Investigation on Metabolites in Structural Diversity from the Deep-Sea Sediment-Derived Bacterium Agrococcus sp. SCSIO 52902 and Their Biosynthesis
Abstract
:1. Introduction
2. Results and Discussion
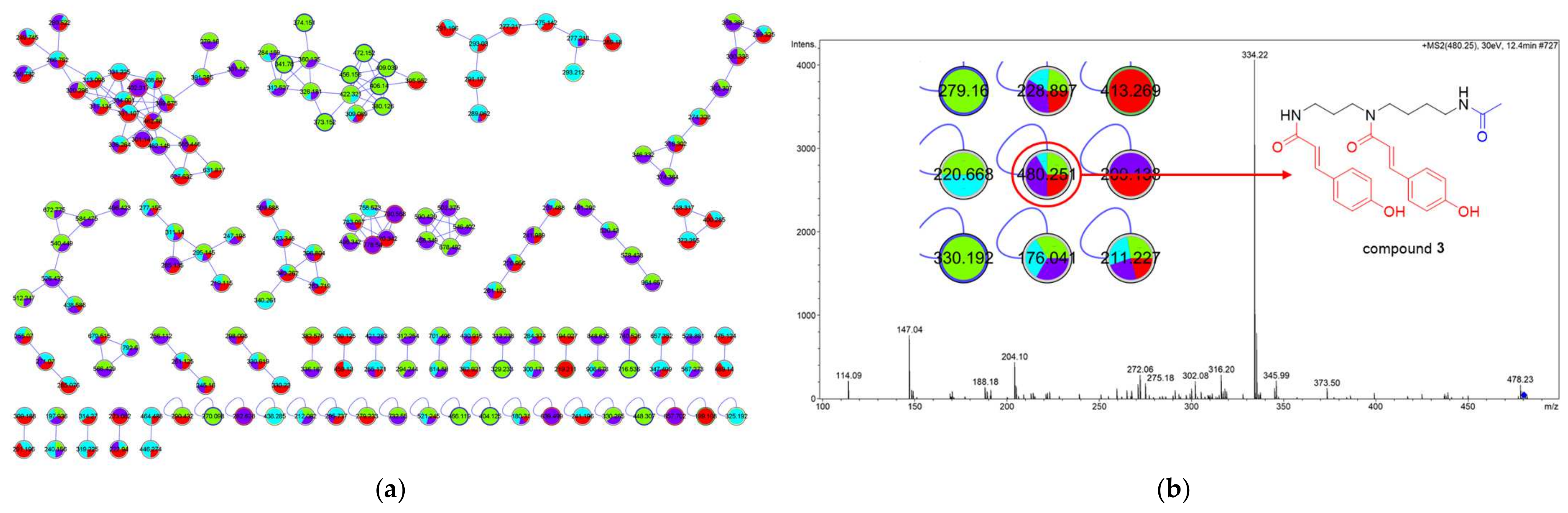
2.1. Analysis of the Molecular Networking
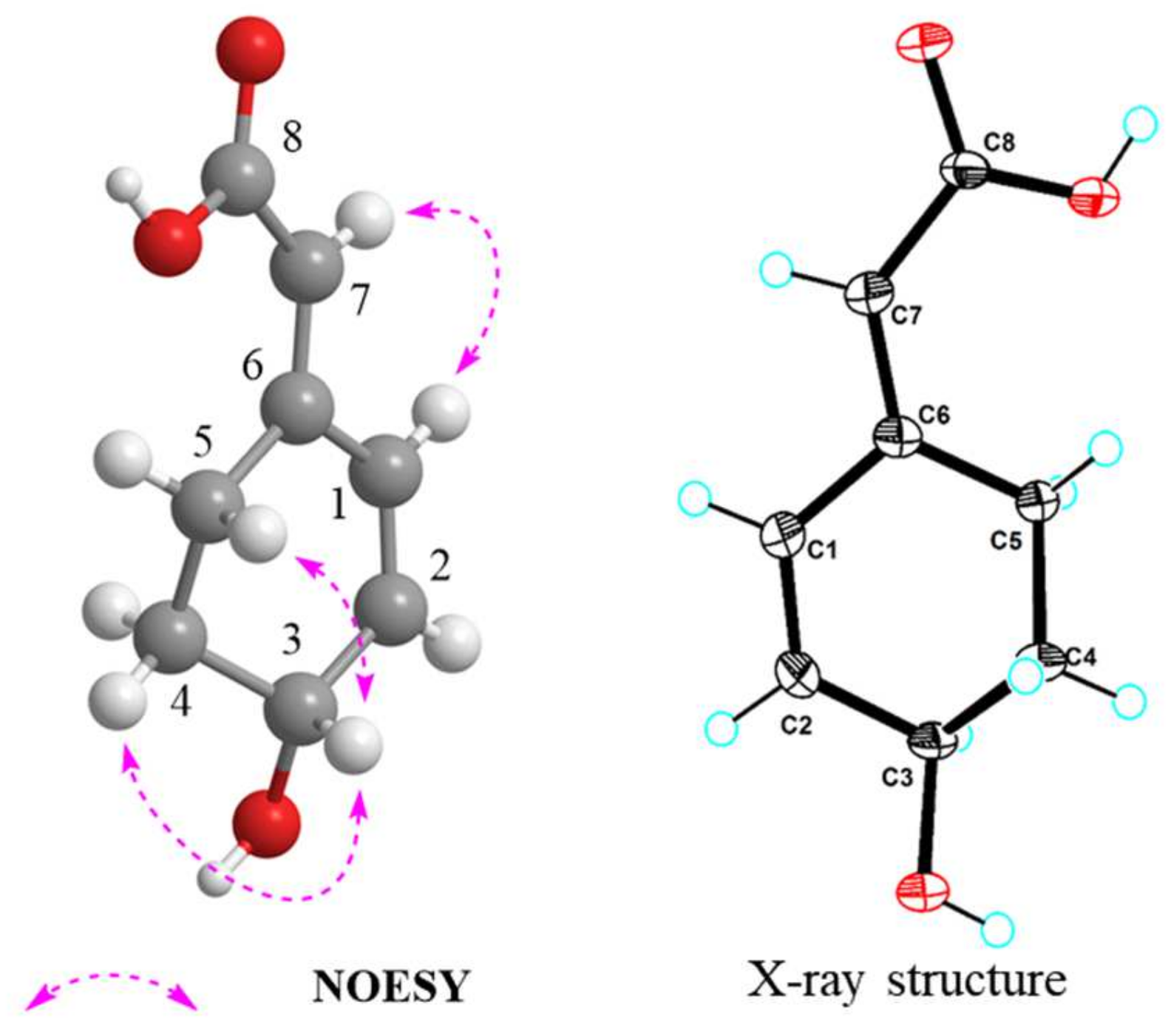
2.2. Structural Elucidation
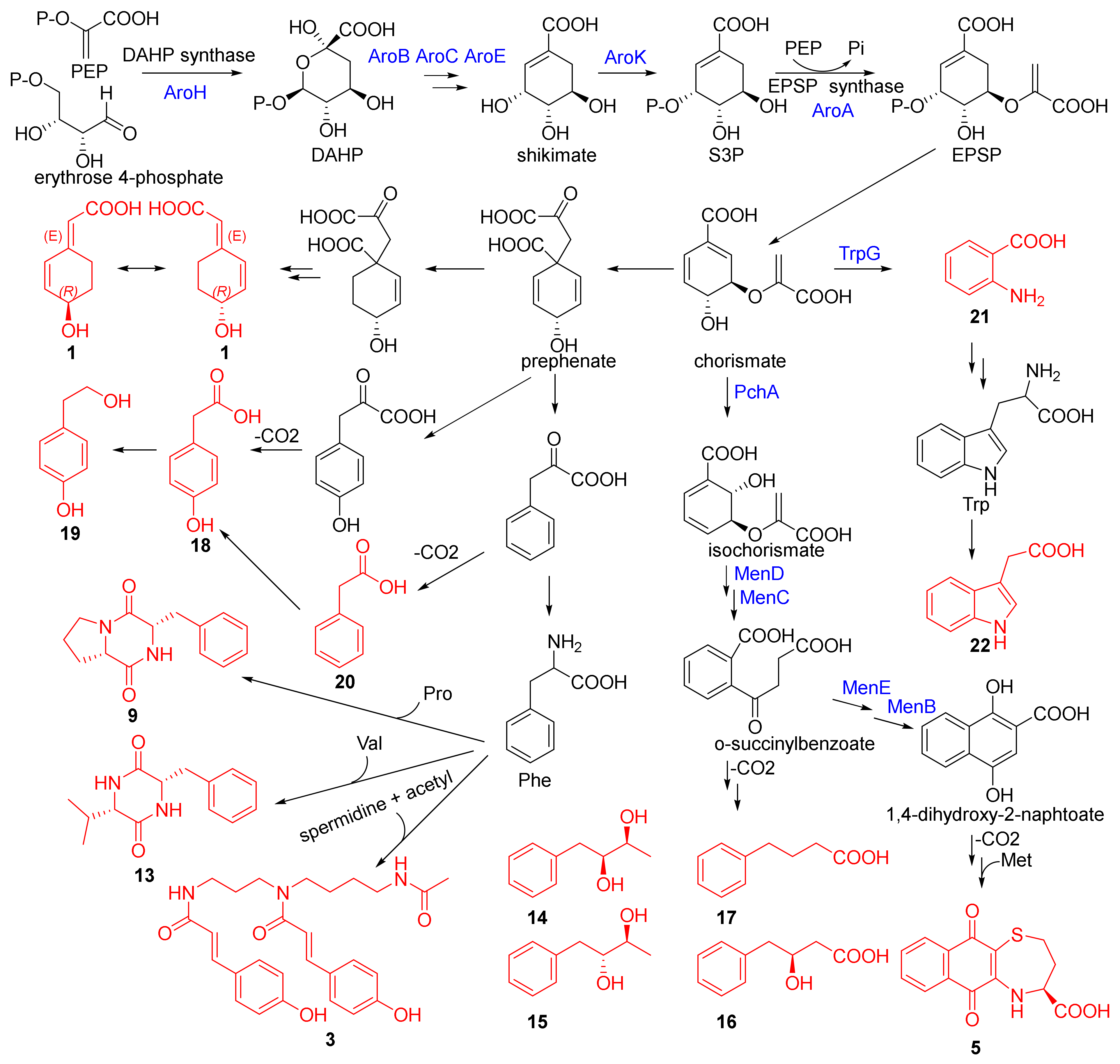
2.3. Putative Biosynthetic Pathway
2.4. Biological Activities
3. Materials and Methods
3.1. General
3.2. Microorganism and Growth Conditions
3.3. Complete Genome Sequence and Bioinformatic Analysis
3.4. Extraction and Isolation
3.5. Molecular Networking
3.6. Marfey’s Analysis
3.7. Chiral HPLC Analysis
3.8. Cytotoxicity Assay
3.9. Antibacterial Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Meng, L.H.; Wang, B.G. Progress in research on bioactive secondary metabolites from deep-sea derived microorganisms. Mar. Drugs 2020, 18, 614. [Google Scholar]
- Yan, L.H.; Li, X.M.; Chi, L.P.; Li, X.; Wang, B.G. Six new antimicrobial metabolites from the deep-sea sediment-derived fungus Aspergillus fumigatus SD-406. Mar. Drugs 2022, 20, 4. [Google Scholar] [CrossRef]
- Limbadri, S.; Luo, X.; Lin, X.; Liao, S.; Wang, J.; Zhou, X.; Yang, B.; Liu, Y. Bioactive novel indole alkaloids and steroids from deep sea-derived fungus Aspergillus fumigatus SCSIO 41012. Molecules 2018, 23, 2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Liu, Z.; Chen, Y.; Tan, H.; Li, S.; Liu, H.; Zhang, W.; Zhu, S. Highly substituted phenol derivatives with nitric oxide inhibitory activities from the deep-sea-derived fungus Trichobotrys effuse FS524. Mar. Drugs 2020, 18, 134. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.S.; Kang, J.S.; Choi, B.K.; Lee, H.S.; Lee, Y.J.; Lee, J.; Shin, H.J. Phenazine derivatives with anti-inflammatory activity from the deep-sea sediment-derived yeast-like fungus Cystobasidium laryngis IV17-028. Mar. Drugs 2019, 17, 482. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.L.; Niu, S.; Xia, J.M.; Peng, K.; Zhang, G.Y.; Yang, X.W. Saccharopolytide A, a new cyclic tetrapeptide with rare 4-hydroxy-proline moieties from the deep-sea derived actinomycete Saccharopolyspora cebuensis MCCC 1A09850. Nat. Prod. Res. 2018, 32, 1627–1631. [Google Scholar] [CrossRef]
- Shin, H.J.; Kim, T.S.; Lee, H.S.; Park, J.Y.; Choi, I.K.; Kwon, H.J. Streptopyrrolidine, an angiogenesis inhibitor from a marine-derived Streptomyces sp. KORDI-3973. Phytochemistry 2008, 69, 2363–2366. [Google Scholar] [CrossRef]
- Niu, S.; Xia, J.M.; Li, Z.; Yang, L.H.; Yi, Z.W.; Xie, C.L.; Peng, G.; Luo, Z.H.; Shao, Z.; Yang, X.W. Aphidicolin chemistry of the deep-sea-derived fungus Botryotinia fuckeliana MCCC 3A00494. J. Nat. Prod. 2019, 82, 2307–2331. [Google Scholar] [CrossRef]
- Chen, S.; Liu, Z.; Tan, H.; Chen, Y.; Zhu, S.; Liu, H.; Zhang, W. Photeroids A and B, unique phenol–sesquiterpene meroterpenoids from the deep-sea-derived fungus Phomopsis tersa. Org. Biomol. Chem. 2020, 18, 642–645. [Google Scholar] [CrossRef]
- Abdel-Mageed, W.M.; Milne, B.F.; Wagner, M.; Schumacher, M.; Sandor, P.; Pathom-aree, W.; Goodfellow, M.; Bull, A.T.; Horikoshi, K.; Ebel, R.; et al. Dermacozines, a new phenazine family from deep-sea dermacocci isolated from a Mariana Trench sediment. Org. Biomol. Chem. 2010, 8, 2352–2362. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Si, L.; Liu, D.; Zhou, A.; Zhang, Z.; Shao, Z.; Wang, S.; Zhang, L.; Zhou, D.; Lin, W. Spiromastilactones: A new class of influenza virus inhibitors from deep-sea fungus. Eur. J. Med. Chem. 2016, 108, 229–244. [Google Scholar] [CrossRef] [PubMed]
- White, R.A.; Grassa, C.J.; Suttle, C.A. First draft genome sequence from a member of the genus Agrococcus, isolated from modern microbialites. Genome Announc. 2013, 1, e00391-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millán-Aguiñaga, N.; Soldatou, S.; Brozio, S.; Munnoch, J.T.; Howe, J.; Hoskisson, P.A.; Duncan, K.R. Awakening ancient polar actinobacteria: Diversity, evolution and specialized metabolite potential. Microbiology 2019, 165, 1169–1180. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A literature review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef] [Green Version]
- Ding, W.; Li, Y.; Chen, M.; Chen, R.; Tian, X.; Yin, H.; Zhang, S. Structures and antitumor activities of ten new and twenty known surfactins from the deep-sea bacterium Limimaricola sp. SCSIO 53532. Bioorganic Chem. 2022, 120, 105589. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, T.; Xing, T.; Ishizaki, T.; Okuda, T.; Oku, N.; Igarashi, Y. Pithohirolide, an antimicrobial tetradepsipeptide from a fungus Pithomyces chartarum. J. Antibiot. 2021, 74, 458–463. [Google Scholar] [CrossRef]
- Werner, C.; Hu, W.; Lorenzi-Riatsch, A.; Hesse, M. Di-coumaroylspermidines and tri-coumaroylspermidines in anthers of different species of the genus Aphelandra. Phytochemistry 1995, 40, 461–465. [Google Scholar] [CrossRef]
- Sobolev, V.S.; Sy, A.A.; Gloer, J.B. Spermidine and flavonoid conjugates from peanut (Arachis hypogaea) flowers. J. Agric. Food Chem. 2008, 56, 2960–2969. [Google Scholar] [CrossRef] [PubMed]
- Al-Busafi, S.; Doncaster, J.R.; Drew, M.G.; Regan, A.C.; Whitehead, R.C. Exploitation of chemical predisposition in synthesis: An approach to the manzamenones. J. Chem. Soc. Perkin Trans. 2002, 476–484. [Google Scholar] [CrossRef]
- Arahiko, E.; Yoshihisa, S.; Takashi, O.; Yasunori, Y.; Tadashi, H.; Osamu, T. Amino Acid Naphthoquinone Derivatives. Japan Patent Application No. JPS5284220A, 13 July 1977. [Google Scholar]
- Mehnaz, S.; Saleem, R.S.Z.; Yameen, B.; Pianet, I.; Schnakenburg, G.; Pietraszkiewicz, H.; Valeriote, F.; Josten, M.; Sahl, H.G.; Franzblau, S.G.; et al. Lahorenoic acids A–C, ortho-dialkyl-substituted aromatic acids from the biocontrol strain Pseudomonas aurantiaca PB-St2. J. Nat. Prod. 2013, 76, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Xiang, W.X.; Liu, Q.; Li, X.M.; Lu, C.H.; Shen, Y.M. Four pairs of proline-containing cyclic dipeptides from Nocardiopsis sp. HT88, an endophytic bacterium of Mallotus nudiflorus L. Nat. Prod. Res. 2020, 34, 2219–2224. [Google Scholar] [CrossRef]
- Young, P.E.; Madison, V.; Blout, E.R. Cyclic peptides. 15. Lanthanide-assisted 13C and 1H NMR analysis of preferred side-chain rotamers in proline-containing cyclic dipeptides. J. Am. Chem. Soc. 1976, 98, 5365–5371. [Google Scholar] [CrossRef]
- Adamczeski, M.; Reed, A.R.; Crews, P. New and known diketopiperazines from the caribbean sponge, Calyx cf. podatypa. J. Nat. Prod. 1995, 58, 201–208. [Google Scholar] [CrossRef]
- Han, B.; Li, W.; Cui, C. Cyclic dipeptides as new cell cycle inhibitors produced by Streptomyces flavoretus 18522. J. Shenyang Pharm. Univ. 2015, 32, 107–110. [Google Scholar]
- Park, A.R.; Jeong, S.I.; Jeon, H.W.; Kim, J.; Kim, N.; Ha, M.T.; Mannaa, M.; Kim, J.; Lee, C.W.; Min, B.S.; et al. A diketopiperazine, cyclo-(L-Pro-L-Ile), derived from Bacillus thuringiensis JCK-1233 controls pine wilt disease by elicitation of moderate hypersensitive reaction. Front. Plant Sci. 2020, 11, 1023. [Google Scholar] [CrossRef]
- Awano, K.I.; Yanai, T.; Watanabe, I.; Takagi, Y.; Kitahara, T.; Mori, K. Synthesis of all four possible stereoisomers of 1-phenyl-2,3-butanediol and both enantiomers of 3-hydroxy-4-phenyl-2-butanone to determine the absolute configuration of the natural constituents. Biosci. Biotechnol. Biochem. 1995, 59, 1251–1254. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.P.; Wang, Y.; Liu, P.P.; Hong, K.; Chen, H.; Yin, X.; Zhu, W.M. Aromatic compounds from the halotolerant fungal strain of Wallemia sebi PXP-89 in a hypersaline medium. Arch. Pharmacal Res. 2011, 34, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Goswami, A.; Mirfakhrae, K.D.; Patel, R.N. Asymmetric acyloin condensation catalyzed by phenylpyruvate decarboxylase. Tetrahedron Asymmetry 1999, 10, 4667–4675. [Google Scholar] [CrossRef]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Kornberg, H.L. The role and control of the glyoxylate cycle in Escherichia coli. Biochem. J. 1966, 99, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, K.M.; Weaver, L.M. The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 473–503. [Google Scholar] [CrossRef]
- Lemke, R.A.S.; Peterson, A.C.; Ziegelhoffer, E.C.; Westphall, M.S.; Tjellström, H.; Coon, J.J.; Donohue, T.J. Synthesis and scavenging role of furan fatty acids. Proc. Natl. Acad. Sci. USA 2014, 111, E3450–E3457. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Müller, M.; Hertweck, C. Formation of the aureothin tetrahydrofuran ring by a bifunctional cytochrome P450 monooxygenase. J. Am. Chem. Soc. 2004, 126, 16742–16743. [Google Scholar] [CrossRef]
- Gaikwad, N.W.; Madyastha, K. Biosynthesis of β-substituted furan skeleton in the lower furanoterpenoids: A model study. Biochem. Biophys. Res. Commun. 2002, 290, 589–594. [Google Scholar] [CrossRef] [Green Version]
- Funa, N.; Ohnishi, Y.; Ebizuka, Y.; Horinouchi, S. Properties and substrate specificity of RppA, a chalcone synthase-related polyketide synthase in Streptomyces griseus. J. Biol. Chem. 2002, 277, 4628–4635. [Google Scholar] [CrossRef] [Green Version]
- Meganathan, R. Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q): A perspective on enzymatic mechanisms. In Vitamins & Hormones; Academic Press: Cambridge, MA, USA, 2001; Volume 61, pp. 173–218. [Google Scholar]
- Balachandran, N.; Heimhalt, M.; Liuni, P.; To, F.; Wilson, D.J.; Junop, M.S.; Berti, P.J. Potent inhibition of 3-deoxy-d-arabinoheptulosonate-7-phosphate (DAHP) synthase by DAHP oxime, a phosphate group mimic. Biochemistry 2016, 55, 6617–6629. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Hilsenbeck, J.L.; Kim, H.J.; Shuttleworth, W.A.; Park, Y.H.; Evans, J.N.; Kang, C. Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state. Mol. Microbiol. 2004, 51, 963–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widhalm, J.R.; Rhodes, D. Biosynthesis and molecular actions of specialized 1, 4-naphthoquinone natural products produced by horticultural plants. Hortic. Res. 2016, 3, 16046. [Google Scholar] [CrossRef]
- Gaille, C.; Reimmann, C.; Haas, D. Isochorismate synthase (PchA), the first and rate-limiting enzyme in salicylate biosynthesis of Pseudomonas aeruginosa. J. Biol. Chem. 2003, 278, 16893–16898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, I.P.; Eberly, L. Structure and regulation of the anthranilate synthase genes in Pseudomonas aeruginosa: I. Sequence of trpG encoding the glutamine amidotransferase subunit. Mol. Biol. Evol. 1986, 3, 436–448. [Google Scholar]
- Mishra, A.K.; Choi, J.; Choi, S.J.; Baek, K.H. Cyclodipeptides: An overview of their biosynthesis and biological activity. Molecules 2017, 22, 1796. [Google Scholar] [CrossRef] [Green Version]
- Sauguet, L.; Moutiez, M.; Li, Y.; Belin, P.; Seguin, J.; Le Du, M.H.; Thai, R.; Masson, C.; Fonvielle, M.; Pernodet, J.L.; et al. Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis. Nucleic Acids Res. 2011, 39, 4475–4489. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, K.; Wavdhane, M.; Haque, S.; Govender, T.; Kruger, H.G.; Mishra, M.K.; Chandra, R.; Tiwari, D. A sensitive WST-8-based bioassay for PEGylated granulocyte colony stimulating factor using the NFS-60 cell line. Pharm. Biol. 2015, 53, 849–854. [Google Scholar] [CrossRef]
- Harathi, K.; Giribabu, D.; Naidu, C.V. Phytochemical evaluation and in vitro antibacterial activity of Sphaeranthus indicus (L.)—An important antijaundice medicinal plant. Am. J. Plant Sci. 2017, 08, 1011–1021. [Google Scholar] [CrossRef] [Green Version]
- Jensen, S.; Ragnarsdottir, O.; Johannsson, R. Marine sources of furan fatty acids. J. Aquat. Food Prod. Technol. 2019, 28, 74–83. [Google Scholar] [CrossRef]




| 1 | 4 | ||||
|---|---|---|---|---|---|
| Position | δH (J in Hz) | δC | Position | δH (J in Hz) | δC |
| 1 | 6.16 (dd, 9.9, 1.5) | 131.7, d | 1 | 173.6, s | |
| 2 | 6.13 (dd, 9.9, 2.5) | 141.2, d | 2 | 3.67 (s) | 33.8, t |
| 3 | 4.31 (ddd, 6.9, 4.9, 2.5) | 67.0, d | 3 | 145.0, s | |
| 4 | 1.55 (tdd, 12.6, 8.8, 4.1), 2.03 (m) | 32.5, t | 4 | 6.12 (d, 3.1) | 108.8, d |
| 5 | 2.54 (dddd, 16.9, 12.1, 4.3, 2.4), 3.41 (dt, 16.9, 4.9) | 24.9, t | 5 | 5.91 (d, 3.1) | 105.5, d |
| 6 | 153.6, s | 6 | 156.5, s | ||
| 7 | 5.66 (s) | 118.0, d | 7 | 2.58 (t, 7.7) | 28.0, t |
| 8 | 170.5, s | 8 | 1.61 (m) | 27.9, t | |
| 9 | 1.33 (m) | 28.8, t | |||
| 10 | 1.29 (overlapped) | 31.6, t | |||
| 11 | 1.29 (overlapped) | 22.6, t | |||
| 12 | 0.88 (t, 6.8) | 14.1, q | |||
| 2 | 3 b | |||||
|---|---|---|---|---|---|---|
| Moiety | Position | δH (J in Hz) | δCa | Position | δH (J in Hz) | δC |
| Hmv | CO | 177.9, s | 2 | 3.32 (m)/3.36 (m) | 37.9/38.2, t | |
| α-C | 4.10 (dd, 9.8, 3.5) | 71.5, d | 3 | 1.85 (m)/1.93 (overlapped) | 28.9/30.7, t | |
| β-C | 1.50 (ddd, 13.1, 9.8, 4.7), 1.56 (ddd, 13.1, 9.3, 3.5) | 45.0, t | 4 | 3.52 (m)/3.56 (m) | 45.7/47.0, t | |
| γ-C | 1.87 (m) | 26.0, d | 6 | 3.46 (m)/3.54 (m) | 47.6/49.0, t | |
| δ1-C | 0.95 (overlapped) | 23.6/24.0, q | 7 | 1.63 (m)/1.68 (m) | 26.3/28.0, t | |
| δ2-C | 0.95 (overlapped) | 21.8, q | 8 | 1.51 (m)/1.55 (m) | 27.7/27.8, t | |
| Val | CO | 172.3, s | 9 | 3.19 (m)/3.21 (m) | 39.9/40.2, t | |
| α-C | 4.46 (d, 6.1) | 58.9, d | 1′ or 1″ | 169.2/169.3/169.4/169.5, s | ||
| β-C | 2.25 (m) | 32.0, d | 2′ or 2″ | 6.42 (d, 15.7)/6.81 (overlapped)/ 6.85 (d, 15.3) | 114.9/115.0/118.2/118.5, d | |
| γ1-C | 0.98 (d, 6.9) | 19.7, q | 3′ or 3″ | 7.45 (overlapped)/7.48 (overlapped)/ 7.51 (d, 15.3)/7.54 (d, 15.3) | 141.9/142.2/144.4/144.5, d | |
| γ2-C | 0.97 (d, 7.0) | 18.4, q | 4′ or 4″ | 127.6/127.7/127.9/128.0, s | ||
| Hiv | CO | 171.4, s | 5′/9′ or 5″/9″ | 7.40 (d, 8.7)/7.41 (d, 8.7)/ 7.44 (d, 8.6)/7.48(d, 8.7) | 130.6/130.7/130.9, d | |
| α-C | 4.86 (overlapped) | 80.1, d | 6′/8′ or 6″/8″ | 6.72 (d, 8.6)/6.79 (d, 8.7)/ 6.80 (overlapped) | 116.8, d | |
| β-C | 2.22 (m) | 31.8, d | 7′ or 7″ | 160.6/160.7/160.8, s | ||
| γ1-C | 1.01 (d, 6.9) | 19.3, q | 1‴ | 173.3/173.4, s | ||
| γ2-C | 0.99 (d, 6.8) | 17.8, q | 2‴ | 1.89 (s)/1.92 (s) | 22.6, q | |
| Leu | CO | 176.4, s | ||||
| α-C | 4.43 (dd, 10.3, 4.4) | 52.5, d | ||||
| β-C | 1.63 (ddd, 13.8, 9.3, 4.5), 1.69 (m) | 41.9, t | ||||
| γ-C | 1.74 (m) | 25.6, d | ||||
| δ1-C | 0.96 (d, 6.4) | 23.6/24.0, q | ||||
| δ2-C | 0.91 (d, 6.5) | 21.8, q | ||||
| Sample | tRLa | tRDb | tRc | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Val | Leu | Hiv | Hmv | Val | Leu | Hiv | Hmv | l-FDAA | |
| Marfey’s derivatives | 21.603 | 32.395 | 31.698 | 41.621 | 15.575 | ||||
| Hydrolysate | 18.122 | 41.889 | 11.14 | 32.568 | |||||
| Compound 2 | 32.355 | 17.651 | 31.677 | 31.811 | 15.607 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, W.; Li, Y.; Tian, X.; Chen, M.; Xiao, Z.; Chen, R.; Yin, H.; Zhang, S. Investigation on Metabolites in Structural Diversity from the Deep-Sea Sediment-Derived Bacterium Agrococcus sp. SCSIO 52902 and Their Biosynthesis. Mar. Drugs 2022, 20, 431. https://doi.org/10.3390/md20070431
Ding W, Li Y, Tian X, Chen M, Xiao Z, Chen R, Yin H, Zhang S. Investigation on Metabolites in Structural Diversity from the Deep-Sea Sediment-Derived Bacterium Agrococcus sp. SCSIO 52902 and Their Biosynthesis. Marine Drugs. 2022; 20(7):431. https://doi.org/10.3390/md20070431
Chicago/Turabian StyleDing, Wenping, Yanqun Li, Xinpeng Tian, Min Chen, Zhihui Xiao, Rouwen Chen, Hao Yin, and Si Zhang. 2022. "Investigation on Metabolites in Structural Diversity from the Deep-Sea Sediment-Derived Bacterium Agrococcus sp. SCSIO 52902 and Their Biosynthesis" Marine Drugs 20, no. 7: 431. https://doi.org/10.3390/md20070431
APA StyleDing, W., Li, Y., Tian, X., Chen, M., Xiao, Z., Chen, R., Yin, H., & Zhang, S. (2022). Investigation on Metabolites in Structural Diversity from the Deep-Sea Sediment-Derived Bacterium Agrococcus sp. SCSIO 52902 and Their Biosynthesis. Marine Drugs, 20(7), 431. https://doi.org/10.3390/md20070431






