Diversity of Bacterial Secondary Metabolite Biosynthetic Gene Clusters in Three Vietnamese Sponges
Abstract
:1. Introduction
2. Results
2.1. Metagenomes of the Sponge-Associated Bacteria
2.2. Diversity of Secondary Metabolite Biosynthetic Gene Clusters from the Metagenomes of the Sponge-Associated Bacteria
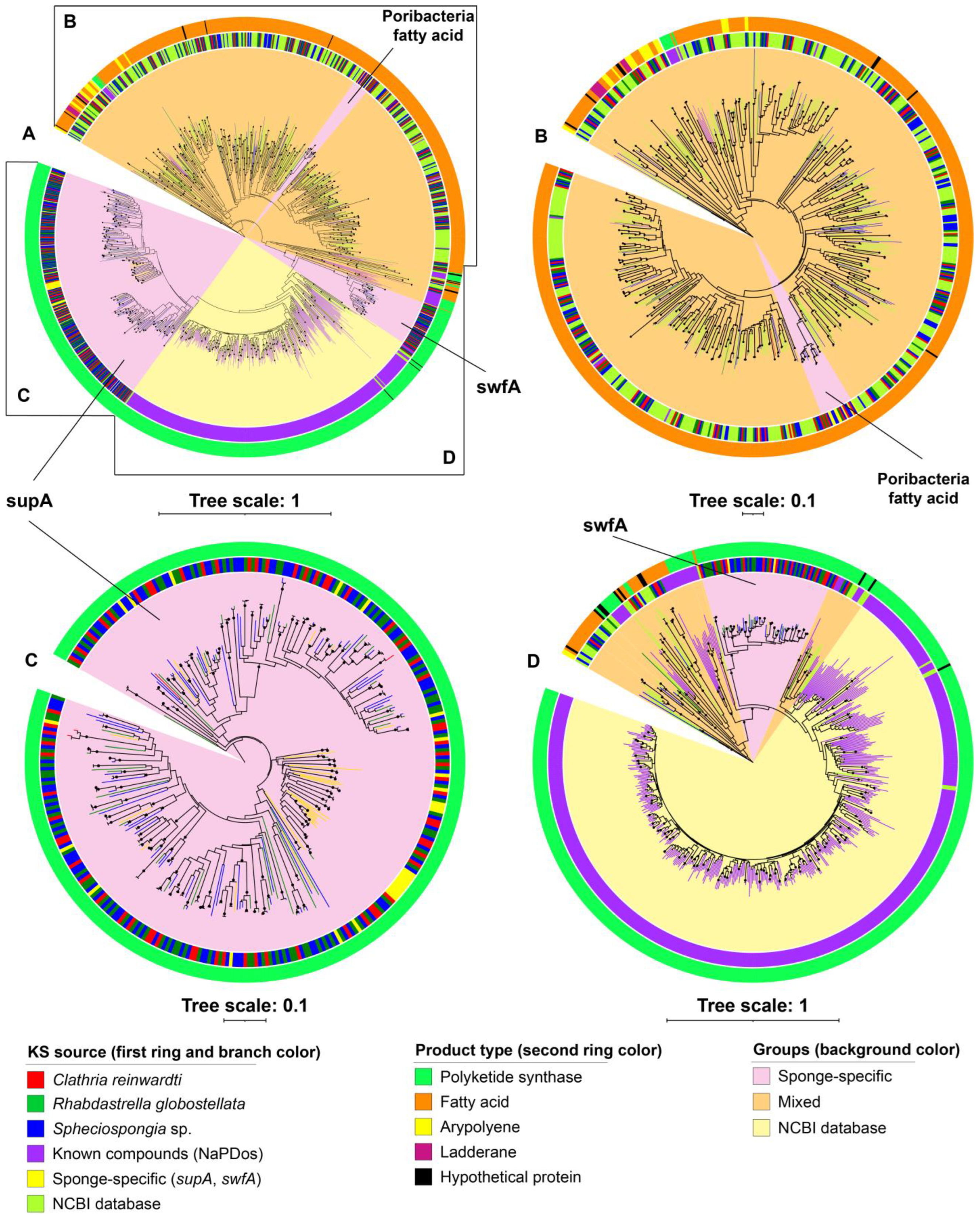
2.3. Ketosynthase Phylogeny of BGCs
3. Discussion
3.1. Diversity of Secondary Metabolite Biosynthetic Gene Clusters from the Metagenomes of Sponge-Associated Bacteria
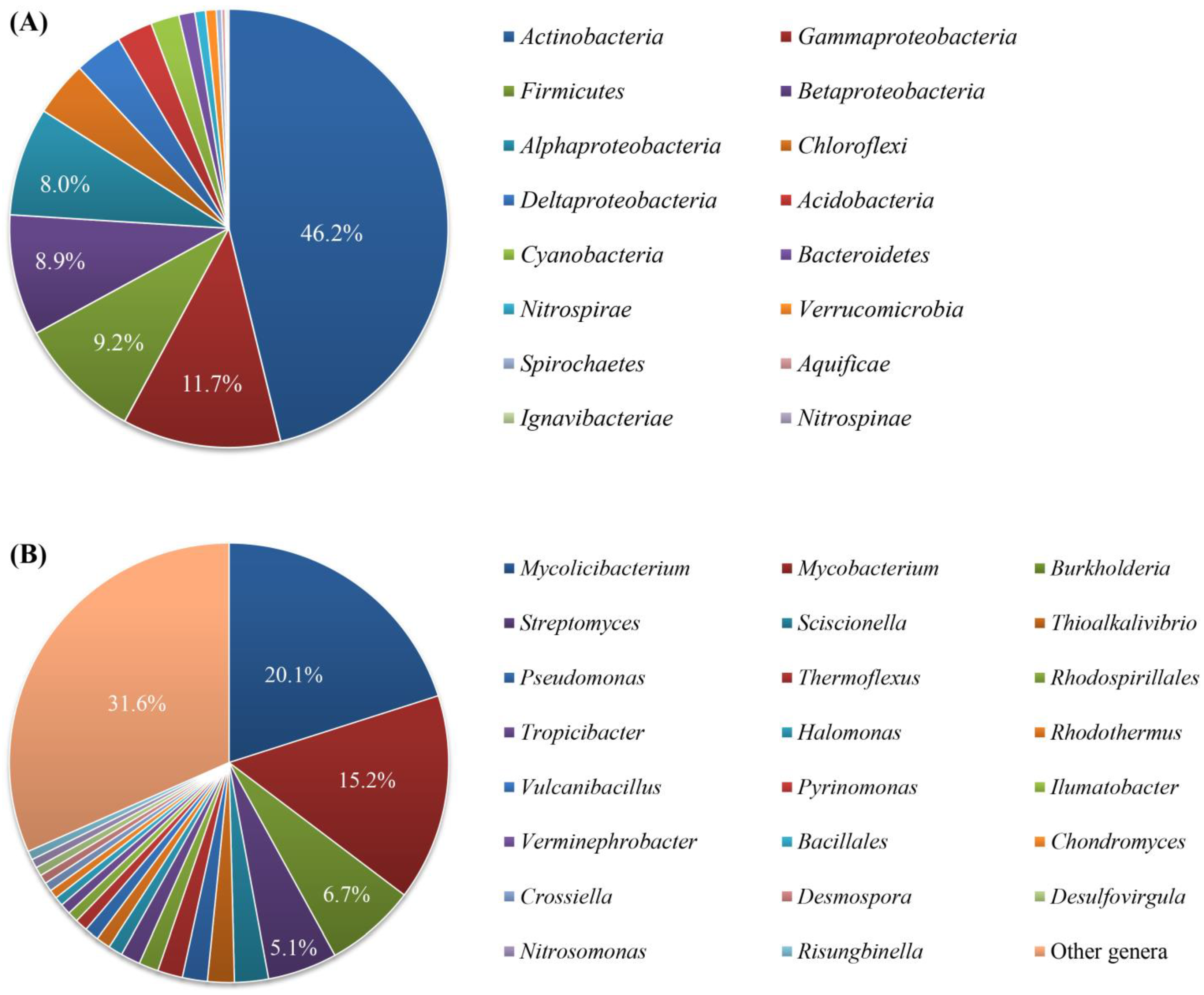
3.2. Predicted Taxa of the Secondary Metabolite Biosynthetic Gene Clusters
3.3. Putative ‘Sponge-Specific’ Clusters
4. Materials and Methods
4.1. Collection and Identification of Sponges
4.2. Enrichment of Sponge-Associated Bacteria
4.3. Metagenomic DNA Extraction, Sequencing and Assembly
4.4. Prediction of Secondary Metabolite Gene Biosynthetic Clusters and Ketosynthase Phylogeny
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, M.S.; Robertson, A.A.B.; Cooper, M.A. Natural product and natural product derived drugs in clinical trials. Nat. Prod. Rep. 2014, 31, 1612–1661. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Kumar, M.; Mittal, A.; Mehta, P.K. Microbial metabolites in nutrition, healthcare and agriculture. 3 Biotech 2017, 7, 15. [Google Scholar] [CrossRef] [Green Version]
- Smanski, M.J.; Zhou, H.; Claesen, J.; Shen, B.; Fischbach, M.A.; Voigt, C.A. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2016, 14, 135–149. [Google Scholar] [CrossRef] [Green Version]
- Osbourn, A. Secondary metabolic gene clusters: Evolutionary toolkits for chemical innovation. Trends Genet. 2010, 26, 449–457. [Google Scholar] [CrossRef]
- Weissman, K.J. The structural biology of biosynthetic megaenzymes. Nat. Chem. Biol. 2015, 11, 660–670. [Google Scholar] [CrossRef]
- Sánchez, C.; Méndez, C.; Salas, J.A. Indolocarbazole natural products: Occurrence, biosynthesis, and biological activity. Nat. Prod. Rep. 2006, 23, 1007–1045. [Google Scholar] [CrossRef]
- Robbins, T.; Liu, Y.-C.; Cane, D.E.; Khosla, C. Structure and mechanism of assembly line polyketide synthases. Curr. Opin. Struct. Biol. 2016, 41, 10–18. [Google Scholar] [CrossRef] [Green Version]
- Miller, B.R.; Gulick, A.M. Structural Biology of Nonribosomal Peptide Synthetases. In Nonribosomal Peptide and Polyketide Biosynthesis; Methods in Molecular Biology; Evans, B., Ed.; Humana Press: New York, NY, USA, 2016; Volume 1401, pp. 3–29. [Google Scholar] [CrossRef]
- Komaki, H.; Ichikawa, N.; Hosoyama, A.; Takahashi-Nakaguchi, A.; Matsuzawa, T.; Suzuki, K.-I.; Fujita, N.; Gonoi, T. Genome based analysis of type-I polyketide synthase and nonribosomal peptide synthetase gene clusters in seven strains of five representative Nocardia species. BMC Genom. 2014, 15, 323. [Google Scholar] [CrossRef] [Green Version]
- Masschelein, J.; Mattheus, W.; Gao, L.-J.; Moons, P.; Van Houdt, R.; Uytterhoeven, B.; Lamberigts, C.; Lescrinier, E.; Rozenski, J.; Herdewijn, P.; et al. A PKS/NRPS/FAS Hybrid Gene Cluster from Serratia plymuthica RVH1 Encoding the Biosynthesis of Three Broad Spectrum, Zeamine-Related Antibiotics. PLoS ONE 2013, 8, e54143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rischer, M.; Raguž, L.; Guo, H.; Keiff, F.; Diekert, G.; Goris, T.; Beemelmanns, C. Biosynthesis, Synthesis, and Activities of Barnesin A, a NRPS-PKS Hybrid Produced by an Anaerobic Epsilonproteobacterium. ACS Chem. Biol. 2018, 13, 1990–1995. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, C.M.; Kimes, N.E.; López-Pérez, M.; Ausó, E.; Rodríguez-Valera, F.; Ghai, R. A Hybrid NRPS-PKS Gene Cluster Related to the Bleomycin Family of Antitumor Antibiotics in Alteromonas macleodii Strains. PLoS ONE 2013, 8, e76021. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Igarashi, Y.; Tamura, T. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters among taxonomically close Streptomyces strains. Sci. Rep. 2018, 8, 6888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rego, A.; Fernandez-Guerra, A.; Duarte, P.; Assmy, P.; Leão, P.N.; Magalhães, C. Secondary metabolite biosynthetic diversity in Arctic Ocean metagenomes. Microb. Genom. 2021, 7, 000731. [Google Scholar] [CrossRef] [PubMed]
- Storey, M.A.; Andreassend, S.K.; Bracegirdle, J.; Brown, A.; Keyzers, R.A.; Ackerley, D.F.; Northcote, P.T.; Owen, J.G. Metagenomic Exploration of the Marine Sponge Mycale hentscheli Uncovers Multiple Polyketide-Producing Bacterial Symbionts. MBio 2020, 11, e02997-19. [Google Scholar] [CrossRef] [Green Version]
- Van Goethem, M.W.; Osborn, A.R.; Bowen, B.P.; Andeer, P.F.; Swenson, T.L.; Clum, A.; Riley, R.; He, G.; Koriabine, M.; Sandor, L.; et al. Long-read metagenomics of soil communities reveals phylum-specific secondary metabolite dynamics. Commun. Biol. 2021, 4, 1302. [Google Scholar] [CrossRef]
- Nguyen, N.A.; Lin, Z.; Mohanty, I.; Garg, N.; Schmidt, E.W.; Agarwal, V. An Obligate Peptidyl Brominase Underlies the Discovery of Highly Distributed Biosynthetic Gene Clusters in Marine Sponge Microbiomes. J. Am. Chem. Soc. 2021, 143, 10221–10231. [Google Scholar] [CrossRef]
- Calcabrini, C.; Catanzaro, E.; Bishayee, A.; Turrini, E.; Fimognari, C. Marine Sponge Natural Products with Anticancer Potential: An Updated Review. Mar. Drugs 2017, 15, 310. [Google Scholar] [CrossRef]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine Sponge Derived Natural Products between 2001 and 2010: Trends and Opportunities for Discovery of Bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [Green Version]
- Mehbub, M.F.; Perkins, M.V.; Zhang, W.; Franco, C.M. New marine natural products from sponges (Porifera) of the order Dictyoceratida (2001 to 2012); a promising source for drug discovery, exploration and future prospects. Biotechnol. Adv. 2016, 34, 473–491. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Hill, R.T. New drugs from marine microbes: The tide is turning. J. Ind. Microbiol. Biotechnol. 2006, 33, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.W.; Bewley, A.C.A.; Faulkner, D.J. Theopalauamide, a Bicyclic Glycopeptide from Filamentous Bacterial Symbionts of the Lithistid Sponge Theonella swinhoei from Palau and Mozambique. J. Org. Chem. 1998, 63, 1254–1258. [Google Scholar] [CrossRef]
- Wilson, M.C.; Mori, T.; Rückert, C.; Uria, A.R.; Helf, M.J.; Takada, K.; Gernert, C.; Steffens, U.A.E.; Heycke, N.; Schmitt, S.; et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014, 506, 58–62. [Google Scholar] [CrossRef] [Green Version]
- Wakimoto, T.; Egami, Y.; Nakashima, Y.; Wakimoto, Y.; Mori, T.; Awakawa, T.; Ito, T.; Kenmoku, H.; Asakawa, Y.; Piel, J.; et al. Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont. Nat. Chem. Biol. 2014, 10, 648–655. [Google Scholar] [CrossRef]
- Ueoka, R.; Uria, A.R.; Reiter, S.; Mori, T.; Karbaum, P.; Peters, E.E.; Helfrich, E.J.N.; Morinaka, B.; Gugger, M.; Takeyama, H.; et al. Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms. Nat. Chem. Biol. 2015, 11, 705–712. [Google Scholar] [CrossRef]
- Tianero, M.D.; Balaich, J.N.; Donia, M.S. Localized production of defence chemicals by intracellular symbionts of Haliclona sponges. Nat. Microbiol. 2019, 4, 1149–1159. [Google Scholar] [CrossRef]
- Lackner, G.; Peters, E.E.; Helfrich, E.J.N.; Piel, J. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc. Natl. Acad. Sci. USA 2017, 114, E347–E356. [Google Scholar] [CrossRef] [Green Version]
- Freeman, M.F.; Vagstad, A.L.; Piel, J. Polytheonamide biosynthesis showcasing the metabolic potential of sponge-associated uncultivated ‘Entotheonella’ bacteria. Curr. Opin. Chem. Biol. 2016, 31, 8–14. [Google Scholar] [CrossRef]
- Mori, T.; Cahn, J.K.B.; Wilson, M.C.; Meoded, R.A.; Wiebach, V.; Martinez, A.F.C.; Helfrich, E.J.N.; Albersmeier, A.; Wibberg, D.; Dätwyler, S.; et al. Single-bacterial genomics validates rich and varied specialized metabolism of uncultivated Entotheonella sponge symbionts. Proc. Natl. Acad. Sci. USA 2018, 115, 1718–1723. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, Y.; Egami, Y.; Kimura, M.; Wakimoto, T.; Abe, I. Metagenomic Analysis of the Sponge Discodermia Reveals the Production of the Cyanobacterial Natural Product Kasumigamide by ‘Entotheonella’. PLoS ONE 2016, 11, e0164468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loureiro, C.; Galani, A.; Gavriilidou, A.; Chaib de Mares, M.; van der Oost, J.; Medema Marnix, H.; Sipkema, D. Comparative Metagenomic Analysis of Biosynthetic Diversity across Sponge Microbiomes Highlights Metabolic Novelty, Conservation, and Diversification. MSystems 2022, 7, e00357-22. [Google Scholar] [CrossRef] [PubMed]
- Balskus, E.P. Sponge symbionts play defense. Nat. Chem. Biol. 2014, 10, 611–612. [Google Scholar] [CrossRef] [PubMed]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Aguila-Ramírez, R.N.; Hernández-Guerrero, C.J.; González-Acosta, B.; Id-Daoud, G.; Hewitt, S.; Pope, J.; Hellio, C. Antifouling activity of symbiotic bacteria from sponge Aplysina gerardogreeni. Int. Biodeterior. Biodegrad. 2014, 90, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Fuerst, J.A. Diversity and biotechnological potential of microorganisms associated with marine sponges. Appl. Microbiol. Biotechnol. 2014, 98, 7331–7347. [Google Scholar] [CrossRef]
- Juliana, F.S.-G.; Marcia, G.-d.; Walter, M.R.O.; Marinella, S.L. Biotechnological Potential of Sponge-Associated Bacteria. Curr. Pharm. Biotechnol. 2014, 15, 143–155. [Google Scholar]
- Indraningrat, A.A.G.; Smidt, H.; Sipkema, D. Bioprospecting Sponge-Associated Microbes for Antimicrobial Compounds. Mar. Drugs 2016, 14, 87. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Z.; Wang, H. Cytotoxic Natural Products from Marine Sponge-Derived Microorganisms. Mar. Drugs 2017, 15, 68. [Google Scholar] [CrossRef] [Green Version]
- Dat, T.T.H.; Steinert, G.; Cuc, N.T.K.; Smidt, H.; Sipkema, D. Bacteria Cultivated from Sponges and Bacteria Not Yet Cul-tivated from Sponges—A Review. Front. Microbiol. 2021, 12, 3427. [Google Scholar] [CrossRef] [PubMed]
- Dat, T.T.H.; Steinert, G.; Thi Kim Cuc, N.; Smidt, H.; Sipkema, D. Archaeal and bacterial diversity and community compo-sition from 18 phylogenetically divergent sponge species in Vietnam. PeerJ 2018, 6, e4970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goud, T.V.; Krishnaiah, P.; Reddy, S.M.; Srinivasulu, M.; Rao, M.R.; Venkateswarlu, Y. Chemical investigation of the marine sponges Clathria reinwardti and Haliclona cribricutis. Indian J. Chem. Sect. B Org. Chem. Incl. Med. Chem. 2005, 44, 607–610. [Google Scholar]
- Tai, B.H.; Hang, D.T.; Trang, D.T.; Yen, P.H.; Huong, P.T.T.; Nhiem, N.X.; Thung, D.C.; Thao, D.T.; Hoai, N.T.; Kiem, P.V. Conjugated Polyene Ketones from the Marine Sponge Clathria (Thalysias) Reinwardti (Vosmaer, 1880) and Their Cytotoxic Activity. Nat. Prod. Commun. 2021, 16, 1934578X211043732. [Google Scholar] [CrossRef]
- Trang, D.T.; Hang, D.T.T.; Dung, D.T.; Bang, N.A.; Huong, P.T.T.; Yen, P.H.; Tai, B.H.; Cuc, N.T.; An, D.H.; Nhiem, N.X.; et al. Chemical constituents of Clathria reinwardti. Vietnam J. Chem. 2022, 60, 21–26. [Google Scholar] [CrossRef]
- Tasdemir, D.; Mangalindan, G.C.; Concepción, G.P.; Verbitski, S.M.; Rabindran, S.; Miranda, M.; Greenstein, M.; Hooper, J.N.A.; Harper, M.K.; Ireland, C.M. Bioactive Isomalabaricane Triterpenes from the Marine Sponge Rhabdastrella globostellata. J. Nat. Prod. 2002, 65, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Clement, J.A.; Li, M.; Hecht, S.M.; Kingston, D.G.I. Bioactive Isomalabaricane Triterpenoids from Rhabdastrella globostellata that Stabilize the Binding of DNA Polymerase β to DNA. J. Nat. Prod. 2006, 69, 373–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouad, M.; Edrada, R.A.; Ebel, R.; Wray, V.; Müller, W.E.G.; Lin, W.H.; Proksch, P. Cytotoxic Isomalabaricane Triterpenes from the Marine Sponge Rhabdastrella globostellata. J. Nat. Prod. 2006, 69, 211–218. [Google Scholar] [CrossRef]
- Hirashima, M.; Tsuda, K.; Hamada, T.; Okamura, H.; Furukawa, T.; Akiyama, S.-i.; Tajitsu, Y.; Ikeda, R.; Komatsu, M.; Doe, M.; et al. Cytotoxic Isomalabaricane Derivatives and a Monocyclic Triterpene Glycoside from the Sponge Rhabdastrella globostellata. J. Nat. Prod. 2010, 73, 1512–1518. [Google Scholar] [CrossRef]
- Trang, D.T.; Dung, D.T.; Hang, D.T.T.; Dung, N.V.; Linh, T.M.; Mai, N.C.; Yen, P.H.; An, D.H.; Nhiem, N.X.; Van Kiem, P.; et al. Diketopiperazines from Rhabdastrella globostellata. Vietnam J. Chem. 2022, 60, 27–31. [Google Scholar] [CrossRef]
- Bourguet-Kondracki, M.-L.; Longeon, A.; Debitus, C.; Guyot, M. New cytotoxic isomalabaricane-type sesterterpenes from the New Caledonian marine sponge Rhabdastrella globostellata. Tetrahedron Lett. 2000, 41, 3087–3090. [Google Scholar] [CrossRef]
- Li, J.; Xu, B.; Cui, J.; Deng, Z.; de Voogd, N.J.; Proksch, P.; Lin, W. Globostelletins A–I, cytotoxic isomalabaricane derivatives from the marine sponge Rhabdastrella globostellata. Bioorg. Med. Chem. 2010, 18, 4639–4647. [Google Scholar] [CrossRef] [PubMed]
- Tawfike, A.; Attia, E.Z.; Desoukey, S.Y.; Hajjar, D.; Makki, A.A.; Schupp, P.J.; Edrada-Ebel, R.; Abdelmohsen, U.R. New bioactive metabolites from the elicited marine sponge-derived bacterium Actinokineospora spheciospongiae sp. nov. AMB Express 2019, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Eltamany, E.E.; Ibrahim, A.K.; Radwan, M.M.; ElSohly, M.A.; Hassanean, H.A.; Ahmed, S.A. Cytotoxic ceramides from the Red Sea sponge Spheciospongia vagabunda. Med. Chem. Res. 2015, 24, 3467–3473. [Google Scholar] [CrossRef]
- Whitson, E.L.; Bugni, T.S.; Chockalingam, P.S.; Concepcion, G.P.; Harper, M.K.; He, M.; Hooper, J.N.A.; Mangalindan, G.C.; Ritacco, F.; Ireland, C.M. Spheciosterol Sulfates, PKCζ Inhibitors from a Philippine Sponge Spheciospongia sp. J. Nat. Prod. 2008, 71, 1213–1217. [Google Scholar] [CrossRef] [Green Version]
- Costantino, V.; Fattorusso, E.; Imperatore, C.; Mangoni, A. Glycolipids from Sponges. 20. J-Coupling Analysis for Stereochemical Assignments in Furanosides: Structure Elucidation of Vesparioside B, a Glycosphingolipid from the Marine Sponge Spheciospongia vesparia. J. Org. Chem. 2008, 73, 6158–6165. [Google Scholar] [CrossRef]
- Costantino, V.; Fattorusso, E.; Imperatore, C.; Mangoni, A. Vesparioside from the Marine Sponge Spheciospongia vesparia, the First Diglycosylceramide with a Pentose Sugar Residue. Eur. J. Org. Chem 2005, 2005, 368–373. [Google Scholar] [CrossRef]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.-H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Podell, S.; Blanton, J.M.; Neu, A.; Agarwal, V.; Biggs, J.S.; Moore, B.S.; Allen, E.E. Pangenomic comparison of globally distributed Poribacteria associated with sponge hosts and marine particles. ISME J. 2019, 13, 468–481. [Google Scholar] [CrossRef] [Green Version]
- Schweizer, E.; Hofmann, J. Microbial Type I Fatty Acid Synthases (FAS): Major Players in a Network of Cellular FAS Systems. Microbiol. Mol. Biol. Rev. 2004, 68, 501–517. [Google Scholar] [CrossRef] [Green Version]
- Yoon, B.K.; Jackman, J.A.; Valle-González, E.R.; Cho, N.-J. Antibacterial Free Fatty Acids and Monoglycerides: Biological Activities, Experimental Testing, and Therapeutic Applications. Int. J. Mol. Sci. 2018, 19, 1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desbois, A.P.; Smith, V.J. Antibacterial free fatty acids: Activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010, 85, 1629–1642. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Lee, J.-H.; Beyenal, H.; Lee, J. Fatty Acids as Antibiofilm and Antivirulence Agents. Trends Microbiol. 2020, 28, 753–768. [Google Scholar] [CrossRef] [PubMed]
- Hess Donavon, J.; Henry-Stanley Michelle, J.; Wells Carol, L. Antibacterial Synergy of Glycerol Monolaurate and Amino-glycosides in Staphylococcus aureus Biofilms. Antimicrob. Agents Chemother. 2014, 58, 6970–6973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, B.C.L.; Han, X.Q.; Lui, S.L.; Wong, C.W.; Wang, T.B.Y.; Cheung, D.W.S.; Cheng, S.W.; Ip, M.; Han, S.Q.B.; Yang, X.-S.; et al. Combating against methicillin-resistant Staphylococcus aureus–two fatty acids from Purslane (Portulaca oleracea L.) exhibit synergistic effects with erythromycin. J. Pharm. Pharmacol. 2015, 67, 107–116. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Lee, J.-H.; Lee, J. Antibiofilm activities of fatty acids including myristoleic acid against Cutibacterium acnes via reduced cell hydrophobicity. Phytomedicine 2021, 91, 153710. [Google Scholar] [CrossRef]
- Casillas-Vargas, G.; Ocasio-Malavé, C.; Medina, S.; Morales-Guzmán, C.; Del Valle, R.G.; Carballeira, N.M.; Sanabria-Ríos, D.J. Antibacterial fatty acids: An update of possible mechanisms of action and implications in the development of the next-generation of antibacterial agents. Prog. Lipid Res. 2021, 82, 101093. [Google Scholar] [CrossRef]
- Park, S.; Lee, J.-H.; Kim, Y.-G.; Hu, L.; Lee, J. Fatty Acids as Aminoglycoside Antibiotic Adjuvants Against Staphylococcus aureus. Front. Microbiol. 2022, 13, 876932. [Google Scholar] [CrossRef]
- Cox, R.J. Polyketides, proteins and genes in fungi: Programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem. 2007, 5, 2010–2026. [Google Scholar] [CrossRef]
- Hertweck, C. The Biosynthetic Logic of Polyketide Diversity. Angew. Chem. Int. Ed. 2009, 48, 4688–4716. [Google Scholar] [CrossRef]
- Staunton, J.; Weissman, K.J. Polyketide biosynthesis: A millennium review. Nat. Prod. Rep. 2001, 18, 380–416. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Du, L. Iterative polyketide biosynthesis by modular polyketide synthases in bacteria. Appl. Microbiol. Biotechnol. 2016, 100, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Fjærvik, E.; Zotchev, S.B. Biosynthesis of the polyene macrolide antibiotic nystatin in Streptomyces noursei. Appl. Microbiol. Biotechnol. 2005, 67, 436–443. [Google Scholar] [CrossRef]
- Rawlings, B.J. Type I polyketide biosynthesis in bacteria (Part A—Erythromycin biosynthesis). Nat. Prod. Rep. 2001, 18, 190–227. [Google Scholar] [CrossRef]
- Choi, S.-S.; Hur, Y.-A.; Sherman, D.H.; Kim, E.-S. Isolation of the biosynthetic gene cluster for tautomycetin, a linear polyketide T cell-specific immunomodulator from Streptomyces sp. CK4412. Microbiology 2007, 153, 1095–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.; Ishida, K.; Jenke-Kodama, H.; Dittmann, E.; Gurgui, C.; Hochmuth, T.; Taudien, S.; Platzer, M.; Hertweck, C.; Piel, J. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat. Biotechnol. 2008, 26, 225–233. [Google Scholar] [CrossRef]
- O’Brien, R.V.; Davis, R.W.; Khosla, C.; Hillenmeyer, M.E. Computational identification and analysis of orphan assembly-line polyketide synthases. J. Antibiot. 2014, 67, 89–97. [Google Scholar] [CrossRef] [Green Version]
- El-Sayed, A.K.; Hothersall, J.; Cooper, S.M.; Stephens, E.; Simpson, T.J.; Thomas, C.M. Characterization of the Mupirocin Biosynthesis Gene Cluster from Pseudomonas fluorescens NCIMB 10586. Chem. Biol. 2003, 10, 419–430. [Google Scholar] [CrossRef] [Green Version]
- Mast, Y.; Weber, T.; Gölz, M.; Ort-Winklbauer, R.; Gondran, A.; Wohlleben, W.; Schinko, E. Characterization of the ‘pristinamycin supercluster’ of Streptomyces pristinaespiralis. Microb. Biotechnol. 2011, 4, 192–206. [Google Scholar] [CrossRef] [Green Version]
- Piel, J.; Hui, D.; Wen, G.; Butzke, D.; Platzer, M.; Fusetani, N.; Matsunaga, S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 2004, 101, 16222–16227. [Google Scholar] [CrossRef] [Green Version]
- Helfrich, E.J.N.; Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2016, 33, 231–316. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, E.J.N.; Ueoka, R.; Chevrette, M.G.; Hemmerling, F.; Lu, X.; Leopold-Messer, S.; Minas, H.A.; Burch, A.Y.; Lindow, S.E.; Piel, J.; et al. Evolution of combinatorial diversity in trans-acyltransferase polyketide synthase assembly lines across bacteria. Nat. Commun. 2021, 12, 1422. [Google Scholar] [CrossRef]
- Hertweck, C.; Luzhetskyy, A.; Rebets, Y.; Bechthold, A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007, 24, 162–190. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.S.; Hopke, J.N. Discovery of a New Bacterial Polyketide Biosynthetic Pathway. ChemBioChem 2001, 2, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.P.; Rouger, C.; Glasser, N.R.; Freitas, S.; de Fraissinette, N.B.; Balskus, E.P.; Tasdemir, D.; Leão, P.N. Chemistry, bioactivity and biosynthesis of cyanobacterial alkylresorcinols. Nat. Prod. Rep. 2019, 36, 1437–1461. [Google Scholar] [CrossRef]
- Kikuchi, H.; Ito, I.; Takahashi, K.; Ishigaki, H.; Iizumi, K.; Kubohara, Y.; Oshima, Y. Isolation, Synthesis, and Biological Activity of Chlorinated Alkylresorcinols from Dictyostelium Cellular Slime Molds. J. Nat. Prod. 2017, 80, 2716–2722. [Google Scholar] [CrossRef]
- Stasiuk, M.; Kozubek, A. Biological activity of phenolic lipids. Cell. Mol. Life Sci. 2010, 67, 841–860. [Google Scholar] [CrossRef]
- Cane, D.E.; Ikeda, H. Exploration and Mining of the Bacterial Terpenome. Acc. Chem. Res. 2012, 45, 463–472. [Google Scholar] [CrossRef] [Green Version]
- Dickschat, J.S. Bacterial terpene cyclases. Nat. Prod. Rep. 2016, 33, 87–110. [Google Scholar] [CrossRef]
- Yamada, Y.; Kuzuyama, T.; Komatsu, M.; Shin-ya, K.; Omura, S.; Cane, D.E.; Ikeda, H. Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 857–862. [Google Scholar] [CrossRef] [Green Version]
- Matobole, R.M.; Van Zyl, L.J.; Parker-Nance, S.; Davies-Coleman, M.T.; Trindade, M. Antibacterial Activities of Bacteria Isolated from the Marine Sponges Isodictya compressa and Higginsia bidentifera Collected from Algoa Bay, South Africa. Mar. Drugs 2017, 15, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi, E.; Keller-Costa, T.; Slaby, B.M.; Cox, C.J.; da Rocha, U.N.; Hentschel, U.; Costa, R. Genomic blueprints of sponge-prokaryote symbiosis are shared by low abundant and cultivatable Alphaproteobacteria. Sci. Rep. 2019, 9, 1999. [Google Scholar] [CrossRef]
- Thomas, T.R.A.; Kavlekar, D.P.; LokaBharathi, P.A. Marine Drugs from Sponge-Microbe Association—A Review. Mar. Drugs 2010, 8, 1417–1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, S.A.; Crossman, L.; Almeida, E.L.; Margassery, L.M.; Kennedy, J.; Dobson, A.D. Diverse and Abundant Secondary Metabolism Biosynthetic Gene Clusters in the Genomes of Marine Sponge Derived Streptomyces spp. Isolates. Mar. Drugs 2018, 16, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gross, H.; König, G.M. Terpenoids from Marine Organisms: Unique Structures and their Pharmacological Potential. Phytochem. Rev. 2006, 5, 115–141. [Google Scholar] [CrossRef]
- Yang, S.-C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial activities of bacteriocins: Application in foods and pharmaceuticals. Front. Microbiol. 2014, 5, 241. [Google Scholar] [CrossRef] [Green Version]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, Antimicrobial Peptides from Bacterial Origin: Overview of Their Biology and Their Impact against Multidrug-Resistant Bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef]
- Zhou, K.; Zhang, R.; Sun, J.; Zhang, W.; Tian, R.-M.; Chen, C.; Kawagucci, S.; Xu, Y. Potential Interactions between Clade SUP05 Sulfur-Oxidizing Bacteria and Phages in Hydrothermal Vent Sponges. Appl. Environ. Microbiol. 2019, 85, e00992-19. [Google Scholar] [CrossRef]
- Phelan, R.W.; Barret, M.; Cotter, P.D.; O’Connor, P.M.; Chen, R.; Morrissey, J.P.; Dobson, A.D.W.; O’Gara, F.; Barbosa, T.M. Subtilomycin: A New Lantibiotic from Bacillus subtilis Strain MMA7 Isolated from the Marine Sponge Haliclona simulans. Mar. Drugs 2013, 11, 1878–1898. [Google Scholar] [CrossRef] [Green Version]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020, 10, 2003. [Google Scholar] [CrossRef] [Green Version]
- Schorn, M.A.; Alanjary, M.M.; Aguinaldo, K.; Korobeynikov, A.; Podell, S.; Patin, N.; Lincecum, T.; Jensen, P.R.; Ziemert, N.; Moore, B.S. Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters. Microbiology 2016, 162, 2075–2086. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Q.; van der Donk, W.A. Insights into the evolution of lanthipeptide biosynthesis. Protein Sci. 2013, 22, 1478–1489. [Google Scholar] [CrossRef] [PubMed]
- Harjes, J.; Ryu, T.; Abdelmohsen, U.R.; Moitinho-Silva, L.; Horn, H.; Ravasi, T.; Hentschel, U. Draft Genome Sequence of the Antitrypanosomally Active Sponge-Associated Bacterium Actinokineospora sp. Strain EG49. Genome Announc. 2014, 2, e00160-14. [Google Scholar] [CrossRef] [Green Version]
- Ian, E.; Malko, D.B.; Sekurova, O.N.; Bredholt, H.; Rückert, C.; Borisova, M.E.; Albersmeier, A.; Kalinowski, J.; Gelfand, M.S.; Zotchev, S.B. Genomics of Sponge-Associated Streptomyces spp. Closely Related to Streptomyces albus J1074: Insights into Marine Adaptation and Secondary Metabolite Biosynthesis Potential. PLoS ONE 2014, 9, e96719. [Google Scholar] [CrossRef] [Green Version]
- Matroodi, S.; Siitonen, V.; Baral, B.; Yamada, K.; Akhgari, A.; Metsä-Ketelä, M. Genotyping-Guided Discovery of Persiamycin A from Sponge-Associated Halophilic Streptomonospora sp. PA3. Front. Microbiol. 2020, 11, 1237. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Garzón, J.F.; Zehl, M.; Schneider, O.; Rückert, C.; Busche, T.; Kalinowski, J.; Bredholt, H.; Zotchev, S.B. Streptomyces spp. from the Marine Sponge Antho dichotoma: Analyses of Secondary Metabolite Biosynthesis Gene Clusters and Some of Their Products. Front. Microbiol. 2020, 11, 437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, H.; O’Connor Paula, M.; Hill, C.; Cotter Paul, D.; Ross, R.P. Analysis of Anti-Clostridium difficile Activity of Thuricin CD, Vancomycin, Metronidazole, Ramoplanin, and Actagardine, both Singly and in Paired Combinations. Antimicrob. Agents Chemother. 2013, 57, 2882–2886. [Google Scholar] [CrossRef] [Green Version]
- Grasemann, H.; Stehling, F.; Brunar, H.; Widmann, R.; Laliberte, T.W.; Molina, L.; Döring, G.; Ratjen, F. Inhalation of Moli1901 in Patients with Cystic Fibrosis. Chest 2007, 131, 1461–1466. [Google Scholar] [CrossRef]
- Donadio, S.; Maffioli, S.; Monciardini, P.; Sosio, M.; Jabes, D. Antibiotic discovery in the twenty-first century: Current trends and future perspectives. J. Antibiot. 2010, 63, 423–430. [Google Scholar] [CrossRef] [Green Version]
- Ghobrial, O.G.; Derendorf, H.; Hillman, J.D. Pharmacokinetic and pharmacodynamic evaluation of the lantibiotic MU1140. J. Pharm. Sci. 2010, 99, 2521–2528. [Google Scholar] [CrossRef]
- Piper, C.; Casey, P.G.; Hill, C.; Cotter, P.D.; Ross, R.P. The Lantibiotic Lacticin 3147 Prevents Systemic Spread of Staphylococcus aureus in a Murine Infection Model. Int. J. Microbiol. 2012, 2012, 806230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabés, D.; Brunati, C.; Candiani, G.; Riva, S.; Romanó, G.; Donadio, S. Efficacy of the New Lantibiotic NAI-107 in Experimental Infections Induced by Multidrug-Resistant Gram-Positive Pathogens. Antimicrob. Agents Chemother. 2011, 55, 1671–1676. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, A.; Gadkari, R.; Reeves, C.D.; Ibrahim, F.; DeLong, E.F.; Hutchinson, C.R. Metagenomic Analysis Reveals Diverse Polyketide Synthase Gene Clusters in Microorganisms Associated with the Marine Sponge Discodermia dissoluta. Appl. Environ. Microbiol. 2005, 71, 4840–4849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunasekera, S.; Abraham, S.; Stegger, M.; Pang, S.; Wang, P.; Sahibzada, S.; O’Dea, M. Evaluating coverage bias in next-generation sequencing of Escherichia coli. PLoS ONE 2021, 16, e0253440. [Google Scholar] [CrossRef]
- Sato, M.P.; Ogura, Y.; Nakamura, K.; Nishida, R.; Gotoh, Y.; Hayashi, M.; Hisatsune, J.; Sugai, M.; Takehiko, I.; Hayashi, T. Comparison of the sequencing bias of currently available library preparation kits for Illumina sequencing of bacterial genomes and metagenomes. DNA Res. 2019, 26, 391–398. [Google Scholar] [CrossRef] [Green Version]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion torrent, pacific biosciences and illumina MiSeq sequencers. BMC Genom. 2012, 13, 341. [Google Scholar] [CrossRef] [Green Version]
- Ghurye, J.S.; Cepeda-Espinoza, V.; Pop, M. Metagenomic Assembly: Overview, Challenges and Applications. Yale J. Biol. Med. 2016, 89, 353–362. [Google Scholar]
- Lapidus, A.L.; Korobeynikov, A.I. Metagenomic Data Assembly–The Way of Decoding Unknown Microorganisms. Front. Microbiol. 2021, 12, 613791. [Google Scholar] [CrossRef]
- Tracanna, V.; de Jong, A.; Medema, M.H.; Kuipers, O.P. Mining prokaryotes for antimicrobial compounds: From diversity to function. FEMS Microbiol. Rev. 2017, 41, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016, 17, 882. [Google Scholar] [CrossRef] [Green Version]
- Challinor, V.L.; Bode, H.B. Bioactive natural products from novel microbial sources. Ann. N. Y. Acad. Sci. 2015, 1354, 82–97. [Google Scholar] [CrossRef]
- Subramani, R.; Sipkema, D. Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar. Drugs 2019, 17, 249. [Google Scholar] [CrossRef] [Green Version]
- Simister, R.L.; Deines, P.; Botté, E.S.; Webster, N.S.; Taylor, M.W. Sponge-specific clusters revisited: A comprehensive phy-logeny of sponge-associated microorganisms. Environ. Microbiol. 2012, 14, 517–524. [Google Scholar] [CrossRef]
- Fieseler, L.; Hentschel, U.; Grozdanov, L.; Schirmer, A.; Wen, G.; Platzer, M.; Hrvatin, S.; Butzke, D.; Zimmermann, K.; Piel, J. Widespread Occurrence and Genomic Context of Unusually Small Polyketide Synthase Genes in Microbial Consortia Associated with Marine Sponges. Appl. Environ. Microbiol. 2007, 73, 2144–2155. [Google Scholar] [CrossRef] [Green Version]
- Della Sala, G.; Hochmuth, T.; Costantino, V.; Teta, R.; Gerwick, W.; Gerwick, L.; Piel, J.; Mangoni, A. Polyketide genes in the marine sponge Plakortis simplex: A new group of mono-modular type I polyketide synthases from sponge symbionts. Environ. Microbiol. Rep. 2013, 5, 809–818. [Google Scholar] [CrossRef] [Green Version]
- Hochmuth, T.; Niederkrüger, H.; Gernert, C.; Siegl, A.; Taudien, S.; Platzer, M.; Crews, P.; Hentschel, U.; Piel, J. Linking Chemical and Microbial Diversity in Marine Sponges: Possible Role for Poribacteria as Producers of Methyl-Branched Fatty Acids. Chembiochem 2010, 11, 2572–2578. [Google Scholar] [CrossRef]
- Borchert, E.; Jackson, S.A.; O’Gara, F.; Dobson, A.D.W. Diversity of Natural Product Biosynthetic Genes in the Microbiome of the Deep Sea Sponges Inflatella pellicula, Poecillastra compressa, and Stelletta normani. Front. Microbiol. 2016, 7, 1027. [Google Scholar] [CrossRef]
- Mohanty, I.; Tapadar, S.; Moore Samuel, G.; Biggs Jason, S.; Freeman Christopher, J.; Gaul David, A.; Garg, N.; Agarwal, V. Presence of Bromotyrosine Alkaloids in Marine Sponges Is Independent of Metabolomic and Microbiome Architectures. MSystems 2021, 6, e01387-20. [Google Scholar] [CrossRef]
- Abe, T.; Sahin, F.P.; Akiyama, K.; Naito, T.; Kishigami, M.; Miyamoto, K.; Sakakibara, Y.; Uemura, D. Construction of a Metagenomic Library for the Marine Sponge Halichondria okadai. Biosci. Biotechnol. Biochem. 2012, 76, 633–639. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ziemert, N.; Podell, S.; Penn, K.; Badger, J.H.; Allen, E.; Jensen, P.R. The Natural Product Domain Seeker NaPDoS: A Phylogeny Based Bioinformatic Tool to Classify Secondary Metabolite Gene Diversity. PLoS ONE 2012, 7, e34064. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]


| Gene Cluster Type | C. reinwardti | R. globostellata | Spheciospongia sp. |
|---|---|---|---|
| Type I PKS | 83 | 112 | 125 |
| Type II PKS | 1 | 1 | 1 |
| Type III PKS | 3 | 6 | 5 |
| Trans-AT type I PKS | 2 | 0 | 2 |
| Trans-AT PKS | 0 | 2 | 0 |
| Terpene | 58 | 66 | 97 |
| Bacteriocin | 12 | 16 | 14 |
| Aryl polyene | 8 | 10 | 9 |
| Ladderane | 3 | 4 | 2 |
| Phosphonate | 5 | 5 | 6 |
| Lantipeptide | 2 | 1 | 3 |
| Cf_fatty_acid 1 | 86 | 119 | 164 |
| Others | 19 | 29 | 35 |
| Number of clusters | 282 | 371 | 463 |
| Number of contigs ≥ 1000 | 93,883 | 118,632 | 154,114 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dat, T.T.H.; Steinert, G.; Cuc, N.T.K.; Cuong, P.V.; Smidt, H.; Sipkema, D. Diversity of Bacterial Secondary Metabolite Biosynthetic Gene Clusters in Three Vietnamese Sponges. Mar. Drugs 2023, 21, 29. https://doi.org/10.3390/md21010029
Dat TTH, Steinert G, Cuc NTK, Cuong PV, Smidt H, Sipkema D. Diversity of Bacterial Secondary Metabolite Biosynthetic Gene Clusters in Three Vietnamese Sponges. Marine Drugs. 2023; 21(1):29. https://doi.org/10.3390/md21010029
Chicago/Turabian StyleDat, Ton That Huu, Georg Steinert, Nguyen Thi Kim Cuc, Pham Viet Cuong, Hauke Smidt, and Detmer Sipkema. 2023. "Diversity of Bacterial Secondary Metabolite Biosynthetic Gene Clusters in Three Vietnamese Sponges" Marine Drugs 21, no. 1: 29. https://doi.org/10.3390/md21010029
APA StyleDat, T. T. H., Steinert, G., Cuc, N. T. K., Cuong, P. V., Smidt, H., & Sipkema, D. (2023). Diversity of Bacterial Secondary Metabolite Biosynthetic Gene Clusters in Three Vietnamese Sponges. Marine Drugs, 21(1), 29. https://doi.org/10.3390/md21010029







