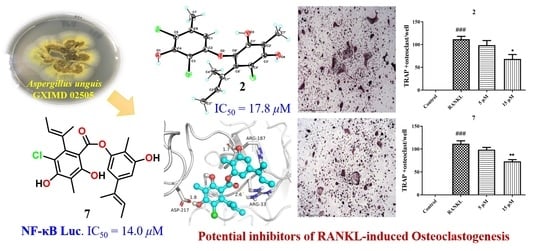
Anti-Osteoclastogenic and Antibacterial Effects of Chlorinated Polyketides from the Beibu Gulf Coral-Derived Fungus Aspergillus unguis GXIMD 02505
Abstract
:1. Introduction
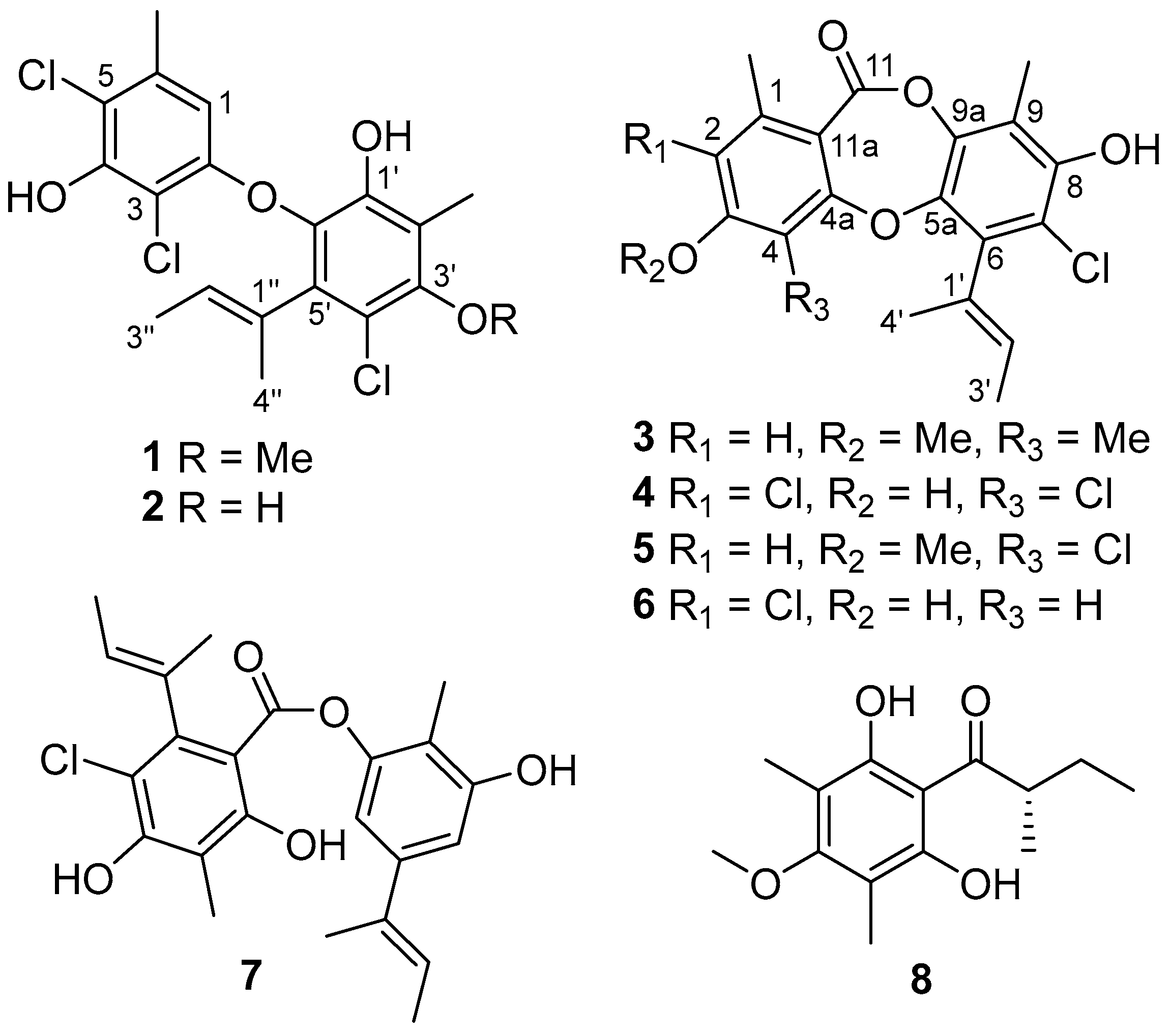
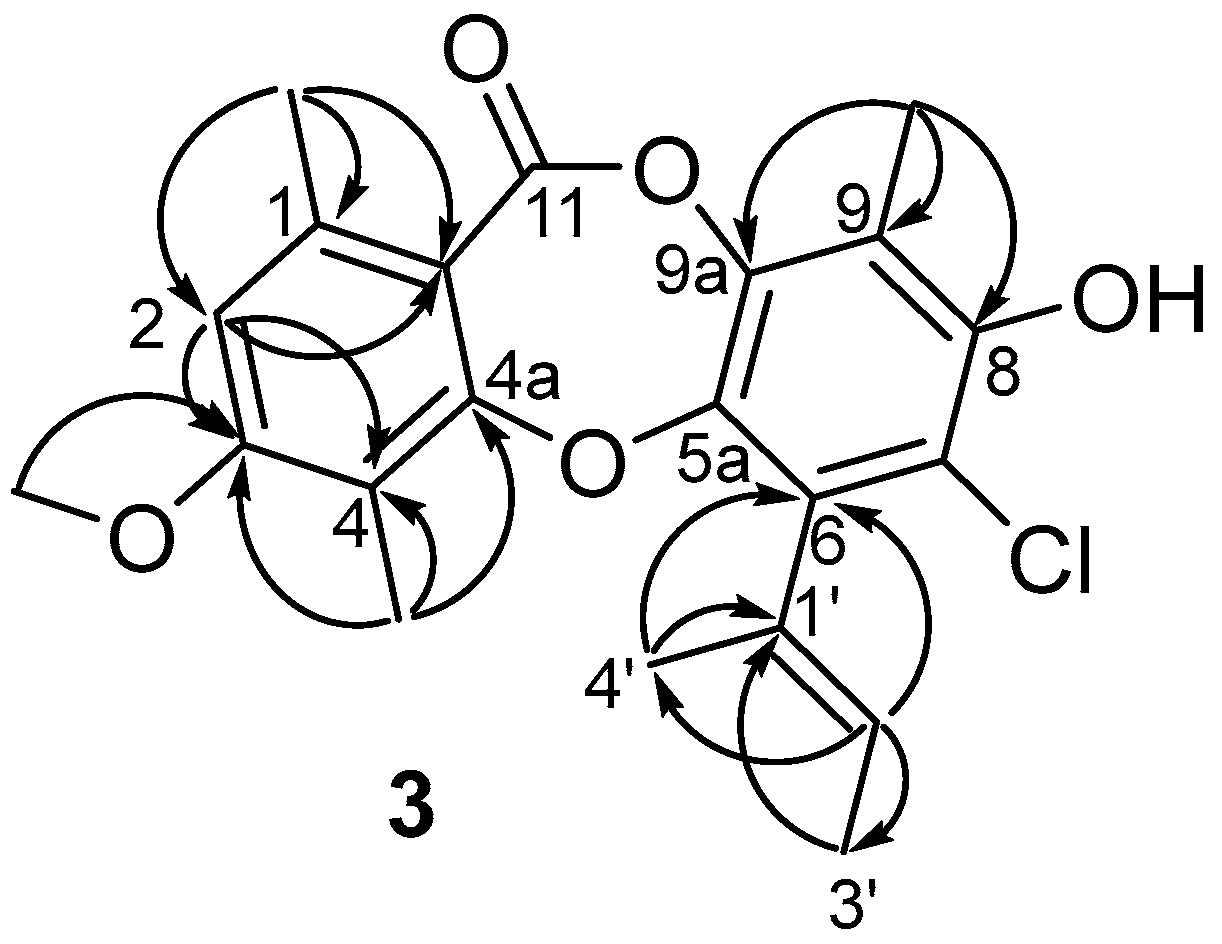
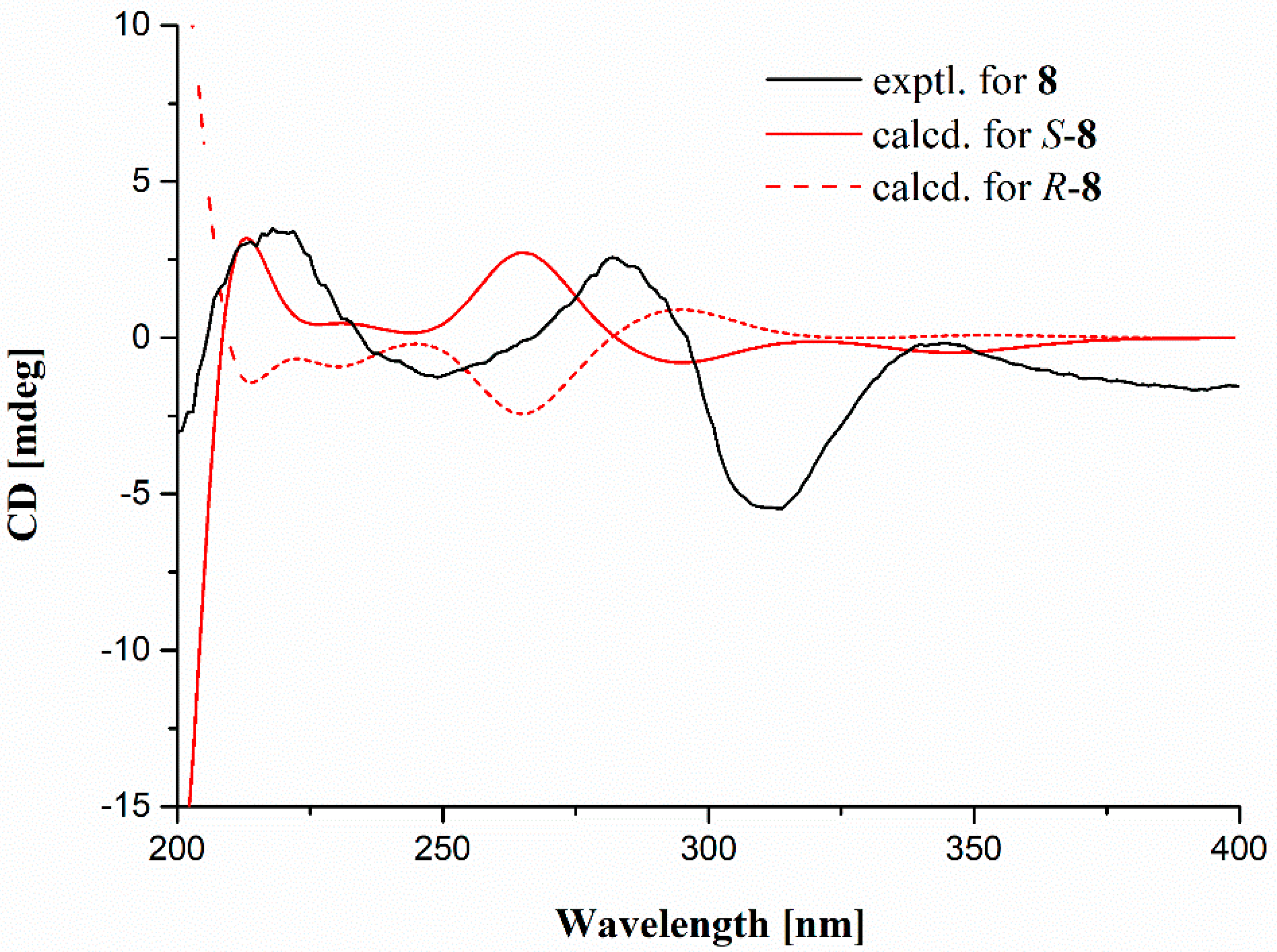
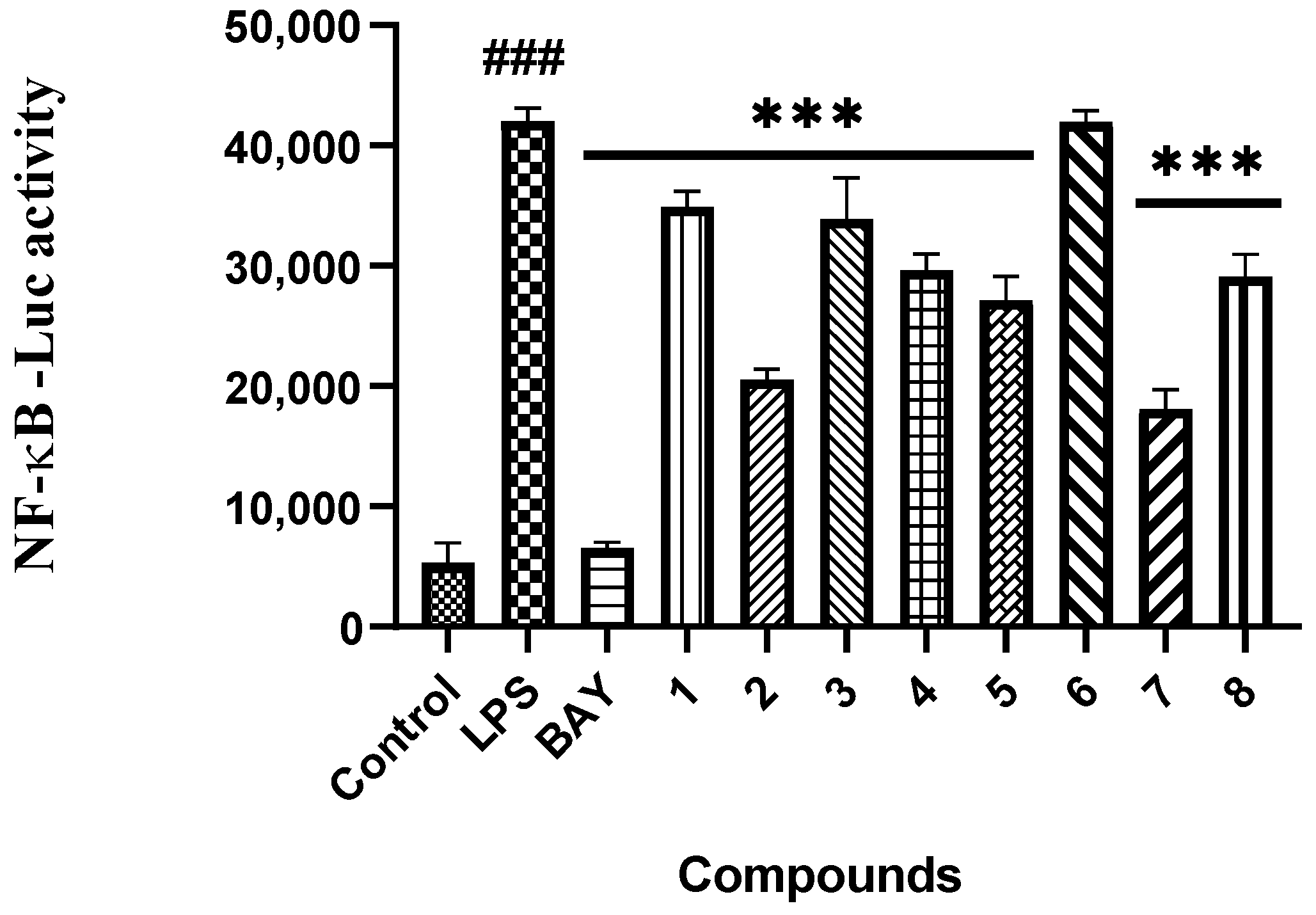
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Strain and Fermentation
3.3. Extraction and Isolation
3.4. Computational Methods
3.5. X-ray Crystallography
3.6. Anti-Osteoclastogenic Assay
3.7. Antibacterial Assay
3.8. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Jacome Galarza, C.E.; Percin, G.I.; Muller, J.T.; Mass, E.; Lazarov, T.; Eitler, J.; Rauner, M.; Yadav, V.K.; Crozet, L.; Bohm, M.; et al. Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature 2019, 568, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Rachner, T.D.; Khosla, S.; Hofbauer, L.C. Osteoporosis: Now and the future. Lancet 2011, 377, 1276–1287. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.H.; Deng, W.D.; Zhang, Y.Y.; Ke, M.H.; Zou, B.H.; Luo, X.W.; Su, J.B.; Wang, Y.Y.; Xu, J.L.; Nandakumar, K.S.; et al. A marine fungus-derived nitrobenzoyl sesquiterpenoid suppresses receptor activator of NF-κB ligand-induced osteoclastogenesis and inflammatory bone destruction. Br. J. Pharmacol. 2020, 177, 4242–4260. [Google Scholar] [CrossRef]
- Wang, X.Y.; Yamauchi, K.; Mitsunaga, T. A review on osteoclast diseases and osteoclastogenesis inhibitors recently developed from natural resources. Fitoterapia 2020, 142, 104482. [Google Scholar] [CrossRef] [PubMed]
- Morshed, M.T.; Vuong, D.; Crombie, A.; Lacey, A.E.; Karuso, P.; Lacey, E.; Piggott, A.M. Expanding antibiotic chemical space around the nidulin pharmacophore. Org. Biomol. Chem. 2018, 16, 3038–3051. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, G.A.; Al Haidari, R.A.; El Kholy, A.A.; Zayed, M.F.; Khayat, M.T. Biologically active fungal depsidones: Chemistry, biosynthesis, structural characterization, and bioactivities. Fitoterapia 2018, 129, 317–365. [Google Scholar] [CrossRef]
- Saetang, P.; Rukachaisirikul, V.; Phongpaichit, S.; Preedanon, S.; Sakayaroj, J.; Hadsadee, S.; Jungsuttiwong, S. Antibacterial and antifungal polyketides from the fungus Aspergillus unguis PSU-MF16. J. Nat. Prod. 2021, 84, 1498–1506. [Google Scholar] [CrossRef]
- Niu, S.W.; Liu, D.; Shao, Z.Z.; Huang, J.; Fan, A.L.; Lin, W.H. Chlorinated metabolites with antibacterial activities from a deep-sea-derived Spiromastix fungus. RSC Adv. 2021, 11, 29661–29667. [Google Scholar] [CrossRef]
- Duong, T.H.; Hang, T.X.H.; Pogam, P.L.; Tran, T.N.; Mac, D.H.; Dinh, M.H.; Sichaem, J. α-Glucosidase inhibitory depsidones from the lichen Parmotrema tsavoense. Planta Med. 2020, 86, 776–781. [Google Scholar] [CrossRef]
- Devi, A.P.; Duong, T.H.; Ferron, S.; Beniddir, M.A.; Dinh, M.H.; Nguyen, V.K.; Pham, N.K.T.; Mac, D.H.; Boustie, J.; Chavasiri, W.; et al. Salazinic acid-derived depsidones and diphenylethers with α-glucosidase inhibitory activity from the lichen Parmotrema dilatatum. Planta Med. 2020, 86, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, L.T.; Yang, H.X.; Li, Z.H.; Liu, J.K.; Ai, H.L.; Wang, G.K.; Feng, T. Depsidones and diaryl ethers from potato endophytic fungus Boeremia exigua. Fitoterapia 2020, 141, 104483. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; An, F.L.; Zhu, X.J.; Yu, H.Y.; Hao, L.L.; Lu, Y.H. Curdepsidones B–G, six depsidones with anti-inflammatory activities from the marine-derived fungus Curvularia sp. IFB-Z10. Mar. Drugs 2019, 17, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bay, M.V.; Nam, P.C.; Quang, D.T.; Mechler, A.; Hien, N.K.; Hoa, N.T.; Vo, Q.V. Theoretical study on the antioxidant activity of natural depsidones. ACS Omega 2020, 5, 7895–7902. [Google Scholar] [CrossRef]
- Zeukang, R.D.; Siwe-Noundou, X.; Fotsing, M.T.; Kuiate, T.T.; Mbafor, J.T.; Krause, R.W.M.; Choudhary, M.I.; de Theodore Atchade, A. Cordidepsine is a potential new anti-HIV depsidone from Cordia millenii, baker. Molecules 2019, 24, 3202. [Google Scholar] [CrossRef] [Green Version]
- Morshed, M.T.; Nguyen, H.T.; Vuong, D.; Crombie, A.; Lacey, E.; Ogunniyi, A.D.; Page, S.W.; Trott, D.J.; Piggott, A.M. Semisynthesis and biological evaluation of a focused library of unguinol derivatives as next-generation antibiotics. Org. Biomol. Chem. 2021, 19, 1022–1036. [Google Scholar] [CrossRef]
- Garlick, J.M.; Sturlis, S.M.; Bruno, P.A.; Yates, J.A.; Peiffer, A.L.; Liu, Y.; Goo, L.; Bao, L.; De Salle, S.N.; Tamayo-Castillo, G.; et al. Norstictic acid is a selective allosteric transcriptional regulator. J. Am. Chem. Soc. 2021, 143, 9297–9302. [Google Scholar] [CrossRef]
- Luo, X.W.; Cai, G.D.; Guo, Y.F.; Gao, C.H.; Huang, W.F.; Zhang, Z.H.; Lu, H.M.; Liu, K.; Chen, J.H.; Xiong, X.F.; et al. Exploring marine-derived ascochlorins as novel human dihydroorotate dehydrogenase inhibitors for treatment of triple-negative breast cancer. J. Med. Chem. 2021, 64, 13918–13932. [Google Scholar] [CrossRef]
- Guo, L.; Luo, X.W.; Yang, P.; Zhang, Y.T.; Huang, J.L.; Wang, H.; Guo, Y.F.; Huang, W.F.; Chen, Z.Q.; Wang, S.S.; et al. Ilicicolin a exerts antitumor effect in castration-resistant prostate cancer via suppressing EZH2 signaling pathway. Front. Pharmacol. 2021, 12, 723729. [Google Scholar] [CrossRef]
- Luo, X.W.; Lin, X.P.; Tao, H.M.; Wang, J.F.; Li, J.Y.; Yang, B.; Zhou, X.F.; Liu, Y.H. Isochromophilones A–F, cytotoxic chloroazaphilones from the marine mangrove endophytic fungus Diaporthe sp. SCSIO 41011. J. Nat. Prod. 2018, 81, 934–941. [Google Scholar] [CrossRef]
- Luo, X.W.; Chen, C.M.; Tao, H.M.; Lin, X.P.; Yang, B.; Zhou, X.F.; Liu, Y.H. Structurally diverse diketopiperazine alkaloids from the marine-derived fungus Aspergillus versicolor SCSIO 41016. Org. Chem. Front. 2019, 6, 736–740. [Google Scholar] [CrossRef]
- Sadorn, K.; Saepua, S.; Bunbamrung, N.; Boonyuen, N.; Komwijit, S.; Rachtawee, P.; Pittayakhajonwut, P. Diphenyl ethers and depsidones from the endophytic fungus Aspergillus unguis BCC54176. Tetrahedron 2022, 105, 132612. [Google Scholar] [CrossRef]
- Sureram, S.; Wiyakrutta, S.; Ngamrojanavanich, N.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. Depsidones, aromatase inhibitors and radical scavenging agents from the marine-derived fungus Aspergillus unguis CRI282-03. Planta Med. 2012, 78, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.; Nielsen, P.H.; Frisvad, J.C. Fungal depside, guisinol, from a marine derived strain of Emericella unguis. Phytochemistry 1999, 50, 263–265. [Google Scholar] [CrossRef]
- Cheng, Q.; Snyder, J.K. Notizen: Two new phloroglucinol derivatives with phosphodiesterase inhibitory activity from the leaves of Eucalyptus robusta. Z. Für Nat. B 1991, 46, 1275–1277. [Google Scholar] [CrossRef]
- Liu, D.H.; Sun, Y.Z.; Kurtan, T.; Mandi, A.; Tang, H.; Li, J.; Su, L.; Zhuang, C.L.; Liu, Z.Y.; Zhang, W. Osteoclastogenesis regulation metabolites from the coral-associated fungus Pseudallescheria boydii TW-1024-3. J. Nat. Prod. 2019, 82, 1274–1282. [Google Scholar] [CrossRef]
- Ding, W.; Ma, C.F.; Zhang, W.P.; Chiang, H.; Tam, C.; Xu, Y.; Zhang, G.Z.; Qian, P.Y. Anti-biofilm effect of a butenolide/polymer coating and metatranscriptomic analyses. Biofouling 2018, 34, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.L.; Wu, C.H.; Qian, P.Y. Marine natural products as antifouling molecules—A mini-review (2014–2020). Biofouling 2020, 36, 1210–1226. [Google Scholar] [CrossRef]
- Wang, K.L.; Wu, Z.H.; Wang, Y.; Wang, C.Y.; Xu, Y. Mini-review: Antifouling natural products from marine microorganisms and their synthetic analogs. Mar. Drugs 2017, 15, 266. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.M.; Chen, W.H.; Tao, H.M.; Yang, B.; Zhou, X.F.; Luo, X.W.; Liu, Y.H. Diversified polyketides and nitrogenous compounds from the mangrove endophytic fungus Penicillium steckii SCSIO 41025. Chin. J. Chem. 2021, 39, 2132–2140. [Google Scholar] [CrossRef]




| Position | δC, Type | δH (J in Hz) | HMBC |
|---|---|---|---|
| 1 | 144.0, C | ||
| 1-Me | 21.6, CH3 | 2.45, s | 1, 2, 11a |
| 2 | 111.2, CH | 6.73, s | 3, 4, 11a |
| 3 | 163.0, C | ||
| 3-OMe | 56.4, CH3 | 3.87, s | 3 |
| 4 | 116.1, C | ||
| 4-Me | 10.2, CH3 | 2.15, s | 3, 4, 4a |
| 4a | 162.3, C | ||
| 5a | 143.6, C | ||
| 6 | 135.7, C | ||
| 7 | 117.3, C | ||
| 8 | 143.7, C | ||
| 9 | 117.8, C | ||
| 9-Me | 9.2, CH3 | 2.24, s | 8, 9a, 9 |
| 9a | 150.2, C | ||
| 11 | 165.3, C | ||
| 11a | 114.2, C | ||
| 1′ | 131.8, C | ||
| 2′ | 128.5, CH | 5.40, q (6.8) | 6, 3′, 4′ |
| 3′ | 14.0, CH3 | 1.82, d (6.8) | 1′ |
| 4′ | 17.7, CH3 | 1.91, s | 6, 1′ |
| Compound | Minimum Inhibitory Concentration (MIC, μg/mL) | |||
|---|---|---|---|---|
| MRSA | M. variabilis | M. jannaschii | Pelagius | |
| 1 | 16 | 32 | 64 | - |
| 2 | 2 | 16 | 32 | - |
| 3 | - | - | - | - |
| 4 | 2 | 8 | 16 | 64 |
| 5 | >128 | 128 | - | - |
| 6 | 32 | 8 | 32 | - |
| 7 | 16 | 64 | >128 | - |
| 8 | >128 | 8 | 32 | - |
| control | 1 d | 1 a 16 b 32 c | 1 a 16 b 8 c | 1 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, Z.; Huang, B.; Liu, K.; Peng, S.; Liu, X.; Gao, C.; Liu, Y.; Tan, Y.; Luo, X. Anti-Osteoclastogenic and Antibacterial Effects of Chlorinated Polyketides from the Beibu Gulf Coral-Derived Fungus Aspergillus unguis GXIMD 02505. Mar. Drugs 2022, 20, 178. https://doi.org/10.3390/md20030178
Zhang Y, Li Z, Huang B, Liu K, Peng S, Liu X, Gao C, Liu Y, Tan Y, Luo X. Anti-Osteoclastogenic and Antibacterial Effects of Chlorinated Polyketides from the Beibu Gulf Coral-Derived Fungus Aspergillus unguis GXIMD 02505. Marine Drugs. 2022; 20(3):178. https://doi.org/10.3390/md20030178
Chicago/Turabian StyleZhang, Yanting, Zhichao Li, Bingyao Huang, Kai Liu, Shuai Peng, Xinming Liu, Chenghai Gao, Yonghong Liu, Yanhui Tan, and Xiaowei Luo. 2022. "Anti-Osteoclastogenic and Antibacterial Effects of Chlorinated Polyketides from the Beibu Gulf Coral-Derived Fungus Aspergillus unguis GXIMD 02505" Marine Drugs 20, no. 3: 178. https://doi.org/10.3390/md20030178