Application of Gene Knockout and Heterologous Expression Strategy in Fungal Secondary Metabolites Biosynthesis
Abstract
:1. Introduction
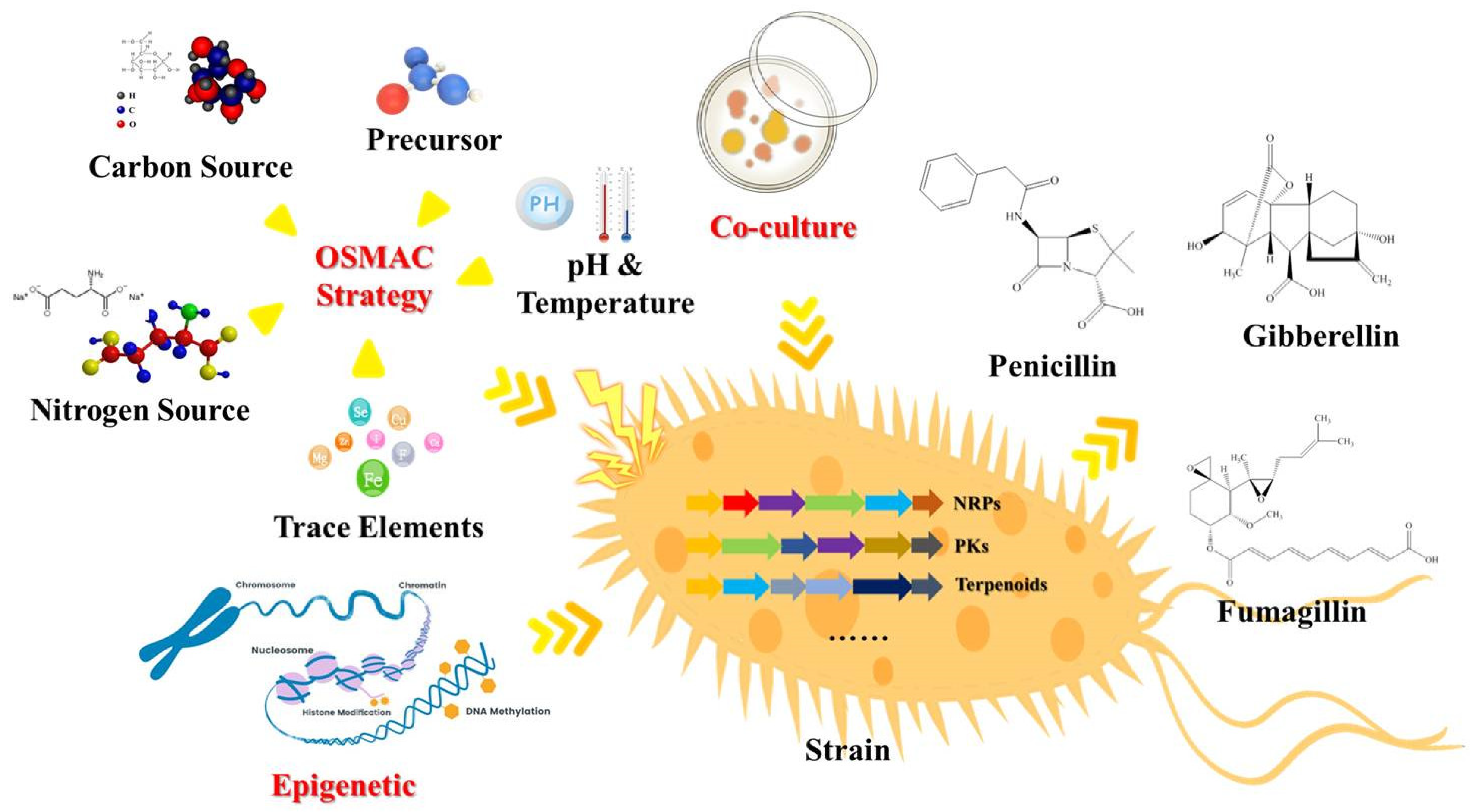
2. Traditional Strategies of Diversity of SMs from Fungi
3. Gene Mining and Bioinformatics Broaden the Discovery of SMs of Fungi
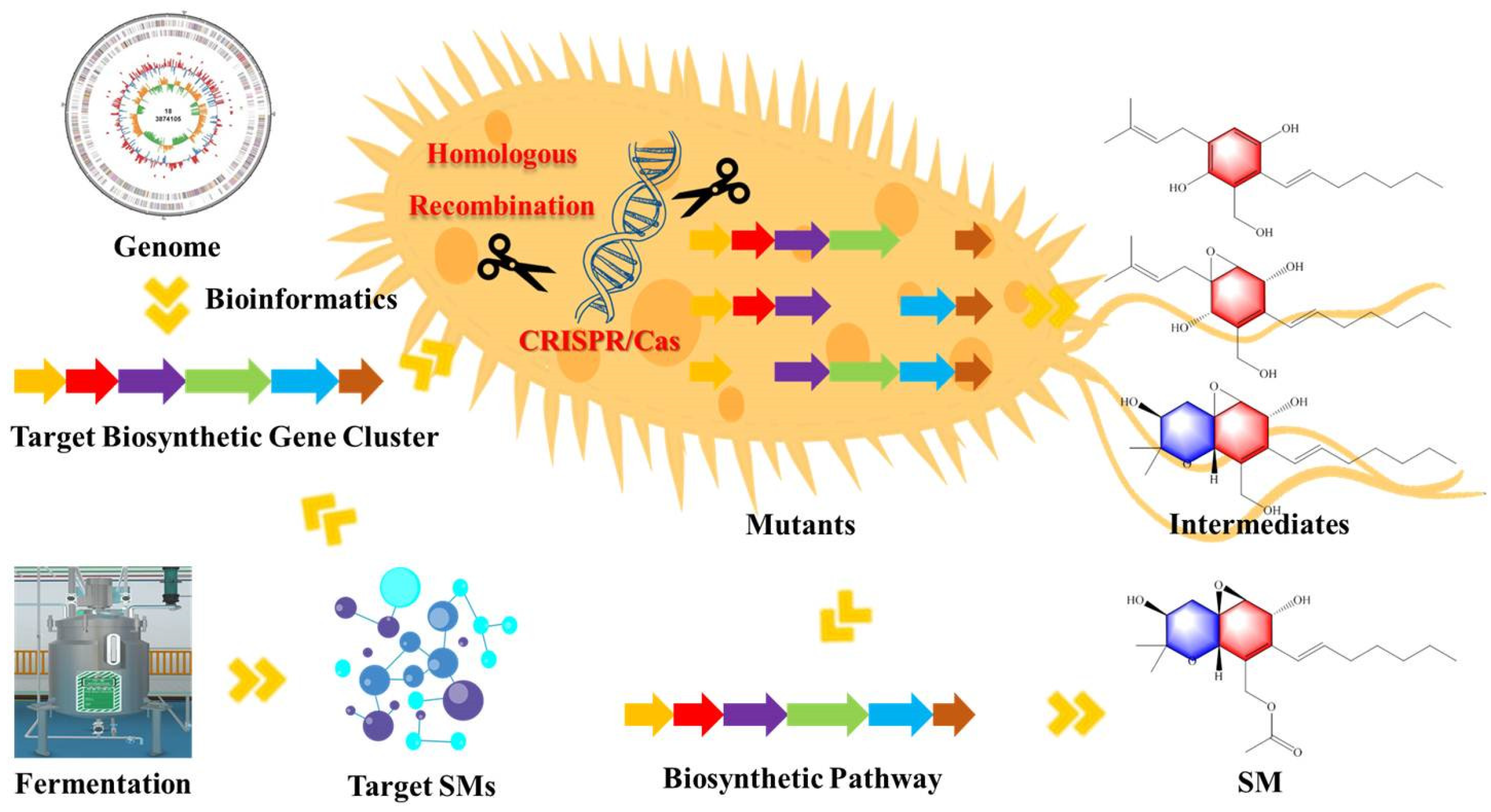
4. Application of Gene Knockout Strategy in Biosynthesis of SMs of Fungi
4.1. Application of Different Gene Knockout Methods
4.1.1. PEG-Mediated Homologous Recombination
4.1.2. PEG-Mediated CRISPR/Cas Technique
4.1.3. Agrobacterium-Mediated Transformation
4.2. Other Applications of Gene Knockout Strategy
4.3. Limitations of Gene Knockout Strategy
5. Application of Heterologous Expression Strategy in Biosynthesis of SMs of Fungi
5.1. Application of Different Heterologous Hosts
5.1.1. Application of Filamentous Fungi as Heterologous Hosts
5.1.2. Application of Saccharomyces cerevisiae as Heterologous Host
5.1.3. Application of Escherichia coli as Heterologous Host
5.2. Application of Heterologous Expression Strategy in Mining Silent BGCs
5.3. Limitations of Heterologous Expression Strategy
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef] [PubMed]
- Aldholmi, M.; Marchand, P.; Ourliac-Garnier, I.; Pape, P.L.; Ganesan, A. A decade of antifungal leads from natural products: 2010–Pharmaceuticals. Pharmaceuticals 2019, 12, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, M.S. Natural products to drugs: Natural product derived compounds in clinical trials. Nat. Prod. Rep. 2005, 22, 162–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tang, S.Y.; Cao, S.G. Antimicrobial compounds from marine fungi. Phytochem. Rev. 2021, 20, 85–117. [Google Scholar] [CrossRef]
- Xu, J.; Yi, M.; Ding, L.; He, S. A review of anti-inflammatory compounds from marine fungi, 2000–2018. Mar. Drugs. 2019, 17, 636. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.Y.; Nielsen, J.; Liu, Z.H. Synthetic biology advanced natural product discovery. Metabolites 2021, 11, 785. [Google Scholar] [CrossRef]
- Alberti, F.; Kaleem, S.; Weaver, J.A. Recent developments of tools for genome and metabolome studies in basidiomycete fungi and their application to natural product research. Biol. Open. 2020, 9, bio056010. [Google Scholar] [CrossRef]
- Vignolle, G.A.; Mach, R.L.; Mach-Aigner, A.R.; Derntl, C. Novel approach in whole genome mining and transcriptome analysis reveal conserved RiPPs in Trichoderma spp. BMC. Genom. 2020, 21, 258. [Google Scholar] [CrossRef] [Green Version]
- Alberti, F.; Foster, G.D.; Bailey, A.M. Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Biot. 2017, 101, 493–500. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yun, K.Y.; Huang, H.M.; Tu, R.; Hua, E.B.; Wang, M. Antisense RNA interference-enhanced CRISPR/Cas9 base editing method for improving base editing efficiency in Streptomyces lividans 66. ACS. Synth. Biol. 2021, 10, 1053–1063. [Google Scholar] [CrossRef]
- Kadooka, C.; Yamaguchi, M.; Okutsu, K.; Yumiko Yoshizaki Takamine, K.; Maruyama, J.; Tamaki, H.; Futagami, T. A CRISPR/Cas9-mediated gene knockout system in Aspergillus luchuensis mut. Kawachii. Biosci. Biotech. Bioch. 2020, 84, 2179–2183. [Google Scholar] [CrossRef]
- Oikawa, H. Heterologous production of fungal natural products: Reconstitution of biosynthetic gene clusters in model host Aspergillus oryzae. Proc. Jpn. Academy. Ser. B Phys. Biol. Sci. 2020, 96, 420–430. [Google Scholar] [CrossRef]
- Rang, J.; Li, Y.L.; Cao, L.; Shuai, L.; Liu, Y.; He, H.C.; Wan, Q.Q.; Luo, Y.W.; Yu, Z.Q.; Zhang, Y.M.; et al. Deletion of a hybrid NRPS-T1PKS biosynthetic gene cluster via Latour gene knockout system in Saccharopolyspora pogona and its effect on butenyl-spinosyn biosynthesis and growth development. Microb. Biotechnol. 2021, 14, 2369–2384. [Google Scholar] [CrossRef]
- Lebar, M.D.; Cary, J.W.; Majumdar, R.; Carter-Wientjes, C.H.; Mack, B.M.; Wei Qj Uka, V.; Saeger, D.D.; Mavungu, J.D.D. Identification and functional analysis of the aspergillic acid gene cluster in Aspergillus flavus. Fungal. Genet. Biol. 2018, 116, 14–23. [Google Scholar] [CrossRef]
- Liu, G.; Qu, Y.B. Engineering of filamentous fungi for efficient conversion of lignocellulose: Tools, recent advances and prospects. Biotechnol. Adv. 2019, 37, 519–529. [Google Scholar] [CrossRef]
- Tomm, H.A.; Ucciferri, L.; Ross, A.C. Advances in microbial culturing conditions to activate silent biosynthetic gene clusters for novel metabolite production. J. Ind. Microbiol. Biot. 2019, 46, 1381–1400. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.L.; Chen, J.W.; Zhang, H.W.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 108, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Pinedo-Rivilla, C.; Aleu, J.; Durán-Patrón, R. Cryptic metabolites from marine-derived microorganisms using OSMAC and epigenetic approaches. Mar. Drugs. 2022, 20, 84. [Google Scholar] [CrossRef]
- Netzker, T.; Fischer, J.; Weber, J.; Mattern, D.J.; Konig, C.C.; Valiante, V.; Schroeckh, V.; Brakhage, A.A. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 2015, 6, 299. [Google Scholar] [CrossRef]
- Li, C.Y.; Chung, Y.M.; Wu, Y.C.; Hunyadi, A.; Wang, C.C.C.; Chang, F.R. Natural products development under epigenetic modulation in fungi. Phytochem. Rev. 2022, 19, 1323–1340. [Google Scholar] [CrossRef]
- Kjærbølling, I.; Mortensen, U.H.; Vesth, T.; Andersen, M.R. Strategies to establish the link between biosynthetic gene clusters and secondary metabolites. Fungal. Genet. Biol. 2019, 130, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Almeida, H.; Palys, S.; Tsang, A.; Diallo, A.B. TOUCAN: A framework for fungal biosynthetic gene cluster discovery. NAR Genom. Bioinform. 2020, 2, lqaa098. [Google Scholar] [CrossRef] [PubMed]
- Alanjary, M.; Kronmiller, B.; Adamek, M.; Blin, K.; Weber, T.; Huson, D.; Philmus, B.; Ziemert, N. The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic. Acids. Res. 2017, 45, W42–W48. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Kautsar, S.A.; Medema, M.H.; Weber, T. The antiSMASH database version 3: Increased taxonomic coverage and new query features for modular enzymes. Nucleic Acids Res. 2021, 49, D639–D643. [Google Scholar] [CrossRef]
- Kloosterman, A.M.; Shelton, K.E.; van Wezel, G.P.; Medema, M.H.; Mitchell, D.A. RRE-Finder: A Genome-Mining Tool for Class-Independent RiPP Discovery. mSystems 2020, 5, e00267-20. [Google Scholar] [CrossRef]
- Nguyen, M.; Ekstrom, A.; Li, X.; Yin, Y. HGT-Finder: A New Tool for Horizontal Gene Transfer Finding and Application to Aspergillus genomes. Toxins 2015, 7, 4035–4053. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Liu, X.B.; Cai, J.M.; Chen, Y.P.; Li, B.X.; Guo, Z.K.; Huang, G.X. Genomic characteristics and comparative genomics analysis of the endophytic fungus Sarocladium brachiariae. BMC Genom. 2019, 20, 782. [Google Scholar] [CrossRef]
- Kumar, A.; Sørensen, J.L.; Hansen, F.T.; Arvas, M.; Syed, M.F.; Hassan, L.; Benz, J.P.; Record, E.; Henrissat, B.; Poggeler, S.; et al. Genome sequencing and analyses of two marine fungi from the North Sea unraveled a plethora of novel biosynthetic gene clusters. Sci. Rep. 2018, 8, 10187. [Google Scholar] [CrossRef]
- Lin, X.J.; Xu, H.; Liu, L.; Li, H.X.; Gao, Z.Z. Draft genome sequence of Neonectria sp. DH2 isolated from Meconopsis grandis Prain in Tibet. 3 Biotech 2020, 10, 346. [Google Scholar] [CrossRef]
- Qi, F.F.; Zhang, W.; Xue, Y.Y.; Geng, C.; Huang, X.N.; Sun, J.; Lu, X.F. Bienzyme-catalytic and dioxygenation-mediated anthraquinone ring opening. J. Am. Chem. Soc. 2021, 143, 16326–16331. [Google Scholar] [CrossRef]
- Won, T.H.; Bok, J.W.; Nadig, N.; Venkatesh, N.; Nickles, G.; Greco, C.; Lim, F.Y.; González, J.B.; Turgeon, B.G.; Keller, N.P.; et al. Copper starvation induces antimicrobial isocyanide integrated into two distinct biosynthetic pathways in fungi. Nat. Commun. 2022, 13, 4828. [Google Scholar] [CrossRef]
- Zheng, L.J.; Wang, H.W.; Fan, A.L.; Li, S.M. Oxepinamide F biosynthesis involves enzymatic D-aminoacyl epimerization, 3H-oxepin formation, and hydroxylation induced double bond migration. Nat. Commun. 2020, 11, 4914. [Google Scholar] [CrossRef]
- Huang, X.N.; Zhang, W.; Tang, S.; Wei, S.H.; Lu, X.F. Collaborative biosynthesis of a class of bioactive azaphilones by two separate gene clusters containing four PKS/NRPSs with transcriptional crosstalk in fungi. Angew. Chem. Int. Ed. 2020, 132, 4379–4383. [Google Scholar] [CrossRef]
- Zheng, L.J.; Yang, Y.L.; Wang, H.W.; Fan, A.L.; Zhang, L.P.; Li, S.M. Ustethylin biosynthesis implies phenethyl derivative formation in Aspergillus ustus. Org. Lett. 2020, 22, 7837–7841. [Google Scholar] [CrossRef]
- Ma, K.; Zhang, P.; Tao, Q.Q.; Keller, N.P.; Yang, Y.L.; Yin, W.B.; Liu, H.W. Characterization and biosynthesis of a rare fungal hopane-type triterpenoid glycoside involved in the antistress property of Aspergillus fumigatus. Org. Lett. 2019, 21, 3252–3256. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, D.; Dun, B.; Yin, M.; Tsega, A.S.; Xie, L.; Li, W.; Yue, Q.; Wang, S.; Gao, H.; et al. Chemometrics and genome mining reveal an unprecedented family of sugar acid–containing fungal nonribosomal cyclodepsipeptides. Proc. Natl. Acad. Sci. USA 2022, 119, e2123379119. [Google Scholar] [CrossRef]
- Liu, S.H.; Sun, J.L.; Hu, Y.L.; Zhang, L.; Zhang, X.; Yan, Z.Y.; Guo, X.; Guo, Z.K.; Jiao, R.H.; Zhang, B. Biosynthesis of Sordarin Revealing a Diels–Alderase for the Formation of the Norbornene Skeleton. Angew. Chem. Int. Ed. 2022, 61, e202205577. [Google Scholar] [CrossRef]
- Liu, H.; Fan, J.; Zhang, P.; Hu, Y.C.; Liu, X.Z.; Li, S.M.; Yin, W.B. New insights into the disulfide bond formation enzymes in epidithiodiketopiperazine alkaloids. Chem. Sci. 2021, 12, 4132–4138. [Google Scholar] [CrossRef]
- Dai, G.Z.; Han, W.B.; Mei, N.Y.; Xu, K.; Jiao, R.H.; Ge, H.M.; Tan, R.X. Pyridoxal-5′-phosphate–dependent bifunctional enzyme catalyzed biosynthesis of indolizidine alkaloids in fungi. Proc. Natl. Acad. Sci. USA. 2019, 117, 1174–1180. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Awakawa, T.; Mori, T.; Ling, M.Q.; Hu, D.; Wu, B.; Abe, I. Heterodimeric non-heme iron enzymes in fungal meroterpenoid biosynthesis. J. Am. Chem. Soc. 2021, 143, 21425–21432. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.F.; Chen, L.X.; Chiang, C.Y.; Lai, C.Y.; Lin, H.C. The biosynthesis of norsesquiterpene aculenes requires three cytochrome P450 enzymes to catalyze a stepwise demethylation process. Angew. Chem. Int. Ed. 2019, 58, 18414–18418. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.W.; Chen, S.H.; Guo, H.; Chen, L.T.; Shen, H.J.; Liu, L.; Gao, Z.Z. Elucidation of the Complete Biosynthetic Pathway of Phomoxanthone A and Identification of a Para–Para Selective Phenol Coupling Dimerase. Org. Lett. 2022, 24, 3069–3074. [Google Scholar] [CrossRef]
- Shu, X.; Wei, G.Z.; Qiao, Y.B.; Zhang, K.X.; Zhang, J.; Ai, G.M.; Tang, M.C.; Zhang, Y.H.; Gao, S.S. TerC Is a Multifunctional and Promiscuous Flavoprotein Monooxygenase That Catalyzes Bimodal Oxidative Transformations. Org. Lett. 2021, 23, 8947–8951. [Google Scholar] [CrossRef]
- Li, B.Q.; Chen, Y.; Zong, Y.Y.; Shang, Y.J.; Zhang, Z.Q.; Xu, X.D.; Wang, X.; Long, M.Y.; Tian, S.P. Dissection of patulin biosynthesis, spatial control and regulation mechanism in Penicillium expansum. Environ. Microbiol. 2019, 21, 1124–1139. [Google Scholar] [CrossRef]
- Zhang, T.; Gu, G.; Liu, G.; Su, J.; Zhan, Z.; Zhao, J.; Qian, J.; Cai, G.; Cen, S.; Zhang, D.; et al. Late-stage cascade of oxidation reactions during the biosynthesis of oxalicine B in Penicillium oxalicum. Acta. Pharm. Sin. B 2022. [Google Scholar] [CrossRef]
- Sun, Y.L.; Chen, B.; Li, X.L.; Yin, Y.; Wang, C.S. Orchestrated Biosynthesis of the Secondary Metabolite Cocktails Enables the Producing Fungus to Combat Diverse Bacteria. mBio 2022, 13, e0180022. [Google Scholar] [CrossRef]
- Wei, Q.; Bai, J.; Yan, D.J.; Bao, X.Q.; Li, W.T.; Liu, B.Y.; Zhang, D.; Qi, X.B.; Yu, D.Q.; Hu, Y.C. Genome mining combined metabolic shunting and OSMAC strategy of an endophytic fungus leads to the production of diverse natural products. Acta. Pharm. Sin. B. 2021, 11, 572–587. [Google Scholar] [CrossRef]
- Qi, F.F.; Zhang, W.; Xue, Y.Y.; Geng, C.; Jin, Z.G.; Li, J.B.; Guo, Q.; Huang, X.N.; Lu, X.F. Microbial production of the plant-derived fungicide physcion. Metab. Eng. 2022, 74, 130–138. [Google Scholar] [CrossRef]
- Xu, D.; Yin, R.Y.; Zhou, Z.Y.; Gu, G.; Zhao, S.J.; Xu, J.R.; Liu, J.F.; Peng, Y.L.; Lai, D.W.; Zhou, L.G. Elucidation of ustilaginoidin biosynthesis reveals a previously unrecognised class of ene-reductases. Chem. Sci. 2021, 12, 14883–14892. [Google Scholar] [CrossRef]
- Tang, Z.J.; Tang, H.Y.; Wang, W.Q.; Xue, Y.F.; Chen, D.D.; Tang, W.H.; Liu, W. Biosynthesis of a New Fusaoctaxin Virulence Factor in Fusarium graminearum Relies on a Distinct Path To Form a Guanidinoacetyl Starter Unit Priming Nonribosomal Octapeptidyl Assembly. J. Am. Chem. Soc. 2021, 143, 19719–19730. [Google Scholar] [CrossRef]
- Zeng, H.C.; Yin, G.P.; Wei, Q.; Li, D.H.; Wang, Y.; Hu, Y.C.; Hu, C.H.; Zou, Y. Unprecedented [5.5.5.6] dioxafenestrane ring construction in fungal insecticidal sesquiterpene biosynthesis. Angew. Chem. Int. Ed. 2019, 131, 6641–6645. [Google Scholar] [CrossRef]
- Ning, Y.D.; Hu, B.; Yu, H.B.; Liu, X.Y.; Jiao, B.H.; Lu, X.L. Optimization of Protoplast Preparation and Establishment of Genetic Transformation System of an Arctic-Derived Fungus Eutypella sp. Front. Microbiol. 2022, 13, 769008. [Google Scholar] [CrossRef]
- Wei, G.Z.; Shu, X.; Yao, Y.P. Heterologous Production of Unnatural Flavipucine Family Products Provides Insights into Flavipucines Biosynthesis. Org. Lett. 2021, 23, 7708–7712. [Google Scholar] [CrossRef]
- Chen, Q.B.; Yuan, G.Y.; Yuan, T.; Zeng, H.T.; Zou, Z.R.; Tu, Z.C.; Gao, J.; Zou, Y. Set of Cytochrome P450s Cooperatively Catalyzes the Synthesis of a Highly Oxidized and Rearranged Diterpene-Class Sordarinane Architecture. J. Am. Chem. Soc. 2022, 144, 3580–3589. [Google Scholar] [CrossRef]
- Wei, Q.; Zeng, H.C.; Zou, Y. Divergent Biosynthesis of Fungal Dioxafenestrane Sesquiterpenes by the Cooperation of Distinctive Baeyer–Villiger Monooxygenases and α-Ketoglutarate-Dependent Dioxygenases. ACS. Catal. 2021, 11, 948–957. [Google Scholar] [CrossRef]
- Zhong, B.F.; Wan, J.; Shang, C.H.; Wen, J.J.; Wang, Y.J.; Bai, J.; Cen, S.; Hu, Y.C. Biosynthesis of rumbrins and inspiration for discovery of HIV inhibitors. Acta. Pharm. Sin. B. 2022, 12, 4193–4203. [Google Scholar] [CrossRef]
- Yee, D.A.; Kakule, T.B.; Cheng, W.; Chen, M.B.; Chong, C.T.Y.; Hai, Y.; Hang, L.F.; Hung, Y.S.; Liu, N.; Ohashi, M. Genome mining of alkaloidal terpenoids from a hybrid terpene and nonribosomal peptide biosynthetic pathway. J. Am. Chem. Soc. 2020, 142, 710–714. [Google Scholar] [CrossRef]
- Xie, L.N.; Zang, X.; Cheng, W.; Zhang, Z.; Zhou, J.H.; Chen, M.B.; Tang, Y. Harzianic acid from Trichoderma afroharzianum is a natural product inhibitor of acetohydroxyacid synthase. J. Am. Chem. Soc. 2021, 143, 9575–9584. [Google Scholar] [CrossRef]
- Liu, L.; Tang, M.C.; Tang, Y. Fungal highly reducing polyketide synthases biosynthesize salicylaldehydes that are precursors to epoxycyclohexenol natural products. J. Am. Chem. Soc. 2019, 141, 19538–19541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qiao, T.Z.; Watanabe, K.; Tang, Y. Concise Biosynthesis of Phenylfuropyridones in Fungi. Angew. Chem. Int. Ed. 2020, 132, 20061–20065. [Google Scholar] [CrossRef]
- Zhang, Z.; Jamieson, C.S.; Zhao, Y.L.; Li, D.H.; Ohashi, M.; Houk, K.N.; Tang, Y. Enzyme-catalyzed inverse-electron demand Diels–Alder reaction in the biosynthesis of antifungal ilicicolin H. J. Am. Chem. Soc. 2019, 141, 5659–5663. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.W.; Jamieson, C.S.; Wang, G.Q.; Yan, Y.; Zhou, J.H.; Houk, K.N.; Tang, Y. A polyketide cyclase that forms medium-ring lactones. J. Am. Chem. Soc. 2020, 143, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.F.; Zhuang, Z.; Liu, T.; Yang, Q.; He, Q.L.; Liu, W.; Lin, G.Q. Unsymmetrically Regioselective Homodimerization Depends on the Subcellular Colocalization of Laccase/Fasciclin Protein in the Biosynthesis of Phlegmacins. ACS. Chem. Biol. 2021, 17, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Matsuda, Y. Genome mining-driven discovery of 5-methylorsellinate-derived meroterpenoids from Aspergillus funiculosus. Org. Lett. 2021, 23, 3211–3215. [Google Scholar] [CrossRef]
- Chen, L.; Wei, X.X.; Matsuda, Y. Depside Bond Formation by the Starter-Unit Acyltransferase Domain of a Fungal Polyketide Synthase. J. Am. Chem. Soc. 2022, 144, 19225–19230. [Google Scholar] [CrossRef]
- Chen, R.; Feng, T.; Li, M.; Zhang, X.Y.; He, J.; Hu, B.; Deng, Z.X.; Liu, T.G.; Liu, J.; Wang, X.H.; et al. Characterization of Tremulane Sesquiterpene Synthase from the Basidiomycete Irpex lacteus. Org. Lett. 2022, 24, 5669–5673. [Google Scholar] [CrossRef]
- Wang, W.G.; Wang, H.; Du, L.Q.; Li, M.; Chen, L.; Yu, J.; Cheng, G.G.; Zhan, M.T.; Hu, Q.F.; Zhang, L.H.; et al. Molecular basis for the biosynthesis of an unusual chain-fused polyketide, gregatin A. J. Am. Chem. Soc. 2020, 142, 8464–8472. [Google Scholar] [CrossRef]
- Murai, K.; Lauterbach, L.; Teramoto, K.; Quan, Z.Y.; Barra, L.; Yamamoto, T.; Nonaka, K.; Shiomi, K.; Nishiyama, M.; Kuzuyama, T.; et al. An unusual skeletal rearrangement in the biosynthesis of the sesquiterpene trichobrasilenol from Trichoderma. Angew. Chem. Int. Ed. 2019, 58, 15046–15050. [Google Scholar] [CrossRef]
- Chen, L.; Tang, J.W.; Liu, Y.Y.; Matsuda, Y. Aspcandine: A Pyrrolobenzazepine Alkaloid Synthesized by a Fungal Nonribosomal Peptide Synthetase-Polyketide Synthase Hybrid. Org. Lett. 2022, 24, 4816–4819. [Google Scholar] [CrossRef]
- Hewage, R.T.; Huang, R.J.; Lai, S.J.; Lien, Y.C.; Weng, S.H.; Li, D.H.; Chen, Y.J.; Wu, S.H.; Chein, R.J.; Lin, H.C. An Enzyme-Mediated Aza-Michael Addition Is Involved in the Biosynthesis of an Imidazoyl Hybrid Product of Conidiogenone B. Org. Lett. 2021, 23, 1904–1909. [Google Scholar] [CrossRef]
- Ye, Y.; Du, L.; Zhang, X.W.; Newmister, S.A.; McCauley, M.; Alegre-Requena, J.V.; Zhang, W.; Mu, S.; Minami, A.; Fraley, A.E.; et al. Fungal-derived brevianamide assembly by a stereoselective semipinacolase. Nat. Catal. 2020, 3, 497–506. [Google Scholar] [CrossRef]
- Yan, D.X.; Matsuda, Y. Biosynthetic Elucidation and Structural Revision of Brevione E: Characterization of the Key Dioxygenase for Pathway Branching from Setosusin Biosynthesis. Angew. Chem. Int. Ed. 2022, e202210938. [Google Scholar] [CrossRef]
- Zhang, K.J.; Zhang, G.J.; Hou, X.W.; Ma, C.; Liu, J.Y.; Che, Q.; Zhu, T.J.; Li, D.H. A Fungal Promiscuous UbiA Prenyltransferase Expands the Structural Diversity of Chrodrimanin-Type Meroterpenoids. Org. Lett. 2022, 24, 2025–2029. [Google Scholar] [CrossRef]
- Chen, Y.R.; Naresh, A.; Liang, S.Y.; Lin, C.H.; Chein, R.J.; Lin, H.C. Discovery of a dual function cytochrome P450 that catalyzes enyne formation in cyclohexanoid terpenoid biosynthesis. Angew. Chem. Int. Ed. 2020, 59, 13537–13541. [Google Scholar] [CrossRef]
- Yao, Y.P.; An, C.Y.; Evans, D.; Liu, W.W.; Wang, W.; Wei, G.Z.; Ding, N.; Houk, K.N.; Gao, S.S. Catalase involved in oxidative cyclization of the tetracyclic ergoline of fungal ergot alkaloids. J. Am. Chem. Soc. 2019, 141, 17517–17521. [Google Scholar] [CrossRef]
- Fujii, I.; Hashimoto, M.; Konishi, K.; Unezawa, A.; Sakuraba, H.; Suzuki, K.; Tsushima, H.; Iwasaki, M.; Yoshida, S.; Kudo, A.; et al. Shimalactone Biosynthesis Involves Spontaneous Double Bicyclo-Ring Formation with 8π-6π Electrocyclization. Angew. Chem. Int. Ed. 2020, 59, 8464–8470. [Google Scholar] [CrossRef]
- Feng, C.; Wei, Q.; Hu, C.H.; Zou, Y. Biosynthesis of diphenyl ethers in fungi. Org. Lett. 2019, 21, 3114–3118. [Google Scholar] [CrossRef]
- Li, H.; Gilchrist, C.L.M.; Phan, C.S.; Lacey, H.J.; Vuong, D.; Moggach, S.A.; Lacey, E.; Piggott, A.M.; Chooi, Y.H. Biosynthesis of a New 237 Benzazepine Alkaloid Nanangelenin A from Aspergillus nanangensis Involves an Unusual l-238 Kynurenine-Incorporating NRPS Catalyzing Regioselective Lactamization. J. Am. Chem. Soc. 2020, 239, 15. [Google Scholar] [CrossRef]
- Yan, D.J.; Chen, Q.B.; Gao, J.; Bai, J.; Liu, B.Y.; Zhang, Y.L.; Zhang, C.; Zou, Y.; Hu, Y.C. Complexity and diversity generation in the biosynthesis of fumiquinazoline-related peptidyl alkaloids. Org. Lett. 2019, 21, 1475–1479. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Sun, Y.L.; Li, S.Q.; Yin, Y.; Wang, C.S. Inductive Production of the Iron-Chelating 2-Pyridones Benefits the Producing Fungus to Compete for Diverse Niches. Mbio 2021, 12, e0327921. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.J.; Cai, Y.S.; Cheng, F.C.; Yang, C.J.; Zhang, W.Q.; Yu, W.L.; Yan, J.J.; Deng, Z.X.; Hong, K. Genome mining reveals a multiproduct sesterterpenoid biosynthetic gene cluster in Aspergillus ustus. Org. Lett. 2021, 23, 1525–1529. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shu, S.; Kalaitzis, J.A.; Shang, Z.; Vuong, D.; Crombie, A.; Lacey, E.; Piggott, A.M.; Chooi, Y.H. Genome mining of Aspergillus hancockii unearths cryptic polyketide hancockinone A featuring a prenylated 6/6/6/5 carbocyclic skeleton. Org. Lett. 2021, 23, 8789–8793. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, K.; Shinki, S.; Kaneko, A.; Murakami, K.; Irie, K.; Murai, M.; Miyoshi, H.; Dan, S.; Kawaji, K.; Hayashi, H.; et al. Synthetic biology based construction of biological activity-related library of fungal decalin-containing diterpenoid pyrones. Nat. Commun. 2020, 11, 1830. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ning, Y.; Xu, Y.; Jiao, B.; Lu, X. Application of Gene Knockout and Heterologous Expression Strategy in Fungal Secondary Metabolites Biosynthesis. Mar. Drugs 2022, 20, 705. https://doi.org/10.3390/md20110705
Ning Y, Xu Y, Jiao B, Lu X. Application of Gene Knockout and Heterologous Expression Strategy in Fungal Secondary Metabolites Biosynthesis. Marine Drugs. 2022; 20(11):705. https://doi.org/10.3390/md20110705
Chicago/Turabian StyleNing, Yaodong, Yao Xu, Binghua Jiao, and Xiaoling Lu. 2022. "Application of Gene Knockout and Heterologous Expression Strategy in Fungal Secondary Metabolites Biosynthesis" Marine Drugs 20, no. 11: 705. https://doi.org/10.3390/md20110705
APA StyleNing, Y., Xu, Y., Jiao, B., & Lu, X. (2022). Application of Gene Knockout and Heterologous Expression Strategy in Fungal Secondary Metabolites Biosynthesis. Marine Drugs, 20(11), 705. https://doi.org/10.3390/md20110705





