71. [Google Scholar]
- Yang, H.; Hu, J.; Long, X.; Liu, Z.; Rengel, Z. Salinity altered root distribution and increased diversity of bacterial communities in the rhizosphere soil of Jerusalem artichoke. Sci. Rep. 2016, 6, 20687. [Google Scholar] [CrossRef]
- Arai, S.; Shibazaki, C.; Shimizu, R.; Adachi, M.; Ishibashi, M.; Tokunaga, H.; Tokunaga, M. Catalytic mechanism and evolutionary characteristics of thioredoxin from Halobacterium salinarum NRC-1. Acta Crystallogr. Sect. D Struct. Biol. 2020, 76, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ventosa, A.; Nieto, J.J.; Oren, A. Biology of Moderately Halophilic Aerobic Bacteria. Microbiol. Mol. Biol. Rev. 1998, 62, 504–544. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, D.C.; Von Der Weid, I.; Vaisman, N.; Seldin, L. Halotolerant Spore-Forming Gram-Positive Bacterial Diversity Associated with Blutaparon portulacoides (St. Hill.) Mears, a Pioneer Species in Brazilian Coastal Dunes. J. Microbiol. Biotechnol. 2006, 16, 193–199. [Google Scholar]
- Stevens, H.; Brinkhoff, T.; Rink, B.; Vollmers, J.; Simon, M. Diversity and abundance of Gram positive bacteria in a tidal flat ecosystem. Environ. Microbiol. 2007, 9, 1810–1822. [Google Scholar] [CrossRef]
- Islam, S.; Tanaka, M. Impacts of pollution on coastal and marine ecosystems including coastal and marine fisheries and approach for management: A review and synthesis. Mar. Pollut. Bull. 2004, 48, 624–649. [Google Scholar] [CrossRef] [PubMed]
- Keesing, J.; Irvine, T. Coastal Biodiversity in the Indian Ocean: The Known, the Unknown and the Unknowable. Int. J. Mol. Sci. 2020, 34, 11–26. [Google Scholar]
- Wafar, M.; Venkataraman, K.; Ingole, B.; Khan, S.A.; LokaBharathi, P. State of Knowledge of Coastal and Marine Biodiversity of Indian Ocean Countries. PLoS ONE 2011, 6, e14613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.-G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 92-93, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fresia, P.; Antelo, V.; Salazar, C.; Giménez, M.; D’Alessandro, B.; Afshinnekoo, E.; Mason, C.; Gonnet, G.H.; Iraola, G. Urban Metagenomics Uncover Antibiotic Resistance Reservoirs in Coastal Beach and Sewage Waters. Microbiome 2019, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.W.; Banks, K.; Gregg, K.; Shedler, S.; Walker, B.K. Antibiotic Resistance in Marine Microbial Communities Proximal to a Florida Sewage Outfall System. Antibiotics 2020, 9, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imchen, M.; Kumavath, R.; Barh, D.; Vaz, A.; Góes-Neto, A.; Tiwari, S.; Ghosh, P.; Wattam, A.R.; Azevedo, V. Comparative mangrove metagenome reveals global prevalence of heavy metals and antibiotic resistome across different ecosystems. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Assar, A.; Abdelraoof, M.I.; Abdel-Maboud, M.; Shaker, K.H.; Menshawy, A.; Swelam, A.H.; Eid, M.; Khalid, R.; Mogahed, M.; Abushouk, A.I.; et al. Knowledge, attitudes, and practices of Egypt’s future physicians towards antimicrobial resistance (KAP-AMR study): A multicenter cross-sectional study. Environ. Sci. Pollut. Res. 2020, 27, 21292–21298. [Google Scholar] [CrossRef]
- Hermsen, E.D.; MacGeorge, E.L.; Andresen, M.-L.; Myers, L.M.; Lillis, C.J.; Rosof, B.M. Decreasing the Peril of Antimicrobial Resistance Through Enhanced Health Literacy in Outpatient Settings: An Underrecognized Approach to Advance Antimicrobial Stewardship. Adv. Ther. 2020, 37, 918–932. [Google Scholar] [CrossRef] [Green Version]
- Osman, O.; Tanguichi, H.; Ikeda, K.; Park, P.; Tanabe-Hosoi, S.; Nagata, S. Copper-resistant halophilic bacterium isolated from the polluted Maruit Lake, Egypt. J. Appl. Microbiol. 2010, 108, 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- Shinde, V.; Thombre, R. Antibiotic resistance profiling of Marine halophilic bacteria and Haloarchaea. J. Appl. Pharm. Sci. 2016, 6, 132–137. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, A.; Chettri, B.; Langpoklakpam, J.S.; Singh, A.K.; Chattopadhyay, D. Draft Genome Sequence of Hydrocarbon-Degrading Enterobacter cloacae Strain S1:CND1, Isolated from Crude Oil-Contaminated Soil from the Noonmati Oil Refinery, Guwahati, Assam, India. Genome Announc. 2016, 4, e00367-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendis, H.C.; Thomas, V.P.; Schwientek, P.; Salamzade, R.; Chien, J.-T.; Waidyarathne, P.; Kloepper, J.; De La Fuente, L. Strain-specific quantification of root colonization by plant growth promoting rhizobacteria Bacillus firmus I-1582 and Bacillus amyloliquefaciens QST713 in non-sterile soil and field conditions. PLoS ONE 2018, 13, e0193119. [Google Scholar] [CrossRef] [PubMed]
- Susič, N.; Žibrat, U.; Sinkovič, L.; Vončina, A.; Razinger, J.; Knapič, M.; Sedlar, A.; Širca, S.; Stare, B.G. From Genome to Field—Observation of the Multimodal Nematicidal and Plant Growth-Promoting Effects of Bacillus firmus I-1582 on Tomatoes Using Hyperspectral Remote Sensing. Plants 2020, 9, 592. [Google Scholar] [CrossRef]
- Logan, N. Bacillus and relatives in foodborne illness. J. Appl. Microbiol. 2011, 112, 417–429. [Google Scholar] [CrossRef]
- Ehlers, S.; Merrill, S. A Staphylococcus saprophyticus; StatPearls Publishing: Treasure Island, FL, USA, 2020; PMID: 29493989. [Google Scholar]
- Vaidya, V.K. Horizontal Transfer of Antimicrobial Resistance by Extended-Spectrum β Lactamase-Producing Enterobacteriaceae. J. Lab. Physicians 2011, 3, 037–042. [Google Scholar] [CrossRef]
- Courvalin, P. Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 1994, 38, 1447–1451. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Lu, J.; Mao, L.; Li, J.; Yuan, Z.; Bond, P.; Guo, J. Antiepileptic drug carbamazepine promotes horizontal transfer of plasmid-borne multi-antibiotic resistance genes within and across bacterial genera. ISME J. 2019, 13, 509–522. [Google Scholar] [CrossRef] [Green Version]
- Furlan, J.P.R.; dos Santos, L.D.R.; Moretto-Altarugio, J.; Ramos, M.S.; Gallo, I.F.L.; Alves, G.D.A.D.; Paulelli, A.C.; Rocha, C.C.D.S.; Cesila, C.A.; Gallimberti, M.; et al. Occurrence and abundance of clinically relevant antimicrobial resistance genes in environmental samples after the Brumadinho dam disaster, Brazil. Sci. Total Environ. 2020, 726, 138100. [Google Scholar] [CrossRef]
- preprocessCore: A Collection of Pre-Processing Functions. R Package Version 1.54.0. Available online: https://github.com/bmbolstad/preprocessCore (accessed on 28 May 2021).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.Rproject.org/ (accessed on 28 May 2021).
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Han, R.; Zhang, X.; Liu, J.; Long, Q.; Chen, L.; Liu, D.; Zhu, D. Microbial community structure and diversity within hypersaline Keke Salt Lake environments. Can. J. Microbiol. 2017, 63, 895–908. [Google Scholar] [CrossRef] [PubMed]
- Oueriaghli, N.; Castro, D.J.; Llamas, I.; Béjar, V.; Martínez-Checa, F. Study of Bacterial Community Composition and Correlation of Environmental Variables in Rambla Salada, a Hypersaline Environment in South-Eastern Spain. Front. Microbiol. 2018, 9, 1377. [Google Scholar] [CrossRef] [PubMed]
- Bora, N.; Dodd, C.; Desmasures, N. (Eds.) Diversity, Dynamics and Functional Role of Actinomycetes on European Smear Ripened Cheeses; Springer International Publishing: Cham, Switzerland, 2015; ISBN 978-3-319-10463-8. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets Brief Communication. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Catalase Test Protocol. Available online: https://asm.org/getattachment/72a871fc-ba92-4128-a194-6f1bab5c3ab7/Catalase-Test-Protocol.pdf (accessed on 28 May 2021).
- Oxidase Test Protocol. Available online: https://asm.org/getattachment/00ce8639-8e76-4acb-8591-0f7b22a347c6/oxidase-test-protocol-3229.pdf (accessed on 28 May 2021).
- Phillips, K.; Zaidan, F.; Elizondo, O.R.; Lowe, K.L. Phenotypic characterization and 16S rDNA identification of culturable non-obligate halophilic bacterial communities from a hypersaline lake, La Sal del Rey, in extreme South Texas (USA). Aquat. Biosyst. 2012, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, J.M.; Sáiz-Jiménez, C. A fluorimetric method for the estimation of G+C mol% content in microorganisms by thermal denaturation temperature. Environ. Microbiol. 2002, 4, 770–773. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]


| Domain | Bacteria | Archaea | ||||||
|---|---|---|---|---|---|---|---|---|
| No. of strains | 22,615 | 925 | ||||||
| crt genes | crtI | crtL | cruF | crtD | crtI | crtL | crtD | cruF |
| No. of genes | 14,795 | 3001 | 397 | 1837 | 427 | 0 | 568 | 415 |
| No. of unique strains | 6766 | 2684 | 382 | 1643 | 292 | 0 | 329 | 377 |
| Serial Number | Ingredients | g/L or Molar Concentration |
|---|---|---|
| 1 | NaCl | 1–6 M |
| 2 | Tryptone | 5 |
| 3 | Yeast extract | 10 |
| 4 | Potassium chloride | 2 |
| 5 | Trisodium citrate | 3 |
| 6 | MgSO4 | 20 |
| Primer Name | Universal Primer | Sequence |
|---|---|---|
| Arch344F | Archeal Forward | 5′-ACGGGGTGCAGGCGCGA-3′ |
| Arch915R | Archeal Reverse | 5′-GTGCTCCCCCGCCAATTCCT-3′ |
| 8F | Bacterial Forward | 5′-AGAGTTTGATCCTGGCTCAG-3′ |
| 518R | Bacterial Reverse | 5′-ATTACCGCGGCTGCTGG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moopantakath, J.; Imchen, M.; Kumavath, R.; Martínez-Espinosa, R.M. Ubiquitousness of Haloferax and Carotenoid Producing Genes in Arabian Sea Coastal Biosystems of India. Mar. Drugs 2021, 19, 442. https://doi.org/10.3390/md19080442
Moopantakath J, Imchen M, Kumavath R, Martínez-Espinosa RM. Ubiquitousness of Haloferax and Carotenoid Producing Genes in Arabian Sea Coastal Biosystems of India. Marine Drugs. 2021; 19(8):442. https://doi.org/10.3390/md19080442
Chicago/Turabian StyleMoopantakath, Jamseel, Madangchanok Imchen, Ranjith Kumavath, and Rosa María Martínez-Espinosa. 2021. "Ubiquitousness of Haloferax and Carotenoid Producing Genes in Arabian Sea Coastal Biosystems of India" Marine Drugs 19, no. 8: 442. https://doi.org/10.3390/md19080442
APA StyleMoopantakath, J., Imchen, M., Kumavath, R., & Martínez-Espinosa, R. M. (2021). Ubiquitousness of Haloferax and Carotenoid Producing Genes in Arabian Sea Coastal Biosystems of India. Marine Drugs, 19(8), 442. https://doi.org/10.3390/md19080442










