Novel Approach for Obtaining Variable Domain of New Antigen Receptor with Different Physicochemical Properties from Japanese Topeshark (Hemitriakis japanica)
Abstract
1. Introduction
2. Results
2.1. Identification of Various Kinds of VNAR under Different Panning Condition
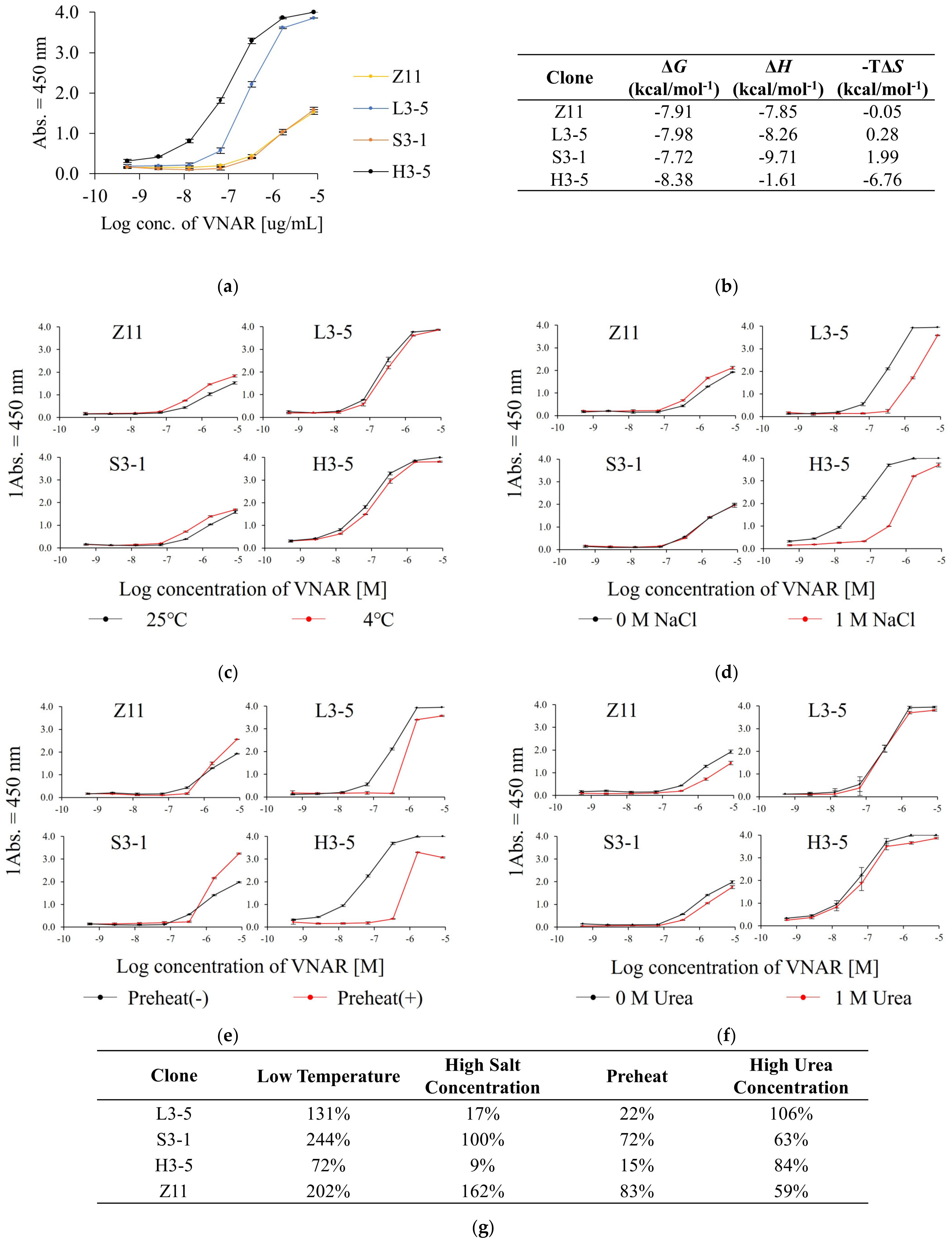
2.2. Comparison of the Reactivity to the Antigen under Each ELISA Condition
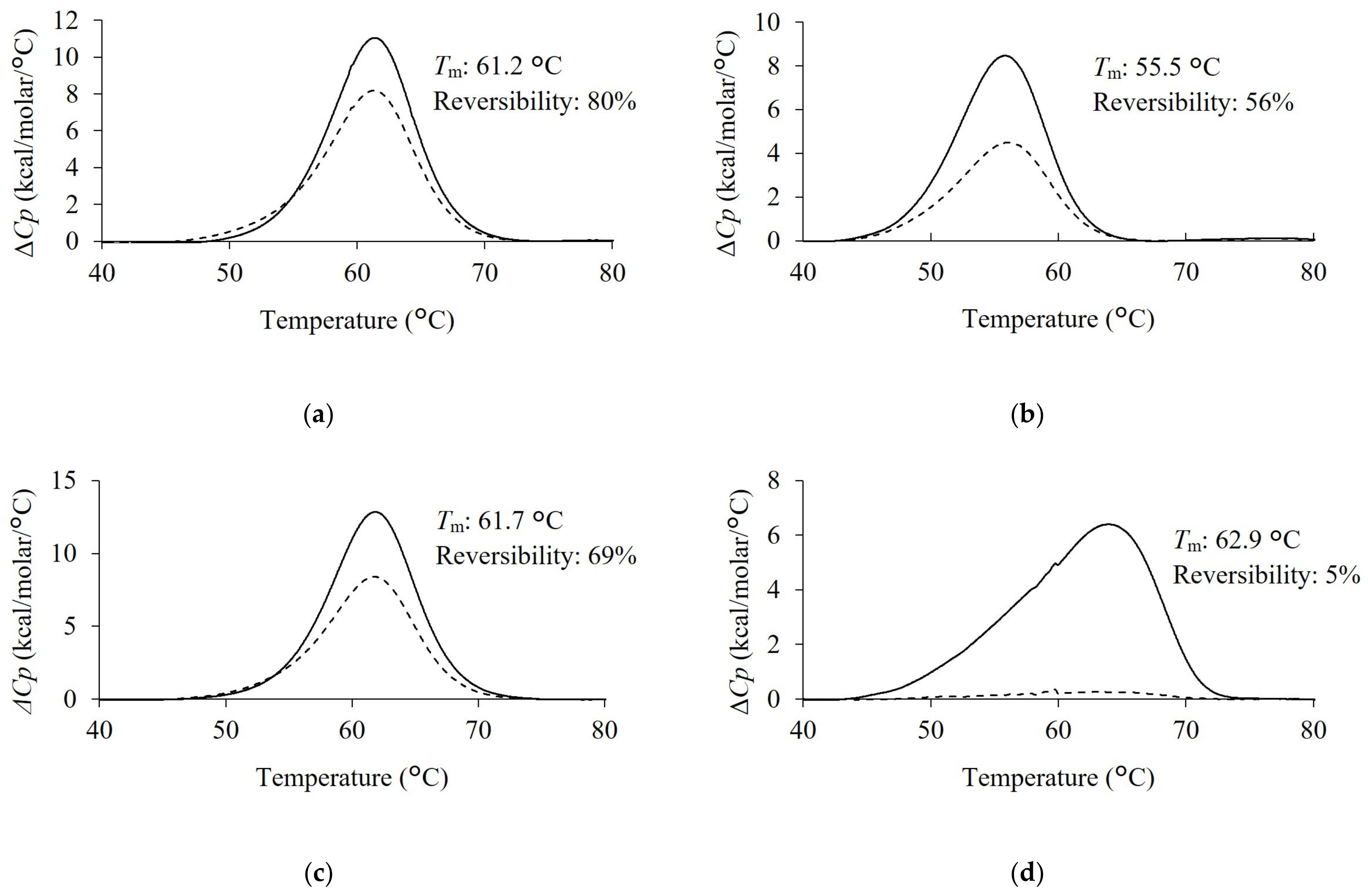
2.3. Analysis of Thermal Stability and Reversibility
3. Discussion
4. Materials and Methods
4.1. Phage Display and Panning Method
4.2. Phage ELISA
4.3. NGS Analysis of VNAR Phage Pool and Clone Selection
4.4. Expression and Purification of VNAR
4.5. Evaluation of Purity by SDS-PAGE
4.6. Evaluation of the Reactivity under Each Condition by ELISA
4.7. Isothermal Titration Calorimetry (ITC)
4.8. Differential Scanning Calorimetry (DSC)
4.9. Surface Plasmon Resonance (SPR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| sdAb | Single-domain antibody |
| hcAb | Heavy-chain antibody |
| VHH | Variable domain of the heavy chain antibody |
| VNAR | Variable domain of new antigen receptor |
| CDR | Complementarity determining region |
| HV2 | Hypervariable region 2 |
| HV4 | Hypervariable region 4 |
| scFv | Single-chain variable fragment |
| NGS | Next generation sequencing |
| N3 | The phage pool in Round 3 under normal condition |
| L3 | The phage pool in Round 3 under low-temperature condition |
| H3 | The phage pool in Round 3 under preheat condition |
| S3 | The phage pool in Round 3 under high salt concentration condition |
| U3 | The phage pool in Round 3 under high urea concentration condition |
| ITC | Isothermal titration calorimetry |
| DSC | Differential scanning calorimetry |
| SPR | Surface plasmon resonance |
| kon | Association rate constant |
| koff | Dissociation rate constant |
| Tm | Melting Temperature |
| ΔG | Gibbs free energy change |
| ΔH | Enthalpy change |
| ΔS | Entropy change |
| ΔCp | The change in heat capacity |
References
- Griffiths, K.; Dolezal, O.; Parisi, K.; Angerosa, J.; Dogovski, C.; Barraclough, M.; Sanalla, A.; Casey, J.; González, I.; Perugini, M.; et al. Shark variable new antigen receptor (VNAR) single domain antibody fragments: Stability and diagnostic applications. Antibodies 2013, 2, 66–81. [Google Scholar] [CrossRef]
- Liu, J.L.; Anderson, G.P.; Delehanty, J.B.; Baumann, R.; Hayhurst, A.; Goldman, E.R. Selection of cholera toxin specific IgNAR single-domain antibodies from a naive shark library. Mol. Immunol. 2007, 44, 1775–1783. [Google Scholar] [CrossRef] [PubMed]
- Wesolowski, J.; Alzogaray, V.; Reyelt, J.; Unger, M.; Juarez, K.; Urrutia, M.; Cauerhff, A.; Danquah, W.; Rissiek, B.; Scheuplein, F.; et al. Single domain antibodies: Promising experimental and therapeutic tools in infection and immunity. Med. Microbiol. Immunol. 2009, 198, 157–174. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Bian, H.; Wu, X.; Fu, T.; Fu, Y.; Hong, J.; Fleming, B.D.; Flajnik, M.F.; Ho, M. Construction and next-generation sequencing analysis of a large phage-displayed VNAR single-domain antibody library from six naïve nurse sharks. Antib. Ther. 2019, 2, 1–11. [Google Scholar] [CrossRef]
- Barelle, C.; Gill, D.S.; Charlton, K. Shark novel antigen receptors—The next generation of biologic therapeutics? Adv. Exp. Med. Biol. 2009, 655, 49–62. [Google Scholar] [CrossRef]
- Cheong, W.S.; Leow, C.Y.; Abdul Majeed, A.B.A.; Leow, C.H. Diagnostic and therapeutic potential of shark variable new antigen receptor (VNAR) single domain antibody. Int. J. Biol. Macromol. 2020, 147, 369–375. [Google Scholar] [CrossRef]
- Zielonka, S.; Weber, N.; Becker, S.; Doerner, A.; Christmann, A.; Christmann, C.; Uth, C.; Fritz, J.; Schäfer, E.; Steinmann, B.; et al. Shark Attack: High affinity binding proteins derived from shark vNAR domains by stepwise in vitro affinity maturation. J. Biotechnol. 2014, 191, 236–245. [Google Scholar] [CrossRef]
- Zielonka, S.; Empting, M.; Grzeschik, J.; Könning, D.; Barelle, C.J.; Kolmar, H. Structural insights and biomedical potential of IgNAR scaffolds from sharks. mAbs 2015, 7, 15–25. Available online: http://www.ncbi.nlm.nih.gov/pubmed/25523873 (accessed on 29 September 2023). [CrossRef]
- Cabanillas-Bernal, O.; Dueñas, S.; Ayala-Avila, M.; Rucavado, A.; Escalante, T.; Licea-Navarro, A.F. Synthetic libraries of shark vNAR domains with different cysteine numbers within the CDR3. PLoS ONE 2019, 14, e0213394. [Google Scholar] [CrossRef]
- Dooley, H.; Flajnik, M.F.; Porter, A.J. Selection and characterization of naturally occurring single-domain (IgNAR) antibody fragments from immunized sharks by phage display. Mol. Immunol. 2003, 40, 25–33. [Google Scholar] [CrossRef]
- Turner, K.B.; Naciri, J.; Liu, J.L.; Anderson, G.P.; Goldman, E.R.; Zabetakis, D. Next-generation sequencing of a single domain antibody repertoire reveals quality of phage display selected candidates. PLoS ONE 2016, 11, e0149393. [Google Scholar] [CrossRef] [PubMed]
- Ljungars, A.; Svensson, C.; Carlsson, A.; Birgersson, E.; Tornberg, U.C.; Frendéus, B.; Ohlin, M.; Mattsson, M. Deep mining of complex antibody phage pools generated by cell panning enables discovery of rare antibodies binding new targets and epitopes. Front. Pharmacol. 2019, 10, 847. [Google Scholar] [CrossRef] [PubMed]
- Rahbarnia, L.; Farajnia, S.; Babaei, H.; Majidi, J.; Veisi, K.; Ahmadzadeh, V.; Akbari, B. Evolution of phage display technology: From discovery to application. J. Drug Target. 2017, 25, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Pershad, K.; Kay, B.K. Generating thermal stable variants of protein domains through phage display. Methods 2013, 60, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Igawa, T.; Ishii, S.; Tachibana, T.; Maeda, A.; Higuchi, Y.; Shimaoka, S.; Moriyama, C.; Watanabe, T.; Takubo, R.; Doi, Y.; et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat. Biotechnol. 2010, 28, 1203–1207. [Google Scholar] [CrossRef]
- Luciani, F.; Bull, R.A.; Lloyd, A.R. Next generation deep sequencing and vaccine design: Today and tomorrow. Trends Biotechnol. 2012, 30, 443–452. [Google Scholar] [CrossRef]
- Zeidel, J.D.; Mathai, J.C.; Campbell, J.D.; Ruiz, W.G.; Apodaca, G.L.; Riordan, J.; Zeidel, M.L. Selective permeability barrier to urea in shark rectal gland. Am. J. Physiol. Ren. Physiol. 2005, 289, 83–89. [Google Scholar] [CrossRef][Green Version]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef]
- Compagno, L.J.V.; Stevens, J.D. Hemitriakis falcata n.sp. and H. abdita n.sp., two new houndsharks (Carcharhiniformes: Triakidae) from Australia. Rec. Aust. Mus. 1993, 45, 195–220. [Google Scholar] [CrossRef]
- Honda, Y.; Kondo, H.; Caipang, C.M.; Hirono, I.; Aoki, T. cDNA cloning of the immunoglobulin heavy chain genes in banded houndshark Triakis scyllium. Fish Shellfish Immunol. 2010, 29, 854–861. [Google Scholar] [CrossRef]
- Ohtani, M.; Hikima, J.; Jung, T.S.; Kondo, H.; Hirono, I.; Aoki, T. Construction of an artificially randomized IgNAR phage display library: Screening of variable regions that bind to hen egg white lysozyme. Mar. Biotechnol. 2013, 15, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Hikima, J.; Jung, T.S.; Kondo, H.; Hirono, I.; Takeyama, H.; Aoki, T. Variable domain antibodies specific for viral hemorrhagic septicemia virus (VHSV) selected from a randomized IgNAR phage display library. Fish Shellfish Immunol. 2013, 34, 724–728. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.S.; Chae, H.D.; Jung, I.; Lee, W.K.; Lee, W.J.; Lee, J.; Gong, Y.; Lee, D.; Kim, B.W.; Kim, J.K.; et al. Isolation and characterization of single domain antibodies from banded houndshark (Triakis scyllium) targeting SARS-CoV-2 spike RBD protein. Fish Shellfish Immunol. 2023, 138, 108807. [Google Scholar] [CrossRef]
- Takeda, H.; Ozawa, T.; Zenke, H.; Ohnuki, Y.; Umeda, Y.; Zhou, W.; Tomoda, H.; Takechi, A.; Narita, K.; Shimizu, T.; et al. VNAR development through antigen immunization of Japanese topeshark (Hemitriakis japanica). Front. Bioeng. Biotechnol. 2023, 11, 1265582. [Google Scholar] [CrossRef]
- O’Connell, M.A.; Belanger, B.A.; Haaland, P.D. Calibration and assay development using the four-parameter logistic model. Intell. Lab. Syst. 1993, 20, 97–114. [Google Scholar] [CrossRef]
- Kim, D.Y.; To, R.; Kandalaft, H.; Ding, W.; van Faassen, H.; Luo, Y.; Schrag, J.D.; St-Amant, N.; Hefford, M.; Hirama, T.; et al. Antibody light chain variable domains and their biophysically improved versions for human immunotherapy. mAbs 2014, 6, 219–235. [Google Scholar] [CrossRef][Green Version]
- Joshi, V.; Shivach, T.; Yadav, N.; Rathore, A.S. Circular dichroism spectroscopy as a tool for monitoring aggregation in monoclonal antibody therapeutics. Anal. Chem. 2014, 86, 11606–11613. [Google Scholar] [CrossRef]
- Brader, M.L.; Estey, T.; Bai, S.; Alston, R.W.; Lucas, K.K.; Lantz, S.; Landsman, P.; Maloney, K.M. Examination of thermal unfolding and aggregation profiles of a series of developable therapeutic monoclonal antibodies. Mol. Pharm. 2015, 12, 1005–1017. [Google Scholar] [CrossRef]
- Navratilova, I.; Papalia, G.A.; Rich, R.L.; Bedinger, D.; Brophy, S.; Condon, B.; Deng, T.; Emerick, A.W.; Guan, H.W.; Hayden, T.; et al. Thermodynamic benchmark study using Biacore technology. Anal. Biochem. 2007, 364, 67–77. [Google Scholar] [CrossRef]
- Prévost, J.; Richard, J.; Gasser, R.; Ding, S.; Fage, C.; Anand, S.P.; Adam, D.; Gupta Vergara, N.G.; Tauzin, A.; Benlarbi, M.; et al. Impact of temperature on the affinity of SARS-CoV-2 Spike glycoprotein for host ACE2. J. Biol. Chem. 2021, 297, 101151. [Google Scholar] [CrossRef]
- Leonard, P.; Hayes, C.J.; O’Kennedy, R. Rapid temperature-dependent antibody ranking using Biacore A100. Anal. Biochem. 2011, 409, 290–292. [Google Scholar] [CrossRef]
- Ferreira, L.A.; Povarova, O.I.; Stepanenko, O.V.; Sulatskaya, A.I.; Madeira, P.P.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B.Y. Effects of low urea concentrations on protein-water interactions. J. Biomol. Struct. Dyn. 2017, 35, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Bata, J.E.; Gyenes, L.; Sehon, A.H. The effect of urea of antibody-antigen reactions. Immunochemistry 1964, 1, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Feige, M.J.; Gräwert, M.A.; Marcinowski, M.; Hennig, J.; Behnke, J.; Ausländer, D.; Herold, E.M.; Peschek, J.; Castro, C.D.; Flajnik, M.; et al. The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins. Proc. Natl. Acad. Sci. USA 2014, 111, 8155–8160. [Google Scholar] [CrossRef] [PubMed]
- Curtis, R.A.; Prausnitz, J.M.; Blanch, H.W. Protein-protein and protein–salt interactions in aqueous protein solutions containing concentrated electrolytes. Biotechnol. Bioeng. 1998, 57, 11–21. [Google Scholar] [CrossRef]
- Valente, J.J.; Verma, K.S.; Manning, M.C.; Wilson, W.W.; Henry, C.S. Second virial coefficient studies of cosolvent-induced protein self-interaction. Biophys. J. 2005, 89, 4211–4218. [Google Scholar] [CrossRef][Green Version]
- Jung, S.; Honegger, A.; Plückthun, A. Selection for improved protein stability by phage display. J. Mol. Biol. 1999, 294, 163–180. [Google Scholar] [CrossRef]
- Bemporad, F.; Taddei, N.; Stefani, M.; Chiti, F. Assessing the role of aromatic residues in the amyloid aggregation of human muscle acylphosphatase. Protein Sci. 2006, 15, 862–870. [Google Scholar] [CrossRef]
- Fellouse, F.A.; Wiesmann, C.; Sidhu, S.S. Synthetic antibodies from a four-amino-acid code: A dominant role for tyrosine in antigen recognition. Proc. Natl. Acad. Sci. USA 2004, 101, 12467–12472. [Google Scholar] [CrossRef]
- Ito, W.; Iba, Y.; Kurosawa, Y. Effects of substitutions of closely related amino acids at the contact surface in an antigen-antibody complex on thermodynamic parameters. J. Biol. Chem. 1993, 268, 16639–16647. [Google Scholar] [CrossRef]
- Tzang, B.S.; Tsay, G.J.; Lee, Y.J.; Li, C.; Sun, Y.S.; Hsu, T.C. The association of VP1 unique region protein in acute parvovirus B19 infection and anti-phospholipid antibody production. Clin. Chim. Acta 2007, 378, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakada-Masuta, T.; Takeda, H.; Uchida, K. Novel Approach for Obtaining Variable Domain of New Antigen Receptor with Different Physicochemical Properties from Japanese Topeshark (Hemitriakis japanica). Mar. Drugs 2023, 21, 550. https://doi.org/10.3390/md21110550
Nakada-Masuta T, Takeda H, Uchida K. Novel Approach for Obtaining Variable Domain of New Antigen Receptor with Different Physicochemical Properties from Japanese Topeshark (Hemitriakis japanica). Marine Drugs. 2023; 21(11):550. https://doi.org/10.3390/md21110550
Chicago/Turabian StyleNakada-Masuta, Tomofumi, Hiroyuki Takeda, and Kazuhisa Uchida. 2023. "Novel Approach for Obtaining Variable Domain of New Antigen Receptor with Different Physicochemical Properties from Japanese Topeshark (Hemitriakis japanica)" Marine Drugs 21, no. 11: 550. https://doi.org/10.3390/md21110550
APA StyleNakada-Masuta, T., Takeda, H., & Uchida, K. (2023). Novel Approach for Obtaining Variable Domain of New Antigen Receptor with Different Physicochemical Properties from Japanese Topeshark (Hemitriakis japanica). Marine Drugs, 21(11), 550. https://doi.org/10.3390/md21110550





