Culturable Diversity of Thraustochytrids from Coastal Waters of Qingdao and Their Fatty Acids
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phylogenetic Diversity of the Isolates
2.2. Season- and Habitat-Specific Culturable Diversity
2.3. Variations of Biomass and Lipid Contents of Cultured Strains
2.4. Fatty Acid Profiles and Their Variations
3. Materials and Methods
3.1. Sample Collection and Isolation of Thraustochytrids
3.2. Sequencing and Phylogenetic Analysis
3.3. Quantification of Biomass and Fatty Acids
3.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deeba, F.; Kumar, K.K.; Gaur, N.A. Overview of Microbial Production of Omega-3-Polyunsaturated Fatty Acid. In Nutraceutical Fatty Acids from Oleaginous Microalgae; John Wiley and Sons: Hoboken, NJ, USA, 2020; pp. 145–163. [Google Scholar]
- Ji, X.J.; Huang, H. Engineering microbes to produce polyunsaturated fatty acids. Trends Biotechnol. 2019, 37, 344–346. [Google Scholar] [CrossRef] [PubMed]
- Leong, W.H.; Lim, J.W.; Lam, M.K.; Uemura, Y.; Ho, Y.C. Third generation biofuels: A nutritional perspective in enhancing microbial lipid production. Renew. Sustain. Energy Rev. 2018, 91, 950–961. [Google Scholar] [CrossRef]
- Adrio, J.L. Oleaginous yeasts: Promising platforms for the production of oleochemicals and biofuels. Biotechnol. Bioeng. 2017, 114, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kamegawa, Y.; Kurita, C.; Kanoh, M.; Murakawa, N.; Sakuradani, E. Efficient production of biolipids by crude glycerol-assimilating fungi. Bioresour. Technol. Rep. 2021, 16, 100861. [Google Scholar] [CrossRef]
- Gosalawit, C.; Imsoonthornruksa, S.; Gilroyed, B.H.; Mcnea, L.; Boontawan, A.; Ketudat-Cairns, M. The potential of the oleaginous yeast Rhodotorula paludigena CM33 to produce biolipids. J. Biotechnol. 2021, 329, 56–64. [Google Scholar] [CrossRef]
- Yao, S.; Lyu, S.; An, Y.; Lu, J.; Gjermansen, C.; Schramm, A. Microalgae–bacteria symbiosis in microalgal growth and biofuel production: A review. J. Appl. Microbiol. 2019, 126, 359–368. [Google Scholar] [CrossRef]
- Bai, M.; Sen, B.; Wang, Q.; Xie, Y.; He, Y.; Wang, G. Molecular detection and spatiotemporal characterization of Labyrinthulomycete protist diversity in the coastal waters along the Pearl River Delta. Microb. Ecol. 2019, 77, 394–405. [Google Scholar] [CrossRef]
- Xie, N.; Sen, B.; Song, Z.; Zhao, Y.; Chen, Z.; Shi, W.; Zhang, Y.; Zhang, J.; Johnson, Z.I.; Wang, G. High phylogenetic diversity and abundance pattern of Labyrinthulomycete protists in the coastal waters of the Bohai Sea. Environ. Microbiol. 2018, 20, 3042–3056. [Google Scholar] [CrossRef]
- Patel, A.; Sarkar, O.; Rova, U.; Christakopoulos, P.; Matsakas, L. Valorization of volatile fatty acids derived from low-cost organic waste for lipogenesis in oleaginous microorganisms-a review. Bioresour. Technol. 2021, 321, 124457. [Google Scholar] [CrossRef]
- Patel, A.; Rova, U.; Christakopoulos, P.; Matsakas, L. Mining of squalene as a value-added byproduct from DHA producing marine thraustochytrid cultivated on food waste hydrolysate. Sci. Total Environ. 2020, 736, 139691. [Google Scholar] [CrossRef]
- Liu, Y.; Singh, P.; Sun, Y.; Luan, S.; Wang, G. Culturable diversity and biochemical features of thraustochytrids from coastal waters of Southern China. Appl. Microbiol. Biotechnol. 2014, 98, 3241–3255. [Google Scholar] [CrossRef] [PubMed]
- Anbu, P.; Kim, D.U.; Jeh, E.J.; Jeong, Y.S.; Hur, B.K. Investigation of the physiological properties and synthesis of PUFAs from thraustochytrids and its electrophoretic karyotypes. Biotechnol. Bioprocess Eng. 2007, 12, 720–729. [Google Scholar] [CrossRef]
- Bagul, V.P.; Annapure, U.S. Isolation of fast-growing thraustochytrids and seasonal variation on the fatty acid composition of thraustochytrids from mangrove regions of Navi Mumbai, India. J. Environ. Manag. 2021, 290, 112597. [Google Scholar] [CrossRef] [PubMed]
- Shene, C.; Paredes, P.; Vergara, D.; Leyton, A.; Garcés, M.; Flores, L.; Rubilar, M.; Bustamante, M.; Armenta, R. Antarctic thraustochytrids: Producers of long-chain omega-3 polyunsaturated fatty acids. Microbiologyopen 2020, 9, e00950. [Google Scholar] [CrossRef]
- Wang, Q.; Ye, H.; Xie, Y.; He, Y.; Sen, B.; Wang, G. Culturable diversity and lipid production profile of Labyrinthulomycete protists isolated from coastal mangrove habitats of China. Mar. Drugs 2019, 17, 268. [Google Scholar] [CrossRef] [Green Version]
- Manikan, V.; Nazir, M.Y.M.; Kalil, M.S.; Isa, M.H.M.; Kader, A.J.A.; Yusoff, W.M.W.; Hamid, A.A. A new strain of docosahexaenoic acid producing microalga from Malaysian coastal waters. Algal Res. 2015, 9, 40–47. [Google Scholar] [CrossRef]
- Ueda, M.; Nomura, Y.; Doi, K.; Nakajima, M.; Honda, D. Seasonal dynamics of culturable thraustochytrids (Labyrinthulomycetes, Stramenopiles) in estuarine and coastal waters. Aquat. Microb. Ecol. 2015, 74, 187–204. [Google Scholar] [CrossRef]
- Xie, N.; Hunt, D.E.; Johnson, Z.I.; He, Y.; Wang, G. Annual partitioning patterns of Labyrinthulomycetes protists reveal their multifaceted role in marine microbial food webs. Appl. Environ. Microbiol. 2021, 87, e01652-20. [Google Scholar] [CrossRef]
- Damare, V.; Raghukumar, S. Morphology and physiology of the marine straminipilan fungi, the aplanochytrids isolated from the equatorial Indian Ocean. Indian J. Geo-Mar. Sci. 2006, 35, 326–340. [Google Scholar]
- Raghukumar, S. Enumeration of thraustochytrids (heterotrophic microorganisms) from the Arabian Sea. Mahasagar 1985, 18, 457–465. [Google Scholar]
- Damare, V.S.; Damare, S.; Ramanujam, P.; Meena, R.M.; Raghukumar, S. Preliminary studies on the association between zooplankton and the stramenopilan fungi, aplanochytrids. Microb. Ecol. 2013, 65, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Leander, C.A.; Porter, D.; Leander, B.S. Comparative morphology and molecular phylogeny of aplanochytrids (Labyrinthulomycota). Eur. J. Protistol. 2004, 40, 317–328. [Google Scholar] [CrossRef]
- Gupta, A.; Singh, D.; Byreddy, A.R.; Thyagarajan, T.; Sonkar, S.P.; Mathur, A.S.; Tuli, D.K.; Barrow, C.J.; Puri, M. Exploring omega-3 fatty acids, enzymes and biodiesel producing thraustochytrids from Australian and Indian marine biodiversity. Biotechnol. J. 2016, 11, 345–355. [Google Scholar] [CrossRef]
- Yadav, M.; Yadav, A.; Kumar, S.; Yadav, J.P. Spatial and seasonal influences on culturable endophytic mycobiota associated with different tissues of Eugenia jambolana Lam. and their antibacterial activity against MDR strains. BMC Microbiol. 2016, 16, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- França, L.; Sannino, C.; Turchetti, B.; Buzzini, P.; Margesin, R. Seasonal and altitudinal changes of culturable bacterial and yeast diversity in Alpine Forest soils. Extremophiles 2016, 20, 855–873. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Gomez, C.; Ramirez-Romero, C.; Cordoba, F.; Raga, G.B.; Salinas, E.; Martinez, L.; Rosas, I.; Quintana, E.T.; Maldonado, L.A.; Rosas, D. Characterization of culturable airborne microorganisms in the Yucatan Peninsula. Atmos. Environ. 2020, 223, 117183. [Google Scholar] [CrossRef]
- Lyu, L.; Wang, Q.; Wang, G. Cultivation and diversity analysis of novel marine thraustochytrids. Mar. Life Sci. Technol. 2021, 3, 263–275. [Google Scholar] [CrossRef]
- Chang, K.J.L.; Dunstan, G.A.; Abell, G.C.; Clementson, L.A.; Blackburn, S.I.; Nichols, P.D.; Koutoulis, A. Biodiscovery of new Australian thraustochytrids for production of biodiesel and long-chain omega-3 oils. Appl. Microbiol. Biotechnol. 2012, 93, 2215–2231. [Google Scholar] [CrossRef]
- Bongiorni, L.; Dini, F. Distribution and abundance of thraustochytrids in different Mediterranean coastal habitats. Aquat. Microb. Ecol. 2002, 30, 49–56. [Google Scholar] [CrossRef]
- Ou, M.C.; Yeong, H.Y.; Pang, K.L.; Phang, S.M. Fatty acid production of tropical thraustochytrids from Malaysian mangroves. Bot. Mar. 2016, 59, 321–338. [Google Scholar] [CrossRef]
- Jaseera, K.; Kaladharan, P.; Vijayan, K.; Sandhya, S.; Antony, M.L.; Pradeep, M. Isolation and phylogenetic identification of heterotrophic thraustochytrids from mangrove habitats along the southwest coast of India and prospecting their PUFA accumulation. J. Appl. Phycol. 2019, 31, 1057–1068. [Google Scholar] [CrossRef]
- Jaritkhuan, S.; Suanjit, S. Species diversity and polyunsaturated fatty acid content of thraustochytrids from fallen mangrove leaves in Chon Buri province, Thailand. Agric. Nat. Resour. 2018, 52, 24–32. [Google Scholar] [CrossRef]
- Gao, M.; Song, X.; Feng, Y.; Li, W.; Cui, Q. Isolation and characterization of Aurantiochytrium species: High docosahexaenoic acid (DHA) production by the newly isolated microalga, Aurantiochytrium sp. SD116. J. Oleo S. 2013, 62, 143–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, R.; Raghukumar, S.; Sambaiah, K.; Kumon, Y.; Nakahara, T. Docosahexaenoic acid accumulation in thraustochytrids: Search for the rationale. Mar. Biol. 2007, 151, 1657–1664. [Google Scholar] [CrossRef]
- Hsu, Y.M.; Yin, M.C. EPA or DHA enhanced oxidative stress and aging protein expression in brain of d-galactose treated mice. Biomedicine 2016, 6, 17. [Google Scholar] [CrossRef]
- Japar, A.S.; Takriff, M.S.; Yasin, N.H.M.; Mahmod, S.S. Optimization of Chlorella biomass harvesting by flocculation and its potential for biofuel production. J. Appl. Phycol. 2021, 33, 1621–1629. [Google Scholar] [CrossRef]
- Patel, A.; Rova, U.; Christakopoulos, P.; Matsakas, L. Simultaneous production of DHA and squalene from Aurantiochytrium sp. grown on forest biomass hydrolysates. Biotechnol. Biofuels 2019, 12, 255. [Google Scholar] [CrossRef]
- Gupta, A.; Wilkens, S.; Adcock, J.L.; Puri, M.; Barrow, C.J. Pollen baiting facilitates the isolation of marine thraustochytrids with potential in omega-3 and biodiesel production. J. Ind. Microbiol. Biotechnol. 2013, 40, 1231–1240. [Google Scholar] [CrossRef]
- Rosa, S.M.; Galvagno, M.A.; Vélez, C.G. Adjusting culture conditions to isolate thraustochytrids from temperate and cold environments in southern Argentina. Mycoscience 2011, 52, 242–252. [Google Scholar] [CrossRef]
- Honda, D.; Yokochi, T.; Nakahara, T.; Raghukumar, S.; Nakagiri, A.; Schaumann, K.; Higashihara, T. Molecular phylogeny of labyrinthulids and thraustochytrids based on the sequencing of 18S ribosomal RNA gene. J. Eukaryot. Microbiol. 1999, 46, 637–647. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Lepage, G.; Roy, C.C. Improved recovery of fatty acid through direct transesterification without prior extraction or purification. J. Lipid Res. 1984, 25, 1391–1396. [Google Scholar] [CrossRef]
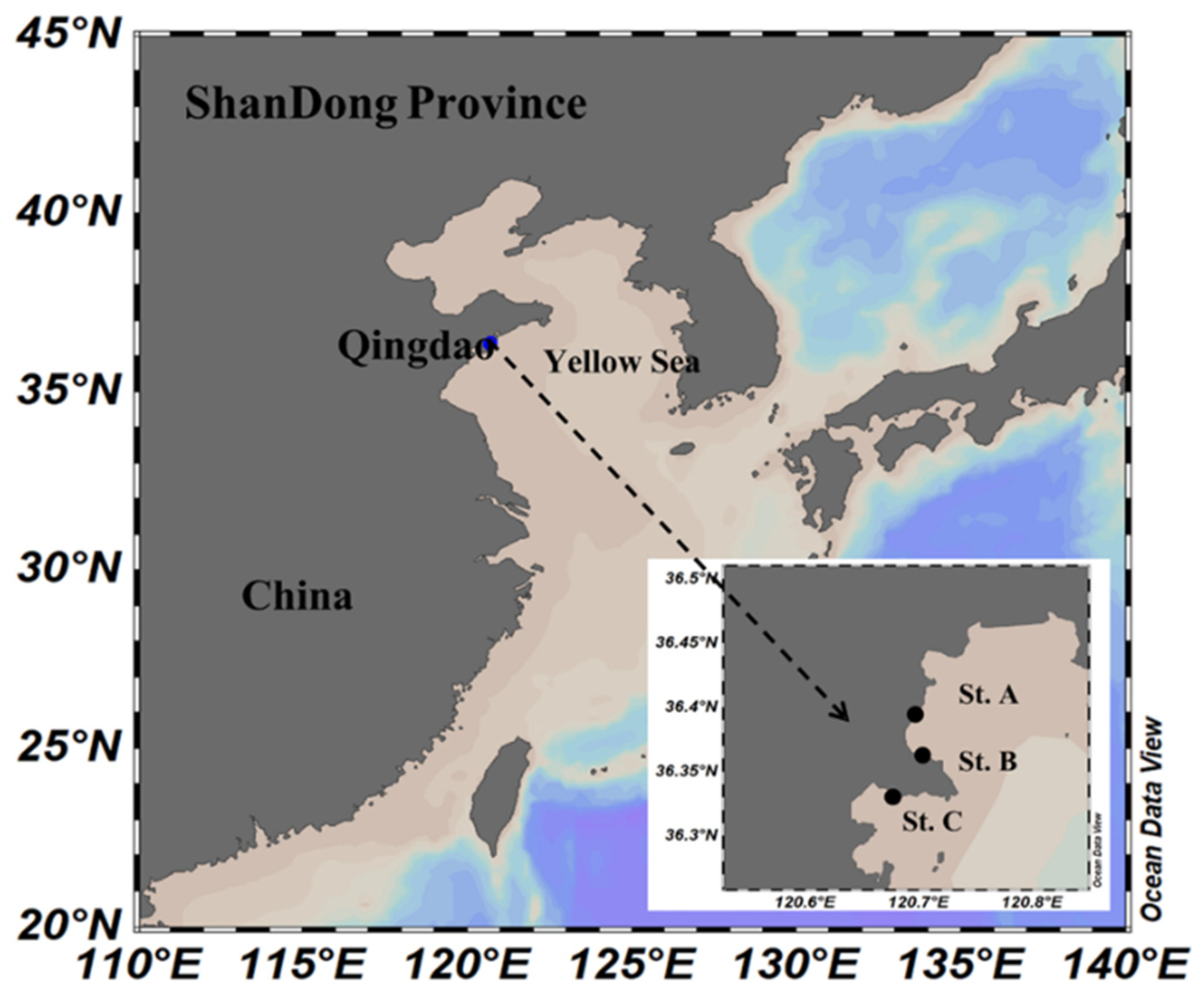
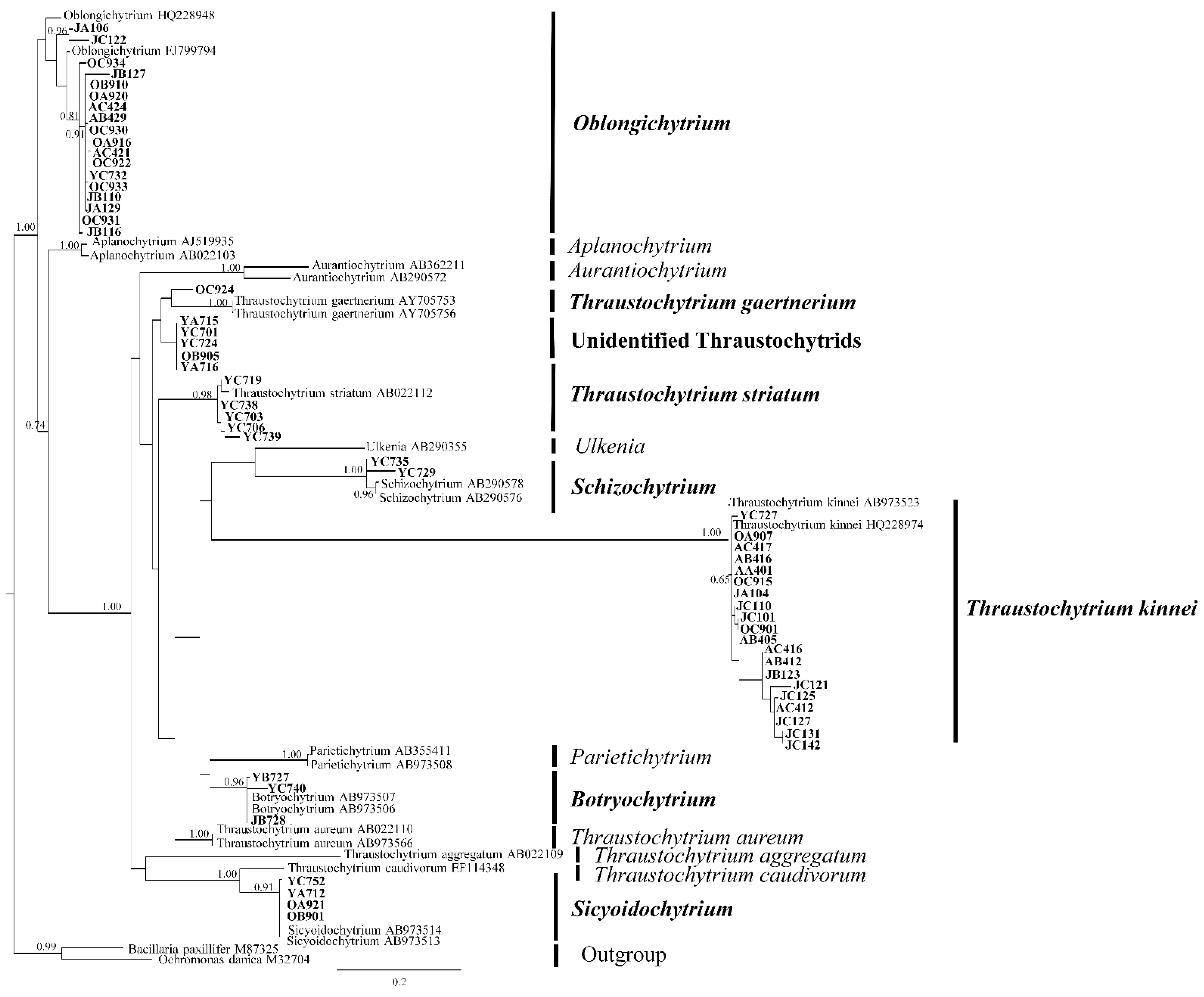
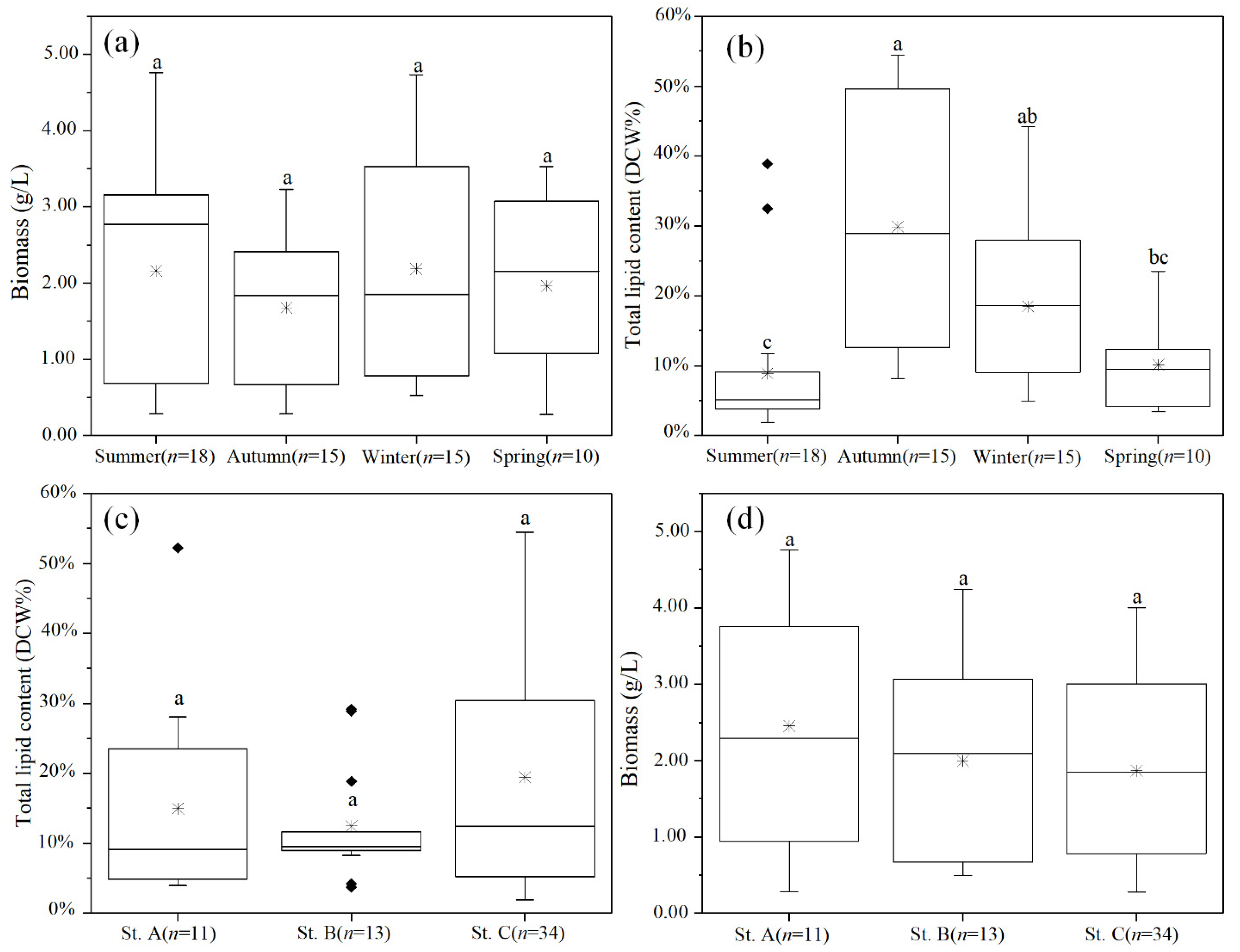

| Station | Genus | Summer | Autumn | Winter | Spring |
|---|---|---|---|---|---|
| St. A | Thraustochytrium | 0 | 1 | 1 | 1 |
| Oblongichytrium | 0 | 2 | 2 | 0 | |
| Sicyoidochytrium | 1 | 1 | 0 | 0 | |
| Schizochytrium | 0 | 0 | 0 | 0 | |
| Botryochytrium | 0 | 0 | 0 | 0 | |
| Unidentified Thraustochytrids | 2 | 0 | 0 | 0 | |
| Total | 3 | 4 | 3 | 1 | |
| St. B | Thraustochytrium | 0 | 0 | 1 | 3 |
| Oblongichytrium | 0 | 1 | 3 | 1 | |
| Sicyoidochytrium | 0 | 1 | 0 | 0 | |
| Schizochytrium | 0 | 0 | 0 | 0 | |
| Botryochytrium | 2 | 0 | 0 | 0 | |
| Unidentified Thraustochytrids | 0 | 1 | 0 | 0 | |
| Total | 2 | 3 | 4 | 4 | |
| St. C | Thraustochytrium | 6 | 3 | 7 | 3 |
| Oblongichytrium | 1 | 5 | 1 | 2 | |
| Sicyoidochytrium | 1 | 0 | 0 | 0 | |
| Schizochytrium | 2 | 0 | 0 | 0 | |
| Botryochytrium | 1 | 0 | 0 | 0 | |
| Unidentified Thraustochytrids | 2 | 0 | 0 | 0 | |
| Total | 13 | 8 | 8 | 5 |
| Fatty Acid | Relative Fatty Acid Content (%) # |
|---|---|
| C12:0 | 1.59 ± 1.78 |
| C14:0 | 2.12 ± 1.76 |
| C15:0 | 5.81 ± 2.88 |
| C16:0 | 18.42 ± 5.57 |
| C17:0 | 1.31 ± 1.84 |
| C18:0 | 7.68 ± 5.03 |
| C18:1 | 4.75 ± 3.84 |
| C20:0 | 3.48 ± 2.66 |
| C20:4, w − 6 | 1.45 ± 1.30 |
| C20:5, w − 3 | 3.89 ± 2.79 |
| C22:5, w − 3 | 3.63 ± 4.79 |
| C22:6, w − 3 | 25.14 ± 9.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, M.; Sen, B.; Wen, S.; Ye, H.; He, Y.; Zhang, X.; Wang, G. Culturable Diversity of Thraustochytrids from Coastal Waters of Qingdao and Their Fatty Acids. Mar. Drugs 2022, 20, 229. https://doi.org/10.3390/md20040229
Bai M, Sen B, Wen S, Ye H, He Y, Zhang X, Wang G. Culturable Diversity of Thraustochytrids from Coastal Waters of Qingdao and Their Fatty Acids. Marine Drugs. 2022; 20(4):229. https://doi.org/10.3390/md20040229
Chicago/Turabian StyleBai, Mohan, Biswarup Sen, Shuai Wen, Huike Ye, Yaodong He, Xiaobo Zhang, and Guangyi Wang. 2022. "Culturable Diversity of Thraustochytrids from Coastal Waters of Qingdao and Their Fatty Acids" Marine Drugs 20, no. 4: 229. https://doi.org/10.3390/md20040229






