The Immune System of Marine Organisms as Source for Drugs against Infectious Diseases
Abstract
:1. The Clinical Arsenal of Antimicrobial Agents Requires an Update
2. Current General Strategies for the Discovery of Antimicrobials
3. Marine Habitats as Promising Sources of New Antimicrobials
4. Antimicrobial Peptides (AMPs)
5. Antimicrobial Metabolites
Funding
Conflicts of Interest
Abbreviations
References
- Giske, C.G.; Monnet, D.L.; Cars, O.; Carmeli, Y. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob. Agents Chemother. 2008, 52, 813–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spellberg, B.; Guidos, R.; Gilbert, D.; Bradley, J.; Boucher, H.W.; Scheld, W.M.; Bartlett, J.G.; Edwards, J., Jr.; America, I.D.S.o. The epidemic of antibiotic-resistant infections: A call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 2008, 46, 155–164. [Google Scholar] [CrossRef]
- Bassetti, M.; Merelli, M.; Temperoni, C.; Astilean, A. New antibiotics for bad bugs: Where are we? Ann. Clin. Microbiol. Antimicrob. 2013, 12, 22. [Google Scholar] [CrossRef] [Green Version]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Coates, A.R.; Halls, G.; Hu, Y. Novel classes of antibiotics or more of the same? Br. J. Pharm. 2011, 163, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Butler, M.S.; Paterson, D.L. Antibiotics in the clinical pipeline in October 2019. J. Antibiot. 2020, 73, 329–364. [Google Scholar] [CrossRef]
- Griffiths, P.D. A perspective on antiviral resistance. J. Clin. Virol. 2009, 46, 3–8. [Google Scholar] [CrossRef]
- Anthony, S.J.; Epstein, J.H.; Murray, K.A.; Navarrete-Macias, I.; Zambrana-Torrelio, C.M.; Solovyov, A.; Ojeda-Flores, R.; Arrigo, N.C.; Islam, A.; Ali Khan, S. A strategy to estimate unknown viral diversity in mammals. MBio 2013, 4, e00598-13. [Google Scholar] [CrossRef] [Green Version]
- Vigant, F.; Santos, N.C.; Lee, B. Broad-spectrum antivirals against viral fusion. Nat. Rev. Microbiol. 2015, 13, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Su, S.; Yang, H.; Jiang, S. Antivirals with common targets against highly pathogenic viruses. Cell 2021, 184, 1604–1620. [Google Scholar] [CrossRef] [PubMed]
- Tompa, D.R.; Immanuel, A.; Srikanth, S.; Kadhirvel, S. Trends and strategies to combat viral infections: A review on FDA approved antiviral drugs. Int. J. Biol. Macromol. 2021, 172, 524–541. [Google Scholar] [CrossRef] [PubMed]
- Cully, M. A tale of two antiviral targets-and the COVID-19 drugs that bind them. Nat. Rev. Drug Discov. 2021, 21, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Findlay, E.G.; Currie, S.M.; Davidson, D.J. Cationic host defence peptides: Potential as antiviral therapeutics. BioDrugs 2013, 27, 479–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawlowski, A.C.; Johnson, J.W.; Wright, G.D. Evolving medicinal chemistry strategies in antibiotic discovery. Curr. Opin. Biotechnol. 2016, 42, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Torres, V.; Encinar, J.A.; Herranz-López, M.; Pérez-Sánchez, A.; Galiano, V.; Barrajón-Catalán, E.; Micol, V. An updated review on marine anticancer compounds: The use of virtual screening for the discovery of small-molecule cancer drugs. Molecules 2017, 22, 1037. [Google Scholar] [CrossRef]
- Bello-Pérez, M.; Falcó, A.; Galiano, V.; Coll, J.; Perez, L.; Encinar, J.A. Discovery of nonnucleoside inhibitors of polymerase from infectious pancreatic necrosis virus (IPNV). Drug Des. Dev. Ther. 2018, 12, 2337. [Google Scholar] [CrossRef] [Green Version]
- Encinar, J.A.; Menendez, J.A. Potential drugs targeting early innate immune evasion of SARS-coronavirus 2 via 2′20-O-methylation of viral RNA. Viruses 2020, 12, 525. [Google Scholar] [CrossRef]
- Neira, J.L.; Palomino-Schätzlein, M.; Hurtado-Gómez, E.; Ortore, M.G.; Falcó, A. An N-terminal half fragment of the histidine phosphocarrier protein, HPr, is disordered but binds to HPr partners and shows antibacterial properties. Biochim. Biophys. Acta (BBA) Gen. Subj. 2021, 1865, 130015. [Google Scholar] [CrossRef]
- Neira, J.L.; Ortega-Alarcón, D.; Rizzuti, B.; Palomino-Schätzlein, M.; Velázquez-Campoy, A.; Falcó, A. Residual Helicity at the Active Site of the Histidine Phosphocarrier, HPr, Modulates Binding Affinity to Its Natural Partners. Int. J. Mol. Sci. 2021, 22, 10805. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cragg, G.M.; Grothaus, P.G.; Newman, D.J. Impact of natural products on developing new anti-cancer agents. Chem. Rev. 2009, 109, 3012–3043. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2011, 28, 196–268. [Google Scholar] [CrossRef] [PubMed]
- Pomponi, S.A. The bioprocess-technological potential of the sea. In Progress in Industrial Microbiology; Elsevier: Amsterdam, The Netherlands, 1999; Volume 35, pp. 5–13. [Google Scholar]
- Williams, P.G. Panning for chemical gold: Marine bacteria as a source of new therapeutics. Trends Biotechnol. 2009, 27, 45–52. [Google Scholar] [CrossRef]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef]
- Kong, D.-X.; Jiang, Y.-Y.; Zhang, H.-Y. Marine natural products as sources of novel scaffolds: Achievement and concern. Drug Discov. Today 2010, 15, 884–886. [Google Scholar] [CrossRef]
- Bergh, Ø.; Børsheim, K.Y.; Bratbak, G.; Heldal, M. High abundance of viruses found in aquatic environments. Nature 1989, 340, 467–468. [Google Scholar] [CrossRef]
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef]
- Otero-Gonzáiez, A.J.; Magalhaes, B.S.; Garcia-Villarino, M.; López-Abarrategui, C.; Sousa, D.A.; Dias, S.C.; Franco, O.L. Antimicrobial peptides from marine invertebrates as a new frontier for microbial infection control. FASEB J. 2010, 24, 1320–1334. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Balasubramanian, S.; Oelschlaeger, T.A.; Grkovic, T.; Pham, N.B.; Quinn, R.J.; Hentschel, U. Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect. Dis. 2017, 17, e30–e41. [Google Scholar] [CrossRef]
- Tassanakajon, A.; Rimphanitchayakit, V.; Visetnan, S.; Amparyup, P.; Somboonwiwat, K.; Charoensapsri, W.; Tang, S. Shrimp humoral responses against pathogens: Antimicrobial peptides and melanization. Dev. Comp. Immunol. 2018, 80, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Falco, A.; Mas, V.; Tafalla, C.; Perez, L.; Coll, J.M.; Estepa, A. Dual antiviral activity of human alpha-defensin-1 against viral haemorrhagic septicaemia rhabdovirus (VHSV): Inactivation of virus particles and induction of a type I interferon-related response. Antivir. Res. 2007, 76, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Blacher, E.; Levy, M.; Tatirovsky, E.; Elinav, E. Microbiome-modulated metabolites at the interface of host immunity. J. Immunol. 2017, 198, 572–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postler, T.S.; Ghosh, S. Understanding the holobiont: How microbial metabolites affect human health and shape the immune system. Cell Metab. 2017, 26, 110–130. [Google Scholar] [CrossRef] [Green Version]
- Bello-Perez, M.; Pereiro, P.; Coll, J.; Novoa, B.; Perez, L.; Falco, A. Zebrafish C-reactive protein isoforms inhibit SVCV replication by blocking autophagy through interactions with cell membrane cholesterol. Sci. Rep. 2020, 10, 1–18. [Google Scholar]
- De Zoysa, M. Medicinal benefits of marine invertebrates: Sources for discovering natural drug candidates. Adv. Food Nutr. Res. 2012, 65, 153–169. [Google Scholar]
- Ponnappan, N.; Budagavi, D.P.; Yadav, B.K.; Chugh, A. Membrane-active peptides from marine organisms-antimicrobials, cell-penetrating peptides and Peptide toxins: Applications and prospects. Probiotics Antimicrob. Proteins 2015, 7, 75–89. [Google Scholar] [CrossRef]
- Castro, R.; Tafalla, C. Overview of fish immunity. In Mucosal Health in Aquaculture; Elsevier: Amsterdam, The Netherlands, 2015; pp. 3–54. [Google Scholar]
- Esteban, M.Á.; Cerezuela, R. Fish mucosal immunity: Skin. In Mucosal Health in Aquaculture; Elsevier: Amsterdam, The Netherlands, 2015; pp. 67–92. [Google Scholar]
- Salinas, I. The mucosal immune system of teleost fish. Biology 2015, 4, 525–539. [Google Scholar] [CrossRef] [Green Version]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef] [Green Version]
- Habbu, P.; Warad, V.; Shastri, R.; Madagundi, S.; Kulkarni, V.H. Antimicrobial metabolites from marine microorganisms. Chin. J. Nat. Med. 2016, 14, 101–116. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, R.E.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321. [Google Scholar] [CrossRef] [PubMed]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Jelinek, R.; Kolusheva, S. Membrane interactions of host-defense peptides studied in model systems. Curr. Protein Pept. Sci. 2005, 6, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Hollmann, A.; Martinez, M.; Maturana, P.; Semorile, L.C.; Maffia, P.C. Antimicrobial peptides: Interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem. 2018, 6, 204. [Google Scholar] [CrossRef] [Green Version]
- Bobone, S.; Piazzon, A.; Orioni, B.; Pedersen, J.Z.; Nan, Y.H.; Hahm, K.S.; Shin, S.Y.; Stella, L. The thin line between cell-penetrating and antimicrobial peptides: The case of Pep-1 and Pep-1-K. J. Pept. Sci. 2011, 17, 335–341. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Falco, A.; Ortega-Villaizan, M.; Chico, V.; Brocal, I.; Perez, L.; Coll, J.M.; Estepa, A. Antimicrobial peptides as model molecules for the development of novel antiviral agents in aquaculture. Mini Rev. Med. Chem. 2009, 9, 1159–1164. [Google Scholar] [CrossRef]
- Balseiro, P.; Falcó, A.; Romero, A.; Dios, S.; Martínez-López, A.; Figueras, A.; Estepa, A.; Novoa, B. Mytilus galloprovincialis myticin C: A chemotactic molecule with antiviral activity and immunoregulatory properties. PLoS ONE 2011, 6, e23140. [Google Scholar] [CrossRef] [Green Version]
- Yu, P.; Gu, H. Bioactive substances from marine fishes, shrimps, and algae and their functions: Present and future. Crit. Rev. Food Sci. Nutr. 2015, 55, 1114–1136. [Google Scholar] [CrossRef] [PubMed]
- Riccio, G.; Ruocco, N.; Mutalipassi, M.; Costantini, M.; Zupo, V.; Coppola, D.; de Pascale, D.; Lauritano, C. Ten-year research update review: Antiviral activities from marine organisms. Biomolecules 2020, 10, 1007. [Google Scholar] [CrossRef] [PubMed]
- Rey-Campos, M.; Novoa, B.; Pallavicini, A.; Gerdol, M.; Figueras, A. Comparative genomics reveals 13 different isoforms of mytimycins (AM) in mytilus galloprovincialis. Int. J. Mol. Sci. 2021, 22, 3235. [Google Scholar] [CrossRef] [PubMed]
- Macedo, M.W.F.S.; Cunha, N.B.D.; Carneiro, J.A.; Costa, R.A.D.; Alencar, S.A.D.; Cardoso, M.H.; Franco, O.L.; Dias, S.C. Marine organisms as a rich source of biologically active peptides. Front. Mar. Sci. 2021, 889, 667764. [Google Scholar] [CrossRef]
- Silphaduang, U.; Colorni, A.; Noga, E.J. Evidence for widespread distribution of piscidin antimicrobial peptides in teleost fish. Dis. Aquat. Organ. 2006, 72, 241–252. [Google Scholar] [CrossRef]
- Sun, B.J.; Xie, H.X.; Song, Y.; Nie, P. Gene structure of an antimicrobial peptide from mandarin fish, Siniperca chuatsi (Basilewsky), suggests that moronecidins and pleurocidins belong in one family: The piscidins. J. Fish Dis. 2007, 30, 335–343. [Google Scholar] [CrossRef]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [Green Version]
- Campoverde, C.; Milne, D.J.; Estévez, A.; Duncan, N.; Secombes, C.J.; Andree, K.B. Ontogeny and modulation after PAMPs stimulation of β-defensin, hepcidin, and piscidin antimicrobial peptides in meagre (Argyrosomus regius). Fish Shellfish Immunol. 2017, 69, 200–210. [Google Scholar] [CrossRef] [Green Version]
- Yi, Y.; You, X.; Bian, C.; Chen, S.; Lv, Z.; Qiu, L.; Shi, Q. High-throughput identification of antimicrobial peptides from amphibious mudskippers. Mar. Drugs 2017, 15, 364. [Google Scholar] [CrossRef] [Green Version]
- Milne, D.J.; Fernández-Montero, Á.; Gundappa, M.K.; Wang, T.; Acosta, F.; Torrecillas, S.; Montero, D.; Zou, J.; Sweetman, J.; Secombes, C.J. An insight into piscidins: The discovery, modulation and bioactivity of greater amberjack, Seriola dumerili, piscidin. Mol. Immunol. 2019, 114, 378–388. [Google Scholar] [CrossRef]
- Barroso, C.; Carvalho, P.; Carvalho, C.; Santarém, N.; Gonçalves, J.F.; Rodrigues, P.N.; Neves, J.V. The diverse piscidin repertoire of the European sea bass (Dicentrarchus labrax): Molecular characterization and antimicrobial activities. Int. J. Mol. Sci. 2020, 21, 4613. [Google Scholar] [CrossRef] [PubMed]
- Zaccone, G.; Capillo, G.; Fernandes, J.M.O.; Kiron, V.; Lauriano, E.R.; Alesci, A.; Lo Cascio, P.; Guerrera, M.C.; Kuciel, M.; Zuwala, K. Expression of the Antimicrobial Peptide Piscidin 1 and Neuropeptides in Fish Gill and Skin: A Potential Participation in Neuro-Immune Interaction. Mar. Drugs 2022, 20, 145. [Google Scholar] [CrossRef] [PubMed]
- Lauth, X.; Babon, J.J.; Stannard, J.A.; Singh, S.; Nizet, V.; Carlberg, J.M.; Ostland, V.E.; Pennington, M.W.; Norton, R.S.; Westerman, M.E. Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J. Biol. Chem. 2005, 280, 9272–9282. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.H.; Chen, J.Y.; Kuo, C.M. Three different hepcidins from tilapia, Oreochromis mossambicus: Analysis of their expressions and biological functions. Mol. Immunol. 2007, 44, 1922–1934. [Google Scholar] [CrossRef] [PubMed]
- Noga, E.J.; Silphaduang, U.; Park, N.G.; Seo, J.K.; Stephenson, J.; Kozlowicz, S. Piscidin 4, a novel member of the piscidin family of antimicrobial peptides. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 152, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.Y.; Chen, J.Y.; Lin, T.L.; Lin, C.H. In vitro activities of three synthetic peptides derived from epinecidin-1 and an anti-lipopolysaccharide factor against Propionibacterium acnes, Candida albicans and Trichomonas vaginalis. Peptides 2009, 30, 1058–1068. [Google Scholar] [CrossRef] [PubMed]
- Falco, A.; Chico, V.; Marroqui, L.; Perez, L.; Coll, J.M.; Estepa, A. Expression and antiviral activity of a beta-defensin-like peptide identified in the rainbow trout (Oncorhynchus mykiss) EST sequences. Mol. Immunol. 2008, 45, 757–765. [Google Scholar] [CrossRef]
- Wang, Y.D.; Rajanbabu, V.; Chen, J.Y. Transcriptome analysis of medaka following epinecidin-1 and TH1-5 treatment of NNV infection. Fish Shellfish Immunol. 2015, 42, 121–131. [Google Scholar] [CrossRef]
- Novoa, B.; Romero, A.; Álvarez, Á.L.; Moreira, R.; Pereiro, P.; Costa, M.M.; Dios, S.; Estepa, A.; Parra, F.; Figueras, A. Antiviral activity of myticin C peptide from mussel: An ancient defense against herpesviruses. J. Virol. 2016, 90, 7692–7702. [Google Scholar] [CrossRef] [Green Version]
- Falco, A.; Medina-Gali, R.M.; Poveda, J.A.; Bello-Perez, M.; Novoa, B.; Encinar, J.A. Antiviral activity of a Turbot (Scophthalmus maximus) NK-lysin peptide by inhibition of low-pH virus-induced membrane fusion. Mar. Drugs 2019, 17, 87. [Google Scholar] [CrossRef] [Green Version]
- Sung, W.S.; Lee, J.; Lee, D.G. Fungicidal effect and the mode of action of piscidin 2 derived from hybrid striped bass. Biochem. Biophys. Res. Commun. 2008, 371, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Colorni, A.; Ullal, A.; Heinisch, G.; Noga, E.J. Activity of the antimicrobial polypeptide piscidin 2 against fish ectoparasites. J. Fish Dis. 2008, 31, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.Q.; Zhang, D.M.; Chen, Y.K.; Wang, Q.J.; Yang, Y.Y. Effects of antimicrobial peptides (AMPs) on blood biochemical parameters, antioxidase activity, and immune function in the common carp (Cyprinus carpio). Fish Shellfish Immunol. 2015, 47, 429–434. [Google Scholar] [CrossRef]
- Kang, S.J.; Park, S.J.; Mishig-Ochir, T.; Lee, B.J. Antimicrobial peptides: Therapeutic potentials. Expert Rev. Anti Infect. 2014, 12, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.L. Antimicrobial peptides stage a comeback. Nat. Biotechnol. 2013, 31, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Silva, O.N.; Mulder, K.C.; Barbosa, A.E.; Otero-Gonzalez, A.J.; Lopez-Abarrategui, C.; Rezende, T.M.; Dias, S.C.; Franco, O.L. Exploring the pharmacological potential of promiscuous host-defense peptides: From natural screenings to biotechnological applications. Front. Microbiol. 2011, 2, 232. [Google Scholar] [CrossRef] [Green Version]
- Pietra, F. Secondary metabolites from marine microorganisms: Bacteria, protozoa, algae and fungi: Achievements and prospects. Nat. Prod. Rep. 1997, 14, 453–464. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Pereiro, P.; Forn-Cuní, G.; Dios, S.; Coll, J.; Figueras, A.; Novoa, B. Interferon-independent antiviral activity of 25-hydroxycholesterol in a teleost fish. Antivir. Res. 2017, 145, 146–159. [Google Scholar] [CrossRef]
- Bello-Perez, M.; Falco, A.; Novoa, B.; Perez, L.; Coll, J. Hydroxycholesterol binds and enhances the anti-viral activities of zebrafish monomeric c-reactive protein isoforms. PLoS ONE 2019, 14, e0201509. [Google Scholar] [CrossRef]
- Adamek, M.; Davies, J.; Beck, A.; Jordan, L.; Becker, A.M.; Mojzesz, M.; Rakus, K.; Rumiac, T.; Collet, B.; Brogden, G. Antiviral actions of 25-hydroxycholesterol in fish vary with the virus-host combination. Front. Immunol. 2021, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A. Unusual amino acids in medicinal chemistry. J. Med. Chem. 2016, 59, 10807–10836. [Google Scholar] [CrossRef] [PubMed]
- Balan, S.S.; Kumar, C.G.; Jayalakshmi, S. Aneurinifactin, a new lipopeptide biosurfactant produced by a marine Aneurinibacillus aneurinilyticus SBP-11 isolated from Gulf of Mannar: Purification, characterization and its biological evaluation. Microbiol. Res. 2017, 194, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Desjardine, K.; Pereira, A.; Wright, H.; Matainaho, T.; Kelly, M.; Andersen, R.J. Tauramamide, a lipopeptide antibiotic produced in culture by Brevibacillus laterosporus isolated from a marine habitat: Structure elucidation and synthesis. J. Nat. Prod. 2007, 70, 1850–1853. [Google Scholar] [CrossRef]
- Wang, P.; Xi, L.; Liu, P.; Wang, Y.; Wang, W.; Huang, Y.; Zhu, W. Diketopiperazine derivatives from the marine-derived actinomycete Streptomyces sp. FXJ7. 328. Mar. Drugs 2013, 11, 1035–1049. [Google Scholar] [CrossRef] [Green Version]
- Yonezawa, K.; Yamada, K.; Kouno, I. New diketopiperazine derivatives isolated from sea urchin-derived Bacillus sp. Chem. Pharm. Bull. 2011, 59, 106–108. [Google Scholar] [CrossRef] [Green Version]
- Song, F.; Liu, X.; Guo, H.; Ren, B.; Chen, C.; Piggott, A.M.; Yu, K.; Gao, H.; Wang, Q.; Liu, M. Brevianamides with antitubercular potential from a marine-derived isolate of Aspergillus versicolor. Org. Lett. 2012, 14, 4770–4773. [Google Scholar] [CrossRef]
- Du, F.-Y.; Li, X.-M.; Li, C.-S.; Shang, Z.; Wang, B.-G. Cristatumins A–D, new indole alkaloids from the marine-derived endophytic fungus Eurotium cristatum EN-220. Bioorg. Med. Chem. Lett. 2012, 22, 4650–4653. [Google Scholar] [CrossRef]
- Park, Y.C.; Gunasekera, S.P.; Lopez, J.V.; McCarthy, P.J.; Wright, A.E. Metabolites from the marine-derived fungus Chromocleista sp. isolated from a deep-water sediment sample collected in the Gulf of Mexico. J. Nat. Prod. 2006, 69, 580–584. [Google Scholar] [CrossRef] [Green Version]
- Schultz, A.W.; Oh, D.-C.; Carney, J.R.; Williamson, R.T.; Udwary, D.W.; Jensen, P.R.; Gould, S.J.; Fenical, W.; Moore, B.S. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J. Am. Chem. Soc. 2008, 130, 4507–4516. [Google Scholar] [CrossRef]
- Li, X.; Kim, S.-K.; Nam, K.W.; Kang, J.S.; Choi, H.D.; Son, B.W. A new antibacterial dioxopiperazine alkaloid related to gliotoxin from a marine isolate of the fungus Pseudallescheria. J. Antibiot. 2006, 59, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Vaz, E.; Fernandez-Suarez, M.; Muñoz, L. Determination of the absolute stereochemistry of etzionin. Tetrahedron Asymmetry 2003, 14, 1935–1942. [Google Scholar] [CrossRef]
- Zampella, A.; D’Auria, M.V.; Paloma, L.G.; Casapullo, A.; Minale, L.; Debitus, C.; Henin, Y. Callipeltin A, an anti-HIV cyclic depsipeptide from the New Caledonian Lithistida sponge Callipelta sp. J. Am. Chem. Soc. 1996, 118, 6202–6209. [Google Scholar] [CrossRef]
- Plaza, A.; Bifulco, G.; Keffer, J.L.; Lloyd, J.R.; Baker, H.L.; Bewley, C.A. Celebesides A–C and theopapuamides B−D, depsipeptides from an Indonesian sponge that inhibit HIV-1 entry. J. Org. Chem. 2009, 74, 504–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zampella, A.; Sepe, V.; Luciano, P.; Bellotta, F.; Monti, M.C.; D’Auria, M.V.; Jepsen, T.; Petek, S.; Adeline, M.-T.R.S.; Laprévôte, O. Homophymine A, an anti-HIV cyclodepsipeptide from the sponge Homophymia sp. J. Org. Chem. 2008, 73, 5319–5327. [Google Scholar] [CrossRef]
- Plaza, A.; Bifulco, G.; Masullo, M.; Lloyd, J.R.; Keffer, J.L.; Colin, P.L.; Hooper, J.N.; Bell, L.J.; Bewley, C.A. Mutremdamide A and koshikamides C−H, peptide inhibitors of HIV-1 entry from different Theonella species. J. Org. Chem. 2010, 75, 4344–4355. [Google Scholar] [CrossRef] [Green Version]
- Rashid, M.A.; Gustafson, K.R.; Cartner, L.K.; Shigematsu, N.; Pannell, L.K.; Boyd, M.R. Microspinosamide, a New HIV-inhibitory cyclic depsipeptide from the marine sponge Sidonops microspinosa. J. Nat. Prod. 2001, 64, 117–121. [Google Scholar] [CrossRef]
- Plaza, A.; Gustchina, E.; Baker, H.L.; Kelly, M.; Bewley, C.A. Mirabamides A–D, depsipeptides from the sponge Siliquariaspongia mirabilis that inhibit HIV-1 fusion. J. Nat. Prod. 2007, 70, 1753–1760. [Google Scholar] [CrossRef]
- Giordano, D.; Costantini, M.; Coppola, D.; Lauritano, C.; Pons, L.N.; Ruocco, N.; di Prisco, G.; Ianora, A.; Verde, C. Biotechnological applications of bioactive peptides from marine sources. Adv. Microb. Physiol. 2018, 73, 171–220. [Google Scholar]
- Lu, Z.; Van Wagoner, R.M.; Harper, M.K.; Baker, H.L.; Hooper, J.N.; Bewley, C.A.; Ireland, C.M. Mirabamides E−H, HIV-inhibitory depsipeptides from the sponge Stelletta clavosa. J. Nat. Prod. 2011, 74, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Oku, N.; Gustafson, K.R.; Cartner, L.K.; Wilson, J.A.; Shigematsu, N.; Hess, S.; Pannell, L.K.; Boyd, M.R.; McMahon, J.B. Neamphamide A, a new HIV-inhibitory depsipeptide from the Papua New Guinea marine sponge Neamphius huxleyi. J. Nat. Prod. 2004, 67, 1407–1411. [Google Scholar] [CrossRef] [PubMed]
- Ford, P.W.; Gustafson, K.R.; McKee, T.C.; Shigematsu, N.; Maurizi, L.K.; Pannell, L.K.; Williams, D.E.; Dilip de Silva, E.; Lassota, P.; Allen, T.M. Papuamides A−D, HIV-inhibitory and cytotoxic depsipeptides from the Sponges Theonella mirabilis and Theonella swinhoei collected in Papua New Guinea. J. Am. Chem. Soc. 1999, 121, 5899–5909. [Google Scholar] [CrossRef]
- Andjelic, C.D.; Planelles, V.; Barrows, L.R. Characterizing the anti-HIV activity of papuamide A. Mar. Drugs 2008, 6, 528–549. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Alonso, M.J.; González-Santiago, L.; Martínez, T.; Losada, A.; Galmarini, C.M.; Muñoz, A. The mechanism of action of plitidepsin. Current Opin. Invest. Drugs 2009, 10, 536–542. [Google Scholar]
- White, K.M.; Rosales, R.; Yildiz, S.; Kehrer, T.; Miorin, L.; Moreno, E.; Jangra, S.; Uccellini, M.B.; Rathnasinghe, R.; Coughlan, L. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 2021, 371, 926–931. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.J.; Rashid, M.A.; Cartner, L.K.; Bokesch, H.R.; Wilson, J.A.; McMahon, J.B.; Gustafson, K.R. Stellettapeptins A and B, HIV-inhibitory cyclic depsipeptides from the marine sponge Stelletta sp. Tetrahedron Lett. 2015, 56, 4215–4219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratnayake, A.S.; Bugni, T.S.; Feng, X.; Harper, M.K.; Skalicky, J.J.; Mohammed, K.A.; Andjelic, C.D.; Barrows, L.R.; Ireland, C.M. Theopapuamide, a cyclic depsipeptide from a Papua New Guinea lithistid sponge Theonella swinhoei. J. Nat. Prod. 2006, 69, 1582–1586. [Google Scholar] [CrossRef] [Green Version]
- Burja, A.M.; Banaigs, B.; Abou-Mansour, E.; Burgessd, J.G.; Wrighta, P.C. Marine cyanobacteria—A prolific source of natural products. Tetrahedron 2001, 57, 9347–9377. [Google Scholar] [CrossRef]
- Kubicki, S.; Bollinger, A.; Katzke, N.; Jaeger, K.-E.; Loeschcke, A.; Thies, S. Marine biosurfactants: Biosynthesis, structural diversity and biotechnological applications. Mar. Drugs 2019, 17, 408. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Zhou, X.; Xu, T.; Yang, X.; Liu, Y. Diketopiperazines from marine organisms. Chem. Biodivers. 2010, 7, 2809–2829. [Google Scholar] [CrossRef]
- Huang, R.-M.; Yi, X.-X.; Zhou, Y.; Su, X.; Peng, Y.; Gao, C.-H. An update on 2, 5-diketopiperazines from marine organisms. Mar. Drugs 2014, 12, 6213–6235. [Google Scholar] [CrossRef] [PubMed]
- Rojas, V.; Rivas, L.; Cárdenas, C.; Guzmán, F. Cyanobacteria and eukaryotic microalgae as emerging sources of antibacterial peptides. Molecules 2020, 25, 5804. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Quan, C.; Hou, X.; Fan, S. Potential pharmacological resources: Natural bioactive compounds from marine-derived fungi. Mar. Drugs 2016, 14, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andavan, G.S.B.; Lemmens-Gruber, R. Cyclodepsipeptides from marine sponges: Natural agents for drug research. Mar. Drugs 2010, 8, 810–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, S.; Acharya, D.; Adholeya, A.; Barrow, C.J.; Deshmukh, S.K. Nonribosomal peptides from marine microbes and their antimicrobial and anticancer potential. Front. Pharmacol. 2017, 8, 828. [Google Scholar] [CrossRef] [PubMed]
- Rinehart, K.L., Jr.; Gloer, J.B.; Hughes, R.G., Jr.; Renis, H.E.; McGovren, J.P.; Swynenberg, E.B.; Stringfellow, D.A.; Kuentzel, S.L.; Li, L.H. Didemnins: Antiviral and antitumor depsipeptides from a Caribbean tunicate. Science 1981, 212, 933–935. [Google Scholar] [CrossRef]
- Rinehart, K.L.; Kishore, V.; Bible, K.C.; Sakai, R.; Sullins, D.W.; Li, K.-M. Didemnins and tunichlorin: Novel natural products from the marine tunicate Trididemnum solidum. J. Nat. Prod. 1988, 51, 1–21. [Google Scholar] [CrossRef]
- Tsukimoto, M.; Nagaoka, M.; Shishido, Y.; Fujimoto, J.; Nishisaka, F.; Matsumoto, S.; Harunari, E.; Imada, C.; Matsuzaki, T. Bacterial production of the tunicate-derived antitumor cyclic depsipeptide didemnin B. J. Nat. Prod. 2011, 74, 2329–2331. [Google Scholar] [CrossRef]
- Xu, Y.; Kersten, R.D.; Nam, S.-J.; Lu, L.; Al-Suwailem, A.M.; Zheng, H.; Fenical, W.; Dorrestein, P.C.; Moore, B.S.; Qian, P.-Y. Bacterial biosynthesis and maturation of the didemnin anti-cancer agents. J. Am. Chem. Soc. 2012, 134, 8625–8632. [Google Scholar] [CrossRef] [Green Version]
- Blunt, J.W.; Munro, M.H. Dictionary of Marine Natural Products with CD-ROM; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]

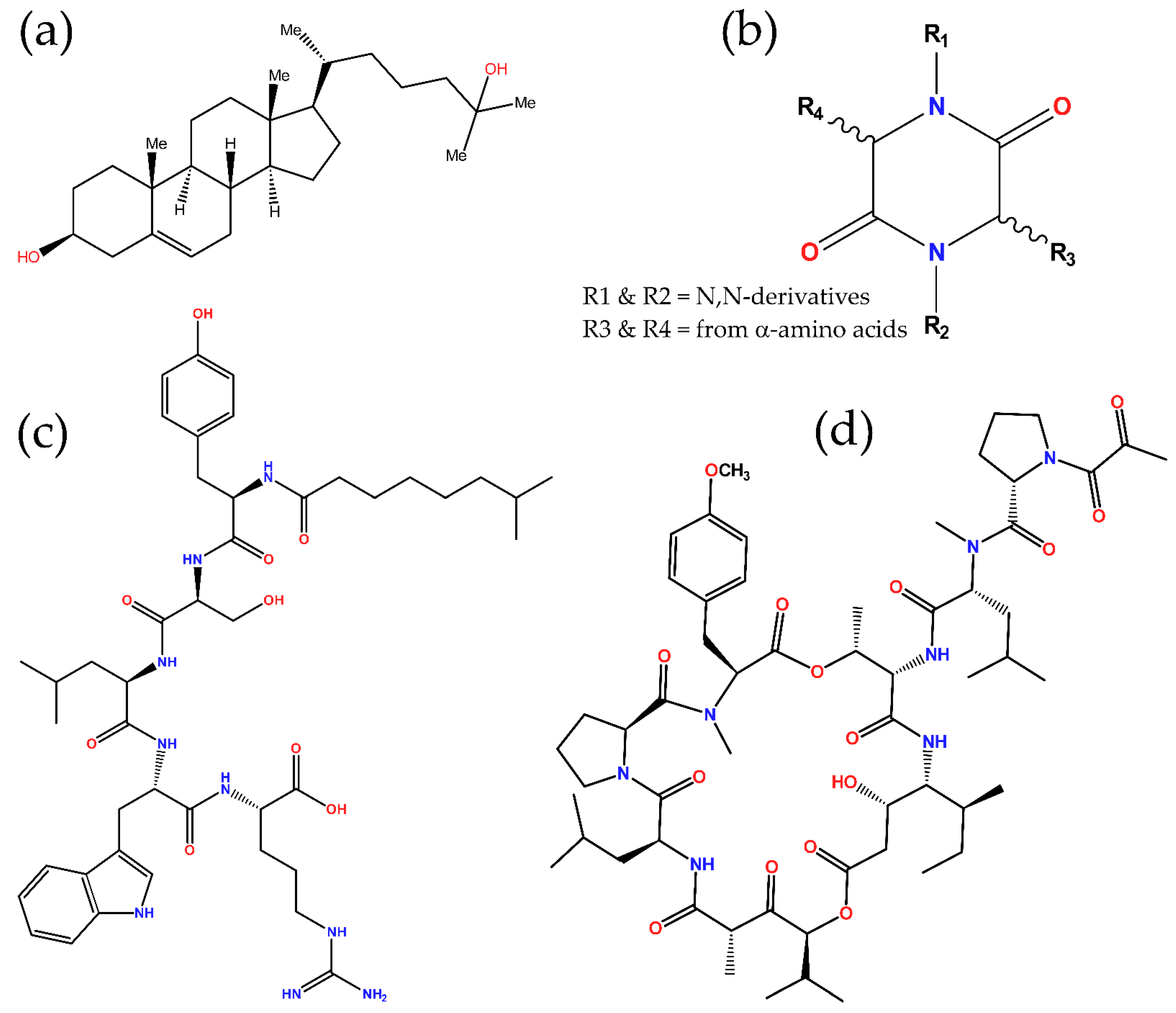
| Antimicrobial Metabolite | Source | Bioactivity | Ref. |
|---|---|---|---|
| Lipopeptides | |||
| Aneurinifactin | Aneurinibacillus aneurinilyticus | Antibacterial a | [86] |
| Tauramamide | Brevibacillus laterosporus | Antibacterial | [87] |
| 2,5-DKPs (cyclodipeptides) | |||
| (3Z,6Z)-3-(4-hydroxybenzylidene)-6- isobutylidenepiperazine-2,5-dione | Streptomyces sp. | Antiviral b | [88] |
| Bacillusamide A | Bacillus sp. | Antibacterial, antifungal | [89] |
| Brevianamide S | Aspergillus versicolor | Antibacterial | [90] |
| Cristatumin A | Eurotium cristatum | Antibacterial | [91] |
| Cyclo(d-6-Hyp-l-Phe), cyclo(l-6-Hyp-l-Phe), and cyclo(6,7-en-Pro-l-Phe) | Chromocleista sp. | Antibacterial | [92] |
| Cyclomarazine A, and cyclomarazine B | Salinispora arenicola | Antibacterial a | [93] |
| Dehydroxybis(dethio)bis(methylthio)gliotoxin | Pseudallescheria sp. | Antibacterial | [94] |
| Etzionin | Unidentified Red Sea tunicate | Antifungal | [95] |
| Cyclodepsipeptides (cyclopeptides) from sponges | |||
| Callipeltin A | Callipelta sp. | Antifungal, antiviral | [96] |
| Celebeside A | Siliquariaspongia mirabilis | Antiviral | [97] |
| Homophymine A | Homophymia sp. | Antiviral | [98] |
| Koshikamides F–H | Theonella cupola, and T. swinhoei | Antiviral | [99] |
| Microspinosamide | Sidonops microspinosa | Antiviral | [100] |
| Mirabamides A–D | S. mirabilis | Antiviral | [101,102] |
| Mirabamides E–H | Stelletta clavosa | Antiviral | [103] |
| Neamphamide A | Neamphius huxleyi | Antiviral | [104] |
| Papuamide A–D | S. mirabilis, and T. swinhoei | Antiviral | [101,105,106] |
| Plitidepsin (dehydrodidemnin B) | Aplidium albicans | Antiviral | [107,108] |
| Stellettapeptins A–B | Stelletta sp. | Antiviral | [109] |
| Theopapuamides A–D | S. mirabilis, and T. swinhoei | Antifungal, antiviral | [97,110] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falco, A.; Adamek, M.; Pereiro, P.; Hoole, D.; Encinar, J.A.; Novoa, B.; Mallavia, R. The Immune System of Marine Organisms as Source for Drugs against Infectious Diseases. Mar. Drugs 2022, 20, 363. https://doi.org/10.3390/md20060363
Falco A, Adamek M, Pereiro P, Hoole D, Encinar JA, Novoa B, Mallavia R. The Immune System of Marine Organisms as Source for Drugs against Infectious Diseases. Marine Drugs. 2022; 20(6):363. https://doi.org/10.3390/md20060363
Chicago/Turabian StyleFalco, Alberto, Mikolaj Adamek, Patricia Pereiro, David Hoole, José Antonio Encinar, Beatriz Novoa, and Ricardo Mallavia. 2022. "The Immune System of Marine Organisms as Source for Drugs against Infectious Diseases" Marine Drugs 20, no. 6: 363. https://doi.org/10.3390/md20060363
APA StyleFalco, A., Adamek, M., Pereiro, P., Hoole, D., Encinar, J. A., Novoa, B., & Mallavia, R. (2022). The Immune System of Marine Organisms as Source for Drugs against Infectious Diseases. Marine Drugs, 20(6), 363. https://doi.org/10.3390/md20060363








