Chemosensory-Related Genes in Marine Copepods
Abstract
:1. Introduction
2. Results
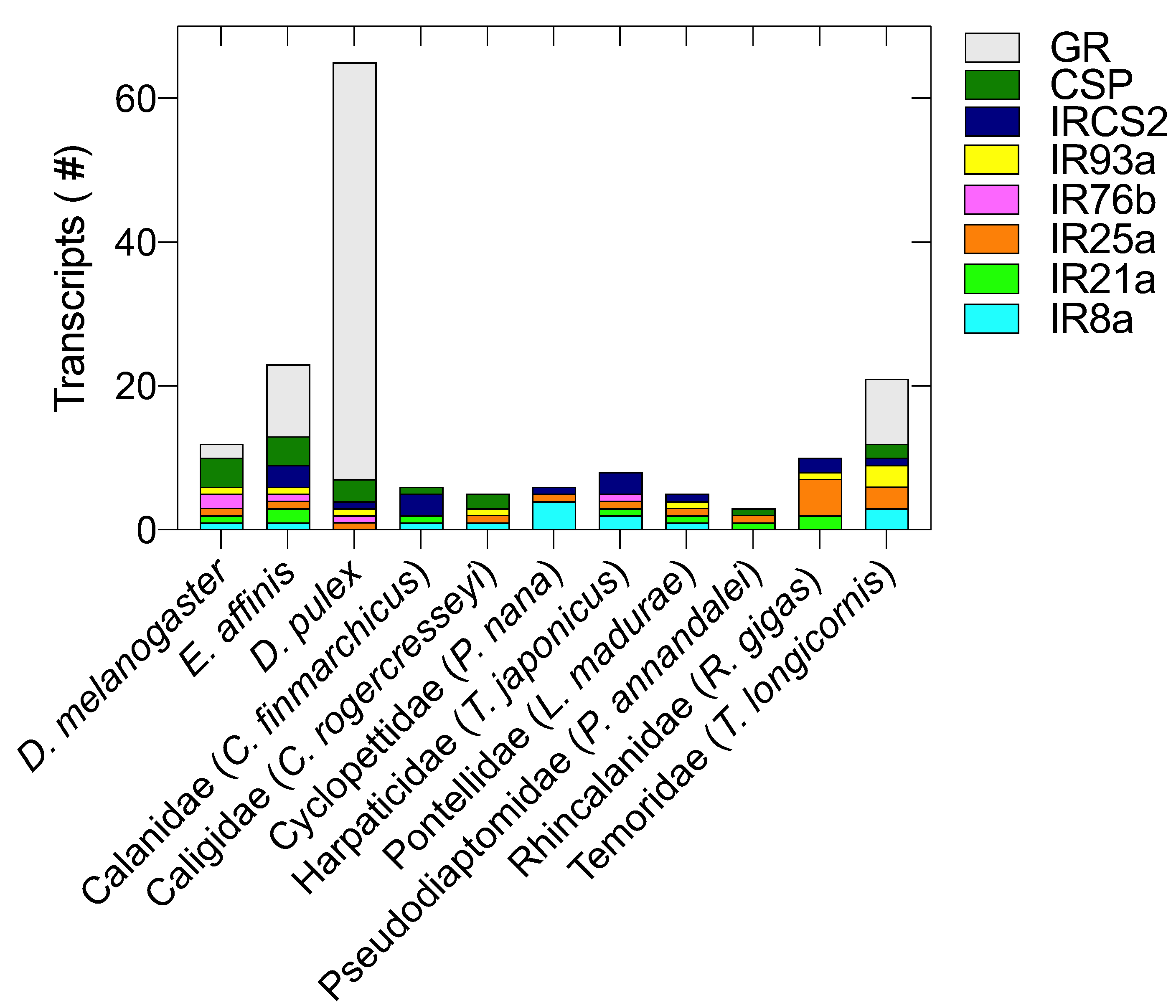
2.1. Identification of Chemosensory Related Genes (CRGs) in Copepods
2.1.1. Ionotropic Receptors (IRs)
2.1.2. Chemosensory Proteins (CSPs), Gustatory Receptors (GRs), Odorant Receptors (ORs) and Odorant-Binding Proteins (OBPs)
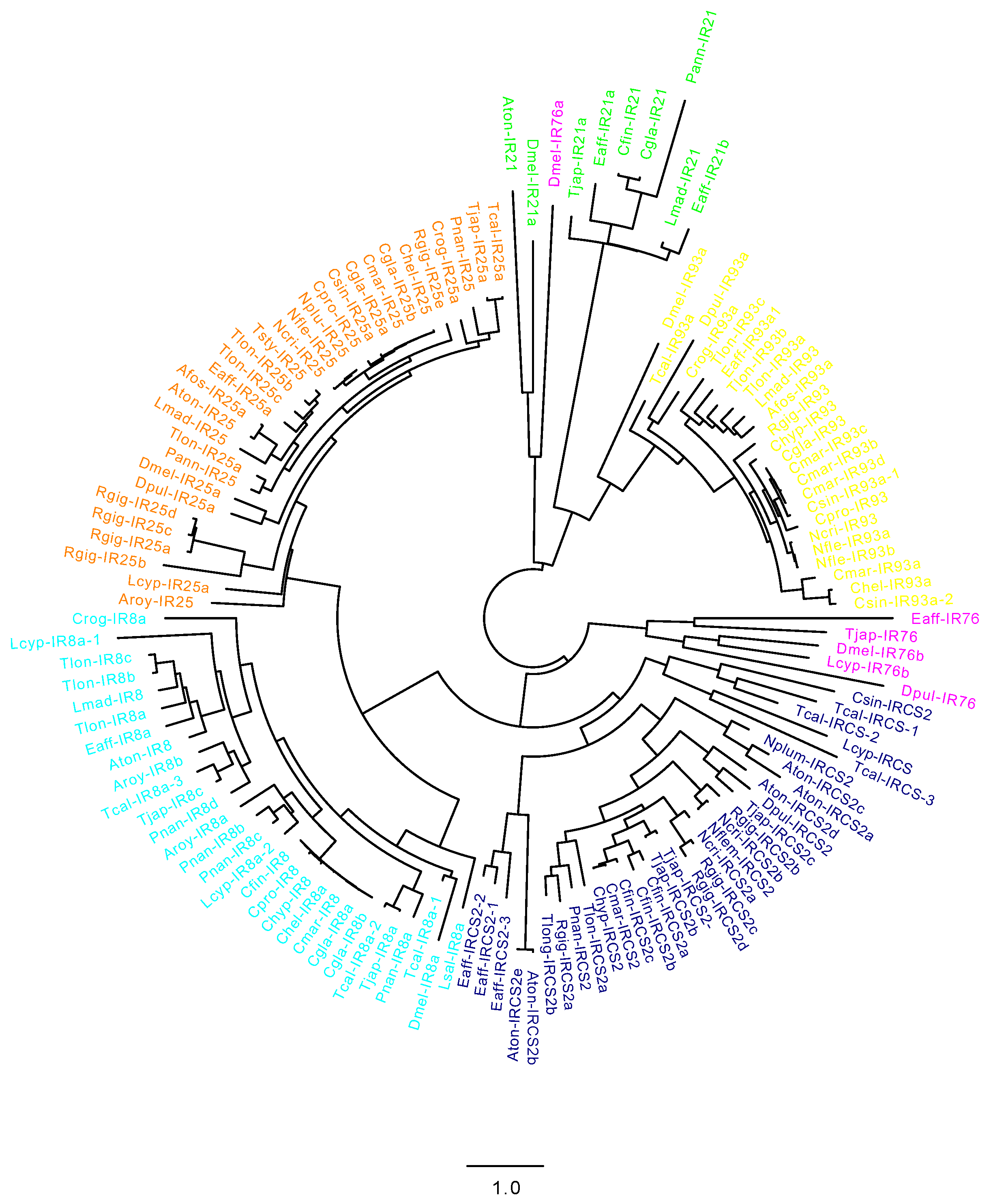
2.2. CRG Diversity and Phylogenetic Analysis
2.3. Relative Expression across Development and When Feeding on Toxic Diets in Calanus Finmarchicus and C. helgolandicus
3. Discussion
4. Materials and Methods
4.1. In Silico Mining, Reciprocal BLAST and Protein Domain Identification
4.2. Cladogram of Copepod Chemosensory-Related Genes
4.3. Relative Expression of Chemosensory Related Genes in Calanus Finmarchicus and C. helgolandicus across Development and When Exposed to Toxic Algae
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Touhara, K.; Vosshall, L.B. Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 2009, 71, 307–332. [Google Scholar] [CrossRef]
- Eyun, S.-I.; Soh, H.Y.; Posavi, M.; Munro, J.B.; Hughes, D.S.; Murali, S.C.; Qu, J.; Dugan, S.; Lee, S.L.; Chao, H. Evolutionary history of chemosensory-related gene families across the Arthropoda. Mol. Biol. Evol. 2017, 34, 1838–1862. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Gracia, A.; Vieira, F.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Ni, L. The structure and function of ionotropic receptors in Drosophila. Front. Mol. Neurosci. 2021, 13, 638839. [Google Scholar] [CrossRef] [PubMed]
- Clyne, P.J.; Warr, C.G.; Carlson, J.R. Candidate taste receptors in Drosophila. Science 2000, 287, 1830–1834. [Google Scholar] [CrossRef]
- Peter, J.C.; Coral, G.W.; Marc, R.F.; Derek, L.; Junhyong, K.; John, R.C. A Novel Family of Divergent Seven-Transmembrane Proteins. Neuron 1999, 22, 327–338. [Google Scholar]
- Croset, V.; Rytz, R.; Cummins, S.F.; Budd, A.; Brawand, D.; Kaessmann, H.; Gibson, T.J.; Benton, R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010, 6, e1001064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abuin, L.; Bargeton, B.; Ulbrich, M.H.; Isacoff, E.Y.; Kellenberger, S.; Benton, R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron 2011, 69, 44–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benton, R. Multigene family evolution: Perspectives from insect chemoreceptors. Trends Ecol. Evol. 2015, 30, 590–600. [Google Scholar] [CrossRef]
- Wicher, D.; Miazzi, F. Functional properties of insect olfactory receptors: Ionotropic receptors and odorant receptors. Cell Tissue Res. 2021, 383, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Jones, W.; Cayirlioglu, P.; Kadow, I.; Vosshall, L. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 2006, 445, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-D.; Löfstedt, C. Moth pheromone receptors: Gene sequences, function, and evolution. Front. Ecol. Evol. 2015, 3, 105. [Google Scholar] [CrossRef] [Green Version]
- Hallem, E.A.; Carlson, J.R. Coding of odors by a receptor repertoire. Cell 2006, 125, 143–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rimal, S.; Lee, Y. The multidimensional ionotropic receptors of Drosophila melanogaster. Insect Mol. Biol. 2018, 27, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Derby, C.D.; Kozma, M.T.; Senatore, A.; Schmidt, M. Molecular mechanisms of reception and perireception in crustacean chemoreception: A comparative review. Chem. Senses 2016, 41, 381–398. [Google Scholar] [CrossRef] [Green Version]
- Kozma, M.T.; Schmidt, M.; Ngo-Vu, H.; Sparks, S.D.; Senatore, A.; Derby, C.D. Chemoreceptor proteins in the Caribbean spiny lobster, Panulirus argus: Expression of ionotropic receptors, gustatory receptors, and TRP channels in two chemosensory organs and brain. PLoS ONE 2018, 13, e0203935. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef] [Green Version]
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef]
- Saha, M.; Berdalet, E.; Carotenuto, Y.; Fink, P.; Harder, T.; John, U.; Not, F.; Pohnert, G.; Potin, P.; Selander, E. Using chemical language to shape future marine health. Front. Ecol. Environ. 2019, 17, 530–537. [Google Scholar] [CrossRef]
- Thomas, O.P.; Bagnères, A.-G. Preface: Aquatic chemical ecology special issue. Aquat. Ecol. 2022, 56, 337–338. [Google Scholar] [CrossRef]
- Mollo, E.; Garson, M.; Polese, G.; Amodeo, P.; Ghiselin, M. Taste and smell in aquatic and terrestrial environments. Nat. Prod. Rep. 2017, 34, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Mollo, E.; Boero, F.; Peñuelas, J.; Fontana, A.; Garson, M.J.; Roussis, V.; Cerrano, C.; Polese, G.; Cattaneo, A.M.; Mudianta, I.W. Taste and Smell: A Unifying Chemosensory Theory. Q. Rev. Biol. 2022, 97, 69–94. [Google Scholar] [CrossRef]
- Yamamoto, H.; Okino, T.; Yoshimura, E.; Tachibana, A.; Shimizu, K.; Fusetani, N. Methyl farnesoate induces larval metamorphosis of the barnacle, Balanus amphitrite via protein kinase C activation. J. Exp. Zool. 1997, 278, 349–355. [Google Scholar] [CrossRef]
- Giordano, G.; Carbone, M.; Ciavatta, M.L.; Silvano, E.; Gavagnin, M.; Garson, M.J.; Cheney, K.L.; Mudianta, I.W.; Russo, G.F.; Villani, G. Volatile secondary metabolites as aposematic olfactory signals and defensive weapons in aquatic environments. Proc. Natl. Acad. Sci. 2017, 114, 3451–3456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinke, M.; Stefels, J.; Stamhuis, E. Dimethyl sulfide triggers search behavior in copepods. Limnol Oceanogr 2006, 51, 1925–1930. [Google Scholar] [CrossRef] [Green Version]
- Bailey, R.J.; Birkett, M.A.; Ingvarsdóttir, A.; Mordue, A.J.; Mordue, W.; O’Shea, B.; Pickett, J.A.; Wadhams, L.J. The role of semiochemicals in host location and non-host avoidance by salmon louse (Lepeophtheirus salmonis) copepodids. Can. J. Fish. Aquat. Sci. 2006, 63, 448–456. [Google Scholar] [CrossRef]
- Brönmark, C.; Hansson, L.-A. Chemical Ecology in Aquatic Systems; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Heuschele, J.; Selander, E. The chemical ecology of copepods. J. Plankton Res. 2014, 36, 895–913. [Google Scholar] [CrossRef] [Green Version]
- Uttieri, M. Trends in copepod studies. In Trends in Copepod Studies—Distribution, Biology and Ecology; Nova Science Publishers Inc.: New York, NY, USA, 2018; pp. 1–11. [Google Scholar]
- Schminke, H.K. Entomology for the copepodologist. J. Plankton Res. 2007, 29 (Suppl. 1), i149–i162. [Google Scholar] [CrossRef] [Green Version]
- Oakley, T.H.; Wolfe, J.M.; Lindgren, A.R.; Zaharoff, A.K. Phylotranscriptomics to bring the understudied into the fold: Monophyletic Ostracoda, fossil placement, and pancrustacean phylogeny. Mol. Biol. Evol. 2013, 30, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Lürling, M. Infodisruption: Pollutants interfering with the natural chemical information conveyance in aquatic systems. In Chemical Ecology in Aquatic Systems; Brönmark, C., Hansson, L.-A., Eds.; Oxford University Press: Oxford, UK, 2012; pp. 250–271. [Google Scholar]
- Amato, A.; Carotenuto, Y. Planktonic calanoids embark into the “omics” era. In Trends in Copepod Studies—Distribution, Biology and Ecology; Uttieri, M., Ed.; Nova Science Publishers, Inc.: New York, NY, USA, 2018; pp. 287–314. [Google Scholar]
- Lenz, P.H.; Lieberman, B.; Cieslak, M.C.; Roncalli, V.; Hartline, D.K. Transcriptomics and metatranscriptomics in zooplankton: Wave of the future? J. Plankton Res. 2021, 43, 3–9. [Google Scholar] [CrossRef]
- Bradford-Grieve, J.M. Colonization of the pelagic realm by calanoid copepods. Hydrobiologia 2002, 485, 223–244. [Google Scholar] [CrossRef]
- Sugier KLaso-Jadart RVacherie BKäfer JBertrand LLabadie, K.; Madoui, M.A. Male differentiation in the marine copepod Oithona nana reveals the development of a new nervous ganglion and Lin12-Notch-Repeat Protein-Associated Proteolysis. Biology 2021, 10, 657. [Google Scholar]
- Peñalva-Arana, D.; Lynch, M.; Robertson, H.M. The chemoreceptor genes of the waterflea Daphnia pulex: Many Grs but no Ors. BMC Evol. Biol. 2009, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ong, J. The fine structure of the mandibular sensory receptors in the brackish water calanoid copepod Gladioferens pectinatus (Brady). Z. Für Zellforsch. Und Mikrosk. Anat. 1969, 97, 178–195. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M.M.; Strickler, J.R. Chemoreceptors and feeding in calanoid copepods (Arthropoda: Crustacea). Proc. Natl. Acad. Sci. USA 1975, 72, 4185–4188. [Google Scholar] [CrossRef] [Green Version]
- Paffenhöfer, G.-A.; Loyd, P. Ultrastructure of setae of the maxilliped of the marine planktonic copepod Temora stylifera. Mar. Ecol. Prog. Ser. 1999, 178, 101–107. [Google Scholar] [CrossRef] [Green Version]
- Gill, C.; Poulet, S. Responses of copepods to dissolved free amino acids. Mar. Ecol. Prog. Ser. 1988, 43, 269–276. [Google Scholar] [CrossRef]
- Paffenhöfer, G.-A.; Strickler, J.; Alcaraz, M. Suspension-feeding by herbivorous calanoid copepods: A cinematographic study. Mar. Biol. 1982, 67, 193–199. [Google Scholar] [CrossRef]
- Montell, C. Gustatory receptors: Not just for good taste. Curr. Biol. 2013, 23, R929–R932. [Google Scholar] [CrossRef] [Green Version]
- Robertson, H.M.; Warr, C.G.; Carlson, J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. 2003, 100 (Suppl. 2), 14537–14542. [Google Scholar] [CrossRef] [Green Version]
- Krång, A.-S.; Knaden, M.; Steck, K.; Hansson, B.S. Transition from sea to land: Olfactory function and constraints in the terrestrial hermit crab Coenobita clypeatus. Proc. R. Soc. B: Biol. Sci. 2012, 279, 3510–3519. [Google Scholar] [CrossRef] [Green Version]
- Russo, E.; Lauritano, C.; d’Ippolito, G.; Fontana, A.; Sarno, D.; von Elert, E.; Ianora, A.; Carotenuto, Y. RNA-Seq and differential gene expression analysis in Temora stylifera copepod females with contrasting non-feeding nauplii survival rates: An environmental transcriptomics study. Bmc Genom. 2020, 21, 1–22. [Google Scholar] [CrossRef]
- Jakobsen, P.J.; Wedekind, C. Copepod reaction to odor stimuli influenced by cestode infection. Behav. Ecol. 1998, 9, 414–418. [Google Scholar] [CrossRef]
- Mollo, E.; Fontana, A.; Roussis, V.; Polese, G.; Amodeo, P.; Ghiselin, M.T. Sensing marine biomolecules: Smell, taste, and the evolutionary transition from aquatic to terrestrial life. Front. Chem. 2014, 2, 92. [Google Scholar] [CrossRef] [Green Version]
- Christie, A.E.; Fontanilla, T.M.; Nesbit, K.T.; Lenz, P.H. Prediction of the protein components of a putative Calanus finmarchicus (Crustacea, Copepoda) circadian signaling systems using a de novo assembled transcriptome. Comp. Biochem. Physiol. D-Genom. Proteom. 2013, 8, 165–193. [Google Scholar] [CrossRef] [Green Version]
- Christie, A.E.; Fontanilla, T.M.; Roncalli, V.; Cieslak, M.C.; Lenz, P.H. Identification and developmental expression of the enzymes responsible for dopamine, histamine, octopamine and serotonin biosynthesis in the copepod crustacean Calanus finmarchicus. Gen. Comp. Endocr. 2014, 195, 28–39. [Google Scholar] [CrossRef] [Green Version]
- Roncalli, V.; Cieslak, M.C.; Passamaneck, Y.; Christie, A.E.; Lenz, P.H. Glutathione S-transferase (GST) gene diversity in the crustacean Calanus finmarchicus–contributors to cellular detoxification. PLoS ONE 2015, 10, e0123322. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Roncalli, V.; Cieslak, M.C.; Lenz, P.H. Transcriptomic responses of the calanoid copepod Calanus finmarchicus to the saxitoxin producing dinoflagellate Alexandrium fundyense. Sci. Rep. 2016, 6, 25708. [Google Scholar] [CrossRef]
- Cieslak, M.C.; Castelfranco, A.M.; Roncalli, V.; Lenz, P.H.; Hartline, D.K. t-Distributed Stochastic Neighbor Embedding (t-SNE): A tool for eco-physiological transcriptomic analysis. Mar. Genom. 2020, 51, 100723. [Google Scholar] [CrossRef]
- Asai, S.; Sanges, R.; Lauritano, C.; Lindeque, P.K.; Esposito, F.; Ianora, A.; Carotenuto, Y. De novo transcriptome assembly and gene expression profiling of the copepod Calanus helgolandicus feeding on the PUA-producing diatom Skeletonema marinoi. Mar. Drugs 2020, 18, 392. [Google Scholar] [CrossRef]
- Morozova, O.; Hirst, M.; Marra, M.A. Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genom. Hum. Genet. 2009, 10, 135–151. [Google Scholar] [CrossRef]

| Genus | Species | Bioproject | Developmental stages |
|---|---|---|---|
| Acartia | tonsa | PRJEB20069 | mix stages (embryo, nauplii, copepodids, preadult, adult) |
| Calanus | finmarchicus | PRJNA236528 | mix stages (embryo, nauplii, copepodids, females) |
| Calanus | glacialis | PRJNA237014 | females |
| Calanus | helgolandicus | PRJNA640515 | females |
| Calanus | hyperboreous | PRJNA744376 | females |
| Calanus | marshallae | PRJNA745090/PRJNA662858 | preadult (CV) |
| Calanus | propinquous | PRJNA669816 | females |
| Labidocera | madurae | PRJNA324849 | mix copepodids (CIII-CV), females |
| Neocalanus | cristatus | PRJNA662858 | preadult (CV) |
| Neocalanus | flemingeri | PRJNA324453 | females |
| Neocalanus | plumchrus | PRJNA662858 | male |
| Pseudodiaptomus | annandalei | PRJNA558682 | embryos, nauplii, copepodids, females, males |
| Rhincalanus | gigas | PRJNA666170 | preadult (CV), adult |
| Temora | longicornis | PRJNA577564 | males and females |
| Temora | stylifera | PRJNA632714 | females |
| Apocyclops | royi | PRJEB28764 | not indicated NCBI |
| Paracyclopina | nana | PRJNA268783 | not indicated NCBI |
| Tigriopus | japonicus | PRJNA274317 | not indicated NCBI |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roncalli, V.; Uttieri, M.; Capua, I.D.; Lauritano, C.; Carotenuto, Y. Chemosensory-Related Genes in Marine Copepods. Mar. Drugs 2022, 20, 681. https://doi.org/10.3390/md20110681
Roncalli V, Uttieri M, Capua ID, Lauritano C, Carotenuto Y. Chemosensory-Related Genes in Marine Copepods. Marine Drugs. 2022; 20(11):681. https://doi.org/10.3390/md20110681
Chicago/Turabian StyleRoncalli, Vittoria, Marco Uttieri, Iole Di Capua, Chiara Lauritano, and Ylenia Carotenuto. 2022. "Chemosensory-Related Genes in Marine Copepods" Marine Drugs 20, no. 11: 681. https://doi.org/10.3390/md20110681
APA StyleRoncalli, V., Uttieri, M., Capua, I. D., Lauritano, C., & Carotenuto, Y. (2022). Chemosensory-Related Genes in Marine Copepods. Marine Drugs, 20(11), 681. https://doi.org/10.3390/md20110681










