Invertebrate C1q Domain-Containing Proteins: Molecular Structure, Functional Properties and Biomedical Potential
Abstract
:1. Introduction
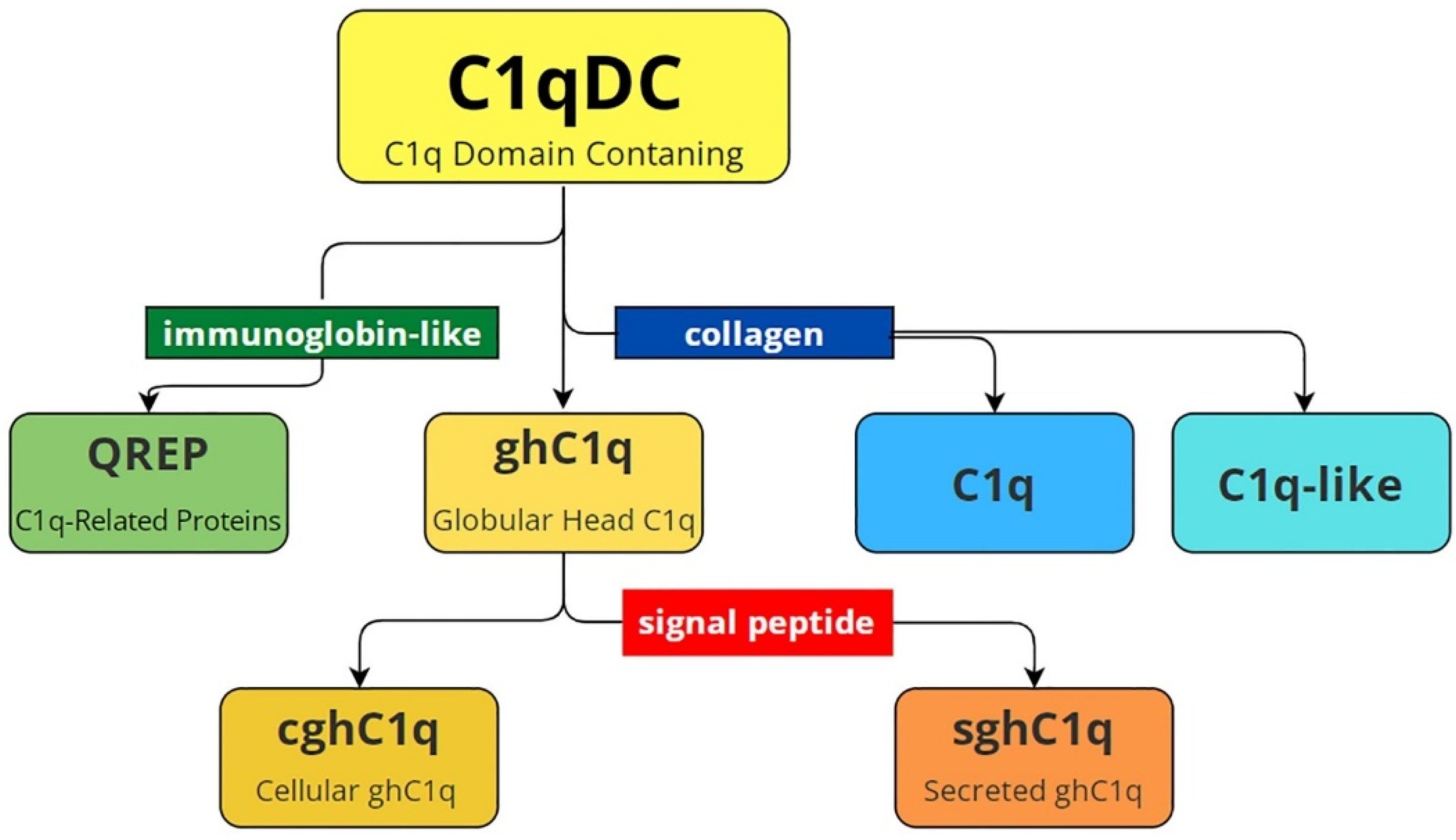
2. C1qDC Proteins’ Structures and Phylogeny
3. Biosynthesis and Tissue Distribution of C1qDC Proteins
4. Antibacterial Properties and Immune Functions of C1qDC Proteins
5. Other Functions of C1qDC Proteins
6. Carbohydrate Specificity of C1qDC Proteins
7. Biomedical Applications of Invertebrate C1qDC Proteins
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharon, N.; Lis, H. Lectins, 2nd ed.; Springer: Dordrecht, The Netherlands, 2007; ISBN 978-1-4020-6605-4. [Google Scholar]
- Taylor, M.E.; Drickamer, K.; Imberty, A.; van Kooyk, Y.; Schnaar, R.L.; Etzler, M.E.; Varki, A. Discovery and Classification of Glycan-Binding Proteins. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2022; ISBN 978-1-62182-421-3. [Google Scholar]
- Bojarová, P.; Křen, V. Sugared Biomaterial Binding Lectins: Achievements and Perspectives. Biomater. Sci. 2016, 4, 1142–1160. [Google Scholar] [CrossRef]
- Gorakshakar, A.C.; Ghosh, K. Use of Lectins in Immunohematology. Asian J. Transfus. Sci. 2016, 10, 12–21. [Google Scholar] [CrossRef]
- Manning, J.C.; Romero, A.; Habermann, F.A.; García Caballero, G.; Kaltner, H.; Gabius, H.-J. Lectins: A Primer for Histochemists and Cell Biologists. Histochem. Cell Biol. 2017, 147, 199–222. [Google Scholar] [CrossRef]
- Mitchell, C.A.; Ramessar, K.; O’Keefe, B.R. Antiviral Lectins: Selective Inhibitors of Viral Entry. Antivir. Res. 2017, 142, 37–54. [Google Scholar] [CrossRef]
- Breitenbach Barroso Coelho, L.C.; Marcelino Dos Santos Silva, P.; Felix de Oliveira, W.; de Moura, M.C.; Viana Pontual, E.; Soares Gomes, F.; Guedes Paiva, P.M.; Napoleão, T.H.; Dos Santos Correia, M.T. Lectins as Antimicrobial Agents. J. Appl. Microbiol. 2018, 125, 1238–1252. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, E.V.M.M.; Oliveira, W.F.; Coelho, L.C.B.B.; Correia, M.T.S. Lectins as Mitosis Stimulating Factors: Briefly Reviewed. Life Sci. 2018, 207, 152–157. [Google Scholar] [CrossRef]
- Devi, R.V.; Basil-Rose, M.R. Lectins as Ligands for Directing Nanostructured Systems. Curr. Drug Deliv. 2018, 15, 448–452. [Google Scholar] [CrossRef]
- Hendrickson, O.D.; Zherdev, A.V. Analytical Application of Lectins. Crit. Rev. Anal. Chem. 2018, 48, 279–292. [Google Scholar] [CrossRef]
- Mazalovska, M.; Kouokam, J.C. Lectins as Promising Therapeutics for the Prevention and Treatment of HIV and Other Potential Coinfections. BioMed Res. Int. 2018, 2018, 3750646. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.S.; Walia, A.K. Lectins from Red Algae and Their Biomedical Potential. J. Appl. Phycol. 2018, 30, 1833–1858. [Google Scholar] [CrossRef]
- Catanzaro, E.; Calcabrini, C.; Bishayee, A.; Fimognari, C. Antitumor Potential of Marine and Freshwater Lectins. Mar. Drugs 2019, 18, 11. [Google Scholar] [CrossRef] [PubMed]
- Laaf, D.; Bojarová, P.; Elling, L.; Křen, V. Galectin-Carbohydrate Interactions in Biomedicine and Biotechnology. Trends Biotechnol. 2019, 37, 402–415. [Google Scholar] [CrossRef] [PubMed]
- Višnjar, T.; Romih, R.; Zupančič, D. Lectins as Possible Tools for Improved Urinary Bladder Cancer Management. Glycobiology 2019, 29, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.M.; Liu, J.H. Lectins and ELLSA as Powerful Tools for Glycoconjugate Recognition Analyses. Glycoconj. J. 2019, 36, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.-U.; Donia, A.; Sial, U.; Zhang, X.; Bokhari, H. Glycoprotein- and Lectin-Based Approaches for Detection of Pathogens. Pathogens 2020, 9, 694. [Google Scholar] [CrossRef]
- Mazalovska, M.; Kouokam, J.C. Plant-Derived Lectins as Potential Cancer Therapeutics and Diagnostic Tools. BioMed Res. Int. 2020, 2020, 1631394. [Google Scholar] [CrossRef]
- Xu, D.; Prestegard, J.H.; Linhardt, R.J.; Esko, J.D. Proteins That Bind Sulfated Glycosaminoglycans. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2022; ISBN 978-1-62182-421-3. [Google Scholar]
- Wang, W.; Song, X.; Wang, L.; Song, L. Pathogen-Derived Carbohydrate Recognition in Molluscs Immune Defense. Int. J. Mol. Sci. 2018, 19, 721. [Google Scholar] [CrossRef]
- De Graaf, D.C.; Brunain, M.; Scharlaken, B.; Peiren, N.; Devreese, B.; Ebo, D.G.; Stevens, W.J.; Desjardins, C.A.; Werren, J.H.; Jacobs, F.J. Two Novel Proteins Expressed by the Venom Glands of Apis Mellifera and Nasonia Vitripennis Share an Ancient C1q-like Domain. Insect Mol. Biol. 2010, 19, 1–10. [Google Scholar] [CrossRef]
- Gerdol, M.; Luo, Y.-J.; Satoh, N.; Pallavicini, A. Genetic and Molecular Basis of the Immune System in the Brachiopod Lingula Anatina. Dev. Comp. Immunol. 2018, 82, 7–30. [Google Scholar] [CrossRef]
- Gerdol, M.; Gomez-Chiarri, M.; Castillo, M.G.; Figueras, A.; Fiorito, G.; Moreira, R.; Novoa, B.; Pallavicini, A.; Ponte, G.; Roumbedakis, K. Immunity in Molluscs: Recognition and Effector Mechanisms, with a Focus on Bivalvia. In Advances in Comparative Immunology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 225–341. [Google Scholar]
- Gorbushin, A.M. Derivatives of the Lectin Complement Pathway in Lophotrochozoa. Dev. Comp. Immunol. 2019, 94, 35–58. [Google Scholar] [CrossRef]
- Hasan, I.; Gerdol, M.; Fujii, Y.; Ozeki, Y. Functional Characterization of OXYL, A SghC1qDC LacNAc-Specific Lectin from The Crinoid Feather Star Anneissia Japonica. Mar. Drugs 2019, 17, 136. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M. First Insights into the Repertoire of Secretory Lectins in Rotifers. Mar. Drugs 2022, 20, 130. [Google Scholar] [CrossRef]
- Peng, J.; Li, Q.; Xu, L.; Wei, P.; He, P.; Zhang, X.; Zhang, L.; Guan, J.; Zhang, X.; Lin, Y.; et al. Chromosome-Level Analysis of the Crassostrea Hongkongensis Genome Reveals Extensive Duplication of Immune-Related Genes in Bivalves. Mol. Ecol. Resour. 2020, 20, 980–994. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Koyanagi, R.; Gyoja, F.; Kanda, M.; Hisata, K.; Fujie, M.; Goto, H.; Yamasaki, S.; Nagai, K.; Morino, Y.; et al. Bivalve-Specific Gene Expansion in the Pearl Oyster Genome: Implications of Adaptation to a Sessile Lifestyle. Zool. Lett. 2016, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; Venier, P.; Pallavicini, A. The Genome of the Pacific Oyster Crassostrea Gigas Brings New Insights on the Massive Expansion of the C1q Gene Family in Bivalvia. Dev. Comp. Immunol. 2015, 49, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Farhat, S.; Bonnivard, E.; Pales Espinosa, E.; Tanguy, A.; Boutet, I.; Guiglielmoni, N.; Flot, J.-F.; Allam, B. Comparative Analysis of the Mercenaria Mercenaria Genome Provides Insights into the Diversity of Transposable Elements and Immune Molecules in Bivalve Mollusks. BMC Genomics 2022, 23, 192. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, Z.; Wang, L.; Yang, C.; Jianga, S.; Song, L. The Immunomodulation of a Novel Tumor Necrosis Factor (CgTNF-1) in Oyster Crassostrea Gigas. Dev. Comp. Immunol. 2014, 45, 291–299. [Google Scholar] [CrossRef]
- Philipp, E.E.R.; Kraemer, L.; Melzner, F.; Poustka, A.J.; Thieme, S.; Findeisen, U.; Schreiber, S.; Rosenstiel, P. Massively Parallel RNA Sequencing Identifies a Complex Immune Gene Repertoire in the Lophotrochozoan Mytilus Edulis. PLoS ONE 2012, 7, e33091. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.; Subramanian, S.; Suwansa-ard, S.; Zhao, M.; O’Connor, W.; Raftos, D.; Elizur, A. The Genome of the Oyster Saccostrea Offers Insight into the Environmental Resilience of Bivalves. DNA Res. 2018, 25, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; Greco, S.; Pallavicini, A. Extensive Tandem Duplication Events Drive the Expansion of the C1q-Domain-Containing Gene Family in Bivalves. Mar. Drugs 2019, 17, 583. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.; Kim, Y.-J.; Markkandan, K.; Shin, W.; Oh, S.; Woo, J.; Yoo, J.; An, H.; Han, K. The Whole-Genome and Transcriptome of the Manila Clam (Ruditapes Philippinarum). Genome Biol. Evol. 2017, 9, 1487–1498. [Google Scholar] [CrossRef]
- Gestal, C.; Pallavicini, A.; Venier, P.; Novoa, B.; Figueras, A. MgC1q, a Novel C1q-Domain-Containing Protein Involved in the Immune Response of Mytilus Galloprovincialis. Dev. Comp. Immunol. 2010, 34, 926–934. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; Manfrin, C.; De Moro, G.; Figueras, A.; Novoa, B.; Venier, P.; Pallavicini, A. The C1q Domain Containing Proteins of the Mediterranean Mussel Mytilus Galloprovincialis: A Widespread and Diverse Family of Immune-Related Molecules. Dev. Comp. Immunol. 2011, 35, 635–643. [Google Scholar] [CrossRef]
- Carland, T.M.; Gerwick, L. The C1q Domain Containing Proteins: Where Do They Come from and What Do They Do? Dev. Comp. Immunol. 2010, 34, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Ahmmed, M.K.; Bhowmik, S.; Giteru, S.G.; Zilani, M.N.H.; Adadi, P.; Islam, S.S.; Kanwugu, O.N.; Haq, M.; Ahmmed, F.; Ng, C.C.W.; et al. An Update of Lectins from Marine Organisms: Characterization, Extraction Methodology, and Potential Biofunctional Applications. Mar. Drugs 2022, 20, 430. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yu, S.; Zhao, J.; Su, X.; Li, T. Cloning and Characterization of a Sialic Acid Binding Lectins (SABL) from Manila Clam Venerupis Philippinarum. Fish Shellfish Immunol. 2011, 30, 1202–1206. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, Y.; Yu, F.; Yu, Z. A Novel Sialic Acid Binding Lectin with Anti-Bacterial Activity from the Hong Kong Oyster (Crassostrea Hongkongensis). Fish Shellfish Immunol. 2011, 31, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, L.; Duan, X.; Cui, L.; Luo, J.; Li, G. Adenovirus Carrying Gene Encoding Haliotis Discus Discus Sialic Acid Binding Lectin Induces Cancer Cell Apoptosis. Mar. Drugs 2014, 12, 3994–4004. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wei, X.; Liu, X.; Xu, J.; Yang, D.; Yang, J.; Fang, J.; Hu, X. Cloning and Transcriptional Analysis of Two Sialic Acid-Binding Lectins (SABLs) from Razor Clam Solen Grandis. Fish Shellfish Immunol. 2012, 32, 578–585. [Google Scholar] [CrossRef]
- Ghosh, S. Sialic Acid Binding Lectins (SABL) from Molluscs, a Review and Insilico Study of SABL from Solen Grandis and Limax Flavus. J. Entomol. Zool. Stud. 2017, 5, 1563–1572. [Google Scholar]
- Peronato, A.; Minervini, G.; Tabarelli, M.; Ballarin, L.; Franchi, N. Characterisation and Functional Role of a Novel C1qDC Protein from a Colonial Ascidian. Dev. Comp. Immunol. 2021, 122, 104077. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Zhang, H.; Zhou, Z.; Siva, V.S.; Song, L. A C1q Domain Containing Protein from Scallop Chlamys Farreri Serving as Pattern Recognition Receptor with Heat-Aggregated IgG Binding Activity. PLoS ONE 2012, 7, e43289. [Google Scholar] [CrossRef]
- Kong, P.; Zhang, H.; Wang, L.; Zhou, Z.; Yang, J.; Zhang, Y.; Qiu, L.; Wang, L.; Song, L. AiC1qDC-1, a Novel gC1q-Domain-Containing Protein from Bay Scallop Argopecten Irradians with Fungi Agglutinating Activity. Dev. Comp. Immunol. 2010, 34, 837–846. [Google Scholar] [CrossRef]
- Lv, Z.; Qiu, L.; Wang, M.; Jia, Z.; Wang, W.; Xin, L.; Liu, Z.; Wang, L.; Song, L. Comparative Study of Three C1q Domain Containing Proteins from Pacific Oyster Crassostrea Gigas. Dev. Comp. Immunol. 2018, 78, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Kong, N.; Sun, J.; Wang, W.; Li, M.; Gong, C.; Dong, M.; Wang, M.; Wang, L.; Song, L. A C1qDC (CgC1qDC-6) with a Collagen-like Domain Mediates Hemocyte Phagocytosis and Migration in Oysters. Dev. Comp. Immunol. 2019, 98, 157–165. [Google Scholar] [CrossRef]
- Zong, Y.; Liu, Z.; Wu, Z.; Han, Z.; Wang, L.; Song, L. A Novel Globular C1q Domain Containing Protein (C1qDC-7) from Crassostrea Gigas Acts as Pattern Recognition Receptor with Broad Recognition Spectrum. Fish Shellfish Immunol. 2019, 84, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Mao, F.; Liu, K.; Bao, Y.; Lin, Y.; Zhang, X.; Xu, D.; Xiang, Z.; Li, J.; Zhang, Y.; Yu, Z. Opsonic Character of the Plasma Proteins in Phagocytosis-Dependent Host Response to Bacterial Infection in a Marine Invertebrate, Crassostrea Gigas. Dev. Comp. Immunol. 2020, 106, 103596. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, W.; Cai, W.; Zhang, Z.; Chen, H.; Ma, S.; Jia, X. Transcriptional Response of Four C1q Domain Containing Protein (C1qDC) Genes from Venerupis Philippinarum Exposed to the Water Soluble Fraction of No.0 Diesel Oil. Ecotoxicol. Environ. Saf. 2016, 132, 40–46. [Google Scholar] [CrossRef]
- Cui, Y.; Wei, Z.; Shen, Y.; Li, C.; Shao, Y.; Zhang, W.; Zhao, X. A Novel C1q-Domain-Containing Protein from Razor Clam Sinonovacula Constricta Mediates G-Bacterial Agglutination as a Pattern Recognition Receptor. Dev. Comp. Immunol. 2018, 79, 166–174. [Google Scholar] [CrossRef]
- Xie, B.; He, Q.; Hao, R.; Zheng, Z.; Du, X. Molecular and Functional Analysis of PmC1qDC in Nacre Formation of Pinctada Fucata Martensii. Fish Shellfish Immunol. 2020, 106, 621–627. [Google Scholar] [CrossRef]
- Liang, X.; Xiong, X.; Cao, Y.; Li, Z.; Chen, J.; Jiao, Y.; Deng, Y.; Du, X. Globular C1q Domain-Containing Protein from Pinctada Fucata Martensii Participates in the Immune Defense Process. Fish Shellfish Immunol. 2022, 123, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Thaimuangphol, W.; Chen, Z.; Li, G.; Gong, X.; Zhao, M.; Chen, Z.; Wang, B.; Wang, Z. A C1q Domain-Containing Protein in Pinctada Fucata Contributes to the Innate Immune Response and Elimination of the Pathogen. Fish Shellfish Immunol. 2022, 131, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.-L.; Jin, M.; Li, X.-C.; Ren, Q.; Lan, J.-F. Four C1q Domain-Containing Proteins Involved in the Innate Immune Response in Hyriopsis cumingii. Fish Shellfish Immunol. 2016, 55, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, W.; Ren, Q. Identification and Function of a Novel C1q Domain-Containing (C1qDC) Protein in Triangle-Shell Pearl Mussel (Hyriopsis Cumingii). Fish Shellfish Immunol. 2016, 58, 612–621. [Google Scholar] [CrossRef]
- Bathige, S.D.N.K.; Umasuthan, N.; Jayasinghe, J.D.H.E.; Godahewa, G.I.; Park, H.-C.; Lee, J. Three Novel C1q Domain Containing Proteins from the Disk Abalone Haliotis Discus Discus: Genomic Organization and Analysis of the Transcriptional Changes in Response to Bacterial Pathogens. Fish Shellfish Immunol. 2016, 56, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-H.; Xiang, L.-X.; Shao, J.-Z. A Novel C1q-Domain-Containing (C1qDC) Protein from Mytilus Coruscus with the Transcriptional Analysis against Marine Pathogens and Heavy Metals. Dev. Comp. Immunol. 2014, 44, 70–75. [Google Scholar] [CrossRef]
- Grinchenko, A.V.; von Kriegsheim, A.; Shved, N.A.; Egorova, A.E.; Ilyaskina, D.V.; Karp, T.D.; Goncharov, N.V.; Petrova, I.Y.; Kumeiko, V.V. A Novel C1q Domain-Containing Protein Isolated from the Mollusk Modiolus Kurilensis Recognizing Glycans Enriched with Acidic Galactans and Mannans. Mar. Drugs 2021, 19, 668. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Kumari, B.; Kumar, R.; Chauhan, V.; Kumar, M. Comparative Functional Analysis of Proteins Containing Low-Complexity Predicted Amyloid Regions. PeerJ 2018, 6, e5823. [Google Scholar] [CrossRef]
- Gaboriaud, C.; Juanhuix, J.; Gruez, A.; Lacroix, M.; Darnault, C.; Pignol, D.; Verger, D.; Fontecilla-Camps, J.C.; Arlaud, G.J. The Crystal Structure of the Globular Head of Complement Protein C1q Provides a Basis for Its Versatile Recognition Properties. J. Biol. Chem. 2003, 278, 46974–46982. [Google Scholar] [CrossRef]
- Shapiro, L.; Scherer, P.E. The Crystal Structure of a Complement-1q Family Protein Suggests an Evolutionary Link to Tumor Necrosis Factor. Curr. Biol. 1998, 8, 335–340. [Google Scholar] [CrossRef]
- Miao, H.; Jia, Y.; Xie, S.; Wang, X.; Zhao, J.; Chu, Y.; Zhou, Z.; Shi, Z.; Song, X.; Li, L. Structural Insights into the C1q Domain of Caprin-2 in Canonical Wnt Signaling. J. Biol. Chem. 2014, 289, 34104–34113. [Google Scholar] [CrossRef]
- Cheng, S.; Seven, A.B.; Wang, J.; Skiniotis, G.; Özkan, E. Conformational Plasticity in the Transsynaptic Neurexin-Cerebellin-Glutamate Receptor Adhesion Complex. Structure 2016, 24, 2163–2173. [Google Scholar] [CrossRef]
- Ressl, S.; Vu, B.K.; Vivona, S.; Martinelli, D.C.; Südhof, T.C.; Brunger, A.T. Structures of C1q-like Proteins Reveal Unique Features among the C1q/TNF Superfamily. Structure 2015, 23, 688–699. [Google Scholar] [CrossRef]
- Tu, X.; Palczewski, K. Crystal Structure of the Globular Domain of C1QTNF5: Implications for Late-Onset Retinal Macular Degeneration. J. Struct. Biol. 2012, 180, 439–446. [Google Scholar] [CrossRef]
- Zhong, C.; Shen, J.; Zhang, H.; Li, G.; Shen, S.; Wang, F.; Hu, K.; Cao, L.; He, Y.; Ding, J. Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions. Cell Rep. 2017, 20, 2328–2340. [Google Scholar] [CrossRef]
- Yuan, H.; Chen, R.; Tariq, M.; Liu, Y.; Sun, Y.; Xia, C. Crystal Structure of Zebrafish Complement 1qA Globular Domain. Protein Sci. 2016, 25, 1883–1889. [Google Scholar] [CrossRef]
- Kvansakul, M.; Bogin, O.; Hohenester, E.; Yayon, A. Crystal Structure of the Collagen A1(VIII) NC1 Trimer. Matrix Biol. 2003, 22, 145–152. [Google Scholar] [CrossRef]
- Bogin, O.; Kvansakul, M.; Rom, E.; Singer, J.; Yayon, A.; Hohenester, E. Insight into Schmid Metaphyseal Chondrodysplasia from the Crystal Structure of the Collagen X NC1 Domain Trimer. Structure 2002, 10, 165–173. [Google Scholar] [CrossRef]
- Kishore, U.; Gaboriaud, C.; Waters, P.; Shrive, A.K.; Greenhough, T.J.; Reid, K.B.M.; Sim, R.B.; Arlaud, G.J. C1q and Tumor Necrosis Factor Superfamily: Modularity and Versatility. Trends Immunol. 2004, 25, 551–561. [Google Scholar] [CrossRef]
- Jones, E.Y.; Stuart, D.I.; Walker, N.P.C. Structure of Tumour Necrosis Factor. Nature 1989, 338, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Kishore, U.; Reid, K.B.M. C1q: Structure, Function, and Receptors. Immunopharmacology 2000, 49, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Kouser, L.; Madhukaran, S.P.; Shastri, A.; Saraon, A.; Ferluga, J.; Al-Mozaini, M.; Kishore, U. Emerging and Novel Functions of Complement Protein C1q. Front. Immunol. 2015, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Son, M. Understanding the Contextual Functions of C1q and LAIR-1 and Their Applications. Exp. Mol. Med. 2022, 54, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.D. Adiponectin: Role in Physiology and Pathophysiology. Int. J. Prev. Med. 2020, 11, 136. [Google Scholar] [CrossRef]
- Khoramipour, K.; Chamari, K.; Hekmatikar, A.A.; Ziyaiyan, A.; Taherkhani, S.; Elguindy, N.M.; Bragazzi, N.L. Adiponectin: Structure, Physiological Functions, Role in Diseases, and Effects of Nutrition. Nutrients 2021, 13, 1180. [Google Scholar] [CrossRef]
- Südhof, T.C. Cerebellin–Neurexin Complexes Instructing Synapse Properties. Curr. Opin. Neurobiol. 2023, 81, 102727. [Google Scholar] [CrossRef]
- Ding, Y.; Xi, Y.; Chen, T.; Wang, J.; Tao, D.; Wu, Z.-L.; Li, Y.; Li, C.; Zeng, R.; Li, L. Caprin-2 Enhances Canonical Wnt Signaling through Regulating LRP5/6 Phosphorylation. J. Cell Biol. 2008, 182, 865–872. [Google Scholar] [CrossRef]
- Lorén, C.E.; Schrader, J.W.; Ahlgren, U.; Gunhaga, L. FGF Signals Induce Caprin2 Expression in the Vertebrate Lens. Differ. Res. Biol. Divers. 2009, 77, 386–394. [Google Scholar] [CrossRef]
- Konopacka, A.; Greenwood, M.; Loh, S.-Y.; Paton, J.; Murphy, D. RNA Binding Protein Caprin-2 Is a Pivotal Regulator of the Central Osmotic Defense Response. eLife 2015, 4, e09656. [Google Scholar] [CrossRef]
- Qiu, L.; Zhou, R.; Zhou, L.; Yang, S.; Wu, J. CAPRIN2 Upregulation by LINC00941 Promotes Nasopharyngeal Carcinoma Ferroptosis Resistance and Metastatic Colonization through HMGCR. Front. Oncol. 2022, 12, 931749. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, L.; Zhang, D.; Jiang, Q.; Sun, R.; Wang, H.; Zhang, H.; Song, L. A Novel Multi-Domain C1qDC Protein from Zhikong Scallop Chlamys Farreri Provides New Insights into the Function of Invertebrate C1qDC Proteins. Dev. Comp. Immunol. 2015, 52, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, L.; Kong, P.; Yang, J.; Zhang, H.; Wang, M.; Zhou, Z.; Qiu, L.; Song, L. A Novel C1qDC Protein Acting as Pattern Recognition Receptor in Scallop Argopecten Irradians. Fish Shellfish Immunol. 2012, 33, 427–435. [Google Scholar] [CrossRef]
- Jiang, S.; Li, H.; Zhang, D.; Zhang, H.; Wang, L.; Sun, J.; Song, L. A C1q Domain Containing Protein from Crassostrea Gigas Serves as Pattern Recognition Receptor and Opsonin with High Binding Affinity to LPS. Fish Shellfish Immunol. 2015, 45, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-Align: A Protein Structure Alignment Algorithm Based on the TM-Score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Reynolds, C.R.; Islam, S.A.; Sternberg, M.J.E. EzMol: A Web Server Wizard for the Rapid Visualization and Image Production of Protein and Nucleic Acid Structures. J. Mol. Biol. 2018, 430, 2244–2248. [Google Scholar] [CrossRef]
- Zhang, H.; Song, L.; Li, C.; Zhao, J.; Wang, H.; Qiu, L.; Ni, D.; Zhang, Y. A Novel C1q-Domain-Containing Protein from Zhikong Scallop Chlamys Farreri with Lipopolysaccharide Binding Activity. Fish Shellfish Immunol. 2008, 25, 281–289. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, L.; Jia, Z.; Sun, J.; Wang, W.; Liu, Z.; Qiu, L.; Wang, M.; Song, L. Hemolymph C1qDC Promotes the Phagocytosis of Oyster Crassostrea Gigas Hemocytes by Interacting with the Membrane Receptor β-Integrin. Dev. Comp. Immunol. 2019, 98, 42–53. [Google Scholar] [CrossRef]
- Li, Y.; Niu, D.; Bai, Y.; Lan, T.; Peng, M.; Dong, Z.; Li, J. Characterization of the ScghC1q-1 Gene in Sinonovacula Constricta and Its Role in Innate Immune Responses. Dev. Comp. Immunol. 2019, 94, 16–21. [Google Scholar] [CrossRef]
- Li, Y.; Niu, D.; Bai, Y.; Lan, T.; Peng, M.; Dong, Z.; Li, J. Identification of a Novel C1q Complement Component in Razor Clam Sinonovacula Constricta and Its Role in Antibacterial Activity. Fish Shellfish Immunol. 2019, 87, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Li, C.; Zheng, Z.; Du, X. Novel Globular C1q Domain-Containing Protein (PmC1qDC-1) Participates in Shell Formation and Responses to Pathogen-Associated Molecular Patterns Stimulation in Pinctada Fucata Martensii. Sci. Rep. 2021, 11, 1105. [Google Scholar] [CrossRef] [PubMed]
- Sokolnikova, Y.; Mokrina, M.; Magarlamov, T.; Grinchenko, A.; Kumeiko, V. Specification of Hemocyte Subpopulations Based on Immune-Related Activities and the Production of the Agglutinin MkC1qDC in the Bivalve Modiolus Kurilensis. Heliyon 2023, 9, e15577. [Google Scholar] [CrossRef]
- Chi, C.; Giri, S.S.; Jun, J.W.; Kim, S.W.; Kim, H.J.; Kang, J.W.; Park, S.C. Detoxification- and Immune-Related Transcriptomic Analysis of Gills from Bay Scallops (Argopecten Irradians) in Response to Algal Toxin Okadaic Acid. Toxins 2018, 10, 308. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, S.; Cheng, H.; Chen, X.; Shen, X.; Cai, Y. Dynamic Transcriptome Response in Meretrix Meretrix to Aroclor 1254 Exposure. Ecotoxicol. Environ. Saf. 2021, 207, 111485. [Google Scholar] [CrossRef]
- Gomes, T.; Chora, S.; Pereira, C.G.; Cardoso, C.; Bebianno, M.J. Proteomic Response of Mussels Mytilus Galloprovincialis Exposed to CuO NPs and Cu2+: An Exploratory Biomarker Discovery. Aquat. Toxicol. Amst. Neth. 2014, 155, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Gorbushin, A.M. Immune Repertoire in the Transcriptome of Littorina Littorea Reveals New Trends in Lophotrochozoan Proto-Complement Evolution. Dev. Comp. Immunol. 2018, 84, 250–263. [Google Scholar] [CrossRef]
- Peng, M.; Li, Z.; Cardoso, J.C.R.; Niu, D.; Liu, X.; Dong, Z.; Li, J.; Power, D.M. Domain-Dependent Evolution Explains Functional Homology of Protostome and Deuterostome Complement C3-Like Proteins. Front. Immunol. 2022, 13, 840861. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, K.; Li, J.; Wang, X.; Ye, Y.; Qi, P. Molecular Characterization of Complement Component 3 (C3) in Mytilus Coruscus Improves Our Understanding of Bivalve Complement System. Fish Shellfish Immunol. 2018, 76, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liang, X.; Li, G.; Liufu, B.; Lin, K.; Li, J.; Wang, J.; Wang, B. Molecular Characterization of Complement Component 3 (C3) in the Pearl Oyster Pinctada Fucata Improves Our Understanding of the Primitive Complement System in Bivalve. Front. Immunol. 2021, 12, 652805. [Google Scholar] [CrossRef]
- Leprêtre, M.; Almunia, C.; Armengaud, J.; Le Guernic, A.; Salvador, A.; Geffard, A.; Palos-Ladeiro, M. Identification of Immune-Related Proteins of Dreissena Polymorpha Hemocytes and Plasma Involved in Host-Microbe Interactions by Differential Proteomics. Sci. Rep. 2020, 10, 6226. [Google Scholar] [CrossRef]
- Moreira, R.; Balseiro, P.; Planas, J.V.; Fuste, B.; Beltran, S.; Novoa, B.; Figueras, A. Transcriptomics of in Vitro Immune-Stimulated Hemocytes from the Manila Clam Ruditapes Philippinarum Using High-Throughput Sequencing. PLoS ONE 2012, 7, e35009. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, L.; Zhu, Y.; Zhang, G.; Guo, X. Transcriptome Analysis Reveals a Rich Gene Set Related to Innate Immunity in the Eastern Oyster (Crassostrea Virginica). Mar. Biotechnol. 2014, 16, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; Venier, P. An Updated Molecular Basis for Mussel Immunity. Fish Shellfish Immunol. 2015, 46, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, R.K.; Ferrier, G.A.; Kim, S.J.; Ogorzalek Loo, R.R.; Zimmer, C.A.; Loo, J.A. Keystone Predation and Molecules of Keystone Significance. Ecology 2017, 98, 1710–1721. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Harimoto, K.; Fuji, R.; Liu, J.; Li, L.; Wang, P.; Akaike, T.; Wang, Z. Pinctada Fucata Mantle Gene 4 (PFMG4) from Pearl Oyster Mantle Enhances Osteoblast Differentiation. Biosci. Biotechnol. Biochem. 2015, 79, 558–565. [Google Scholar] [CrossRef]
- Qin, C.; Pan, Q.; Qi, Q.; Fan, M.; Sun, J.; Li, N.; Liao, Z. In-Depth Proteomic Analysis of the Byssus from Marine Mussel Mytilus Coruscus. J. Proteomics 2016, 144, 87–98. [Google Scholar] [CrossRef]
- Tahtouh, M.; Croq, F.; Vizioli, J.; Sautiere, P.-E.; Van Camp, C.; Salzet, M.; Daha, M.R.; Pestel, J.; Lefebvre, C. Evidence for a Novel Chemotactic C1q Domain-Containing Factor in the Leech Nerve Cord. Mol. Immunol. 2009, 46, 523–531. [Google Scholar] [CrossRef]
- Tahtouh, M.; Garçon-Bocquet, A.; Croq, F.; Vizioli, J.; Sautière, P.-E.; Van Camp, C.; Salzet, M.; Nagnan-le Meillour, P.; Pestel, J.; Lefebvre, C. Interaction of HmC1q with Leech Microglial Cells: Involvement of C1qBP-Related Molecule in the Induction of Cell Chemotaxis. J. Neuroinflammation 2012, 9, 37. [Google Scholar] [CrossRef]
- Matsumoto, R.; Shibata, T.F.; Kohtsuka, H.; Sekifuji, M.; Sugii, N.; Nakajima, H.; Kojima, N.; Fujii, Y.; Kawsar, S.M.A.; Yasumitsu, H.; et al. Glycomics of a Novel Type-2 N-Acetyllactosamine-Specific Lectin Purified from the Feather Star, Oxycomanthus Japonicus (Pelmatozoa: Crinoidea). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 158, 266–273. [Google Scholar] [CrossRef]
- Mahla, R.S.; Reddy, M.C.; Prasad, D.V.R.; Kumar, H. Sweeten PAMPs: Role of Sugar Complexed PAMPs in Innate Immunity and Vaccine Biology. Front. Immunol. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed]
- Silva-Gomes, S.; Decout, A.; Nigou, J. Pathogen-Associated Molecular Patterns (PAMPs). In Encyclopedia of Inflammatory Diseases; Parnham, M., Ed.; Springer: Basel, Switzerland, 2015; pp. 1–16. ISBN 978-3-0348-0620-6. [Google Scholar]
- Han, B.; Baruah, K.; Cox, E.; Vanrompay, D.; Bossier, P. Structure-Functional Activity Relationship of β-Glucans From the Perspective of Immunomodulation: A Mini-Review. Front. Immunol. 2020, 11, 658. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 28513415. [Google Scholar] [CrossRef] [PubMed]
- Raina, S. Lipopolysaccharides: Regulated Biosynthesis and Structural Diversity. Int. J. Mol. Sci. 2023, 24, 7498. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.D.; Vollmer, W.; Foster, S.J. Different Walls for Rods and Balls: The Diversity of Peptidoglycan. Mol. Microbiol. 2014, 91, 862–874. [Google Scholar] [CrossRef]
- Garde, S.; Chodisetti, P.K.; Reddy, M. Peptidoglycan: Structure, Synthesis, and Regulation. EcoSal Plus 2021, 9. [Google Scholar] [CrossRef]
- Whitfield, C.; Williams, D.M.; Kelly, S.D. Lipopolysaccharide O-Antigens-Bacterial Glycans Made to Measure. J. Biol. Chem. 2020, 295, 10593–10609. [Google Scholar] [CrossRef]
- Mishra, A.; Behura, A.; Mawatwal, S.; Kumar, A.; Naik, L.; Mohanty, S.S.; Manna, D.; Dokania, P.; Mishra, A.; Patra, S.K.; et al. Structure-Function and Application of Plant Lectins in Disease Biology and Immunity. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2019, 134, 110827. [Google Scholar] [CrossRef]
- Patlay, A.A.; Belousov, A.S.; Silant’ev, V.E.; Shatilov, R.A.; Shmelev, M.E.; Kovalev, V.V.; Perminova, I.V.; Baklanov, I.N.; Kumeiko, V.V. Preparation and Characterization of Hydrogel Films and Nanoparticles Based on Low-Esterified Pectin for Anticancer Applications. Polymers 2023, 15, 3280. [Google Scholar] [CrossRef]
- Silant’ev, V.E.; Shmelev, M.E.; Belousov, A.S.; Patlay, A.A.; Shatilov, R.A.; Farniev, V.M.; Kumeiko, V.V. How to Develop Drug Delivery System Based on Carbohydrate Nanoparticles Targeted to Brain Tumors. Polymers 2023, 15, 2516. [Google Scholar] [CrossRef]
- Dennis, J.W.; Laferté, S.; Waghorne, C.; Breitman, M.L.; Kerbel, R.S. Β1-6 Branching of Asn-Linked Oligosaccharides Is Directly Associated with Metastasis. Science 1986, 236, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Dennis, J.W.; Laferte, S. Tumor Cell Surface Carbohydrate and the Metastatic Phenotype. Cancer Metastasis Rev. 1987, 5, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Mei, S.; Cui, L.; Zhao, Z.; Chen, J.; Wu, T.; Li, G. Marine Lectins DlFBL and HddSBL Fused with Soluble Coxsackie-Adenovirus Receptor Facilitate Adenovirus Infection in Cancer Cells BUT Have Different Effects on Cell Survival. Mar. Drugs 2017, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Mei, S.; Cheng, J.; Wu, T.; Luo, J. Haliotis Discus Discus Sialic Acid-Binding Lectin Reduces the Oncolytic Vaccinia Virus Induced Toxicity in a Glioblastoma Mouse Model. Mar. Drugs 2018, 16, 141. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global View of Human Protein Glycosylation Pathways and Functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in Cancer: Mechanisms and Clinical Implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Mereiter, S.; Balmaña, M.; Campos, D.; Gomes, J.; Reis, C.A. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell 2019, 36, 6–16. [Google Scholar] [CrossRef]
- Peixoto, A.; Relvas-Santos, M.; Azevedo, R.; Santos, L.L.; Ferreira, J.A. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front. Oncol. 2019, 9, 380. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, J.; Lubman, D.M.; Gao, C. Aberrant Glycosylation and Cancer Biomarker Discovery: A Promising and Thorny Journey. Clin. Chem. Lab. Med. 2019, 57, 407–416. [Google Scholar] [CrossRef]
- Costa, A.F.; Campos, D.; Reis, C.A.; Gomes, C. Targeting Glycosylation: A New Road for Cancer Drug Discovery. Trends Cancer 2020, 6, 757–766. [Google Scholar] [CrossRef]
- Thomas, D.; Rathinavel, A.K.; Radhakrishnan, P. Altered Glycosylation in Cancer: A Promising Target for Biomarkers and Therapeutics. Biochim. Biophys. Acta BBA Rev. Cancer 2021, 1875, 188464. [Google Scholar] [CrossRef] [PubMed]
- De Leoz, M.L.A.; Young, L.J.T.; An, H.J.; Kronewitter, S.R.; Kim, J.; Miyamoto, S.; Borowsky, A.D.; Chew, H.K.; Lebrilla, C.B. High-Mannose Glycans Are Elevated during Breast Cancer Progression. Mol. Cell. Proteomics MCP 2011, 10, M110.002717. [Google Scholar] [CrossRef] [PubMed]
- Butler, W.; Huang, J. Glycosylation Changes in Prostate Cancer Progression. Front. Oncol. 2021, 11, 809170. [Google Scholar] [CrossRef] [PubMed]
- Boyaval, F.; Dalebout, H.; Van Zeijl, R.; Wang, W.; Fariña-Sarasqueta, A.; Lageveen-Kammeijer, G.S.M.; Boonstra, J.J.; McDonnell, L.A.; Wuhrer, M.; Morreau, H.; et al. High-Mannose N-Glycans as Malignant Progression Markers in Early-Stage Colorectal Cancer. Cancers 2022, 14, 1552. [Google Scholar] [CrossRef] [PubMed]
- Park, D.D.; Phoomak, C.; Xu, G.; Olney, L.P.; Tran, K.A.; Park, S.S.; Haigh, N.E.; Luxardi, G.; Lert-itthiporn, W.; Shimoda, M.; et al. Metastasis of Cholangiocarcinoma Is Promoted by Extended High-Mannose Glycans. Proc. Natl. Acad. Sci. USA 2020, 117, 7633–7644. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Taniguchi, N. Enzymes for N-Glycan Branching and Their Genetic and Nongenetic Regulation in Cancer. Biomolecules 2016, 6, 25. [Google Scholar] [CrossRef]
- Chen, Q.; Tan, Z.; Guan, F.; Ren, Y. The Essential Functions and Detection of Bisecting GlcNAc in Cell Biology. Front. Chem. 2020, 8, 511. [Google Scholar] [CrossRef]



| Species | Protein | Hm | Dg | Ms | Mn | Gl | Gn | Other | References |
|---|---|---|---|---|---|---|---|---|---|
| V. philippinarum (R. philippinarum) | VpSABL | 1 | 40 | 1 | 160 | 20 | [40] | ||
| S. grandis | SgSABL-1 | 1 | 221 | 1 | 2 | 209 | 3 | [43] | |
| C. hongkongensis (M. hongkongensis) | Ch-salectin | 1 | 2 | 3 | 1 | 6 | 6 | heart: 2 | [41] |
| C. farreri | CfC1qDC | 1 | 16 | 16 | 16 | kidney: 23 | [92] | ||
| CfC1qDC-2 | 1 | 5972 | 67 | 46 | 12 | 425 | kidney: 62 | [86] | |
| A. irradians | AiC1qDC-1 | 1 | 5888 | 20 | 8 | 4 | heart: 14 | [47] | |
| AiC1qDC-2 | 483 | 1 | 51 | 22 | 78 | [87] | |||
| M. galloprovincialis | MgC1q | 4000 | 2 | 1 | 1 | 2 | 450 | [36] | |
| M. coruscus (M. unguiculatus) | McC1qDC | 670 | 1 | 1 | 3 | 7 | 4 | foot: 1 | [60] |
| C. gigas (M. gigas) | CgC1qDC-1 | 81 | 4 | 1 | 82 | 3 | 2 | [88] | |
| CgC1qDC-2 | 70 | 38 | 58 | 0 | 5 | 1 | [48] | ||
| CgC1qDC-3 | 20 | 3 | 8 | 260 | 45 | 1 | |||
| CgC1qDC-4 | 10 | 1 | 1 | 30 | 1 | 11 | |||
| CgC1qDC-5 | 15 | 1 | 1 | 13 | 1 | 21 | [93] | ||
| CgC1qDC-6 | 7 | 1 | 4 | 4 | 1 | 1 | [49] | ||
| CgC1qDC-7 | 8 | 1 | 1 | 2 | 5 | 2 | labial palp: 1 | [50] | |
| p1-CgC1q | 70 | 80 | 10 | 20 | 10 | 20 | heart: 1 | [51] | |
| S. constricta | ScC1qDC | 4 | 1500 | 3 | foot: 1 siphon: 4 | [53] | |||
| ScghC1q-1 | 8 | 58000 | 6 | 9 | 12 | foot: 1 siphon: 10 | [94] | ||
| Sc-ghC1q | 5 | 15000 | 2 | 4 | 17 | foot: 2 siphon: 3 | [95] | ||
| P. fucata | PmC1qDC | 70 | 1 | 60 | 60 | foot: 10 | [54] | ||
| PmC1qDC-1 | 1 | 2 | 4–47 | 4 | foot: 15 | [96] | |||
| Pf-ghC1q | 2 | 1 | 3 | 2 | 4 | [56] | |||
| H. cumingii | HcC1qDC5 | 1 | 12 | 8 | 3 | [58] | |||
| H. discus discus | AbC1qDC1 | 1 | 2100 | 150 | 100 | 5 | digestive tract: 0 | [59] | |
| AbC1qDC2 | 1 | 55 | 60 | 60 | 2 | digestive tract: 1 | |||
| AbC1qDC3 | 1 | 15 | 20 | 7 | 0 | digestive tract: 4 |
| Species | Proteins | PAMPs | G- Bacteria | G+ Bacteria | Other | References |
|---|---|---|---|---|---|---|
| V. philippinarum (R. philippinarum) | VpSABL | V. anguillarum | [40] | |||
| VpC1qDC1 VpC1qDC2 VpC1qDC3 VpC1qDC4 | soluble fraction of No.0 diesel oil | [52] | ||||
| S. grandis | SgSABL-1 | LPS PGN GLU | [43] | |||
| C. hongkongensis (M. hongkongensis) | Ch-salectin | V. anguillarum | [41] | |||
| C. farreri | CfC1qDC | LPS PGN GLU polyI:C |
Listonella
anguillarum | [46,92] | ||
| CfC1qDC-2 |
LPS PGN GLU polyI:C | [86] | ||||
| A. irradians | AiC1qDC-1 | L. anguillarum | M. luteus | Fungi Pichia pastoris | [47,87] | |
| M. galloprovincialis | MgC1q | V. anguillarum | M. lysodeikticus | [36] | ||
| M. coruscus (M. unguiculatus) | McC1qDC | V. alginolyticus Vibrio harveyi | Cu2+ Cd2+ | [60] | ||
| C. gigas (M. gigas) | CgC1qDC-1 | V. splendidus | [88] | |||
| CgC1qDC-2 CgC1qDC-3 CgC1qDC-4 | V. splendidus V. anguillarum | [48] | ||||
| CgC1qDC-5 | LPS | V. splendidus V. anguillarum | [93] | |||
| p1-CgC1q | V. alginolyticus | [51] | ||||
| S. constricta | ScghC1q-1 | V. anguillarum | S. aureus | [94] | ||
| Sc-ghC1q | V. anguillarum | S. aureus | [95] | |||
| P. fucata | PmC1qDC-1 | LPS PGN polyI:C | [55,96] | |||
| Pf-ghC1q | V. alginolyticus | [56] | ||||
| H. cumingii | HcC1qDC1 HcC1qDC2 HcC1qDC3 HcC1qDC4 HcC1qDC5 | A. hydrophila | S. aureus | [57,58] | ||
| H. discus discus | AbC1qDC1 AbC1qDC2 AbC1qDC3 | V. parahaemolyticus | L. monocytogenes | [59] | ||
| B. schlosseri | BsC1qDC * | Bacillus clausii | Fungi Saccharomyces cerevisiae | [45] |
| Species | Proteins | Functions or Involving Process | References |
|---|---|---|---|
| C. farreri | CfC1qDC CfC1qDC-2 | embryonic development | [46] [86] |
| M. galloprovincialis | MgC1q | embryonic development | [36] |
| B. schlosseri | BsC1qDC | embryonic development | [45] |
| P. fucata martensii | PmC1qDC-1 | embryonic development shell formation and recovery | [96] [54,96] |
| M. californianus, M. galloprovincialis | KEYSTONEin | shell formation and recovery chemoattractant for predatory starfish | [109] |
| M. coruscus (M. unguiculatus) | C1qDCs | byssus filaments formation | [111] |
| H. medicinalis | HmC1q | microglia activation and nerve system development | [112,113] |
| A. mellifera N. vitripennis | AmC1q-VP NvC1q-VP | toxin transporters | [21] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grinchenko, A.; Buriak, I.; Kumeiko, V. Invertebrate C1q Domain-Containing Proteins: Molecular Structure, Functional Properties and Biomedical Potential. Mar. Drugs 2023, 21, 570. https://doi.org/10.3390/md21110570
Grinchenko A, Buriak I, Kumeiko V. Invertebrate C1q Domain-Containing Proteins: Molecular Structure, Functional Properties and Biomedical Potential. Marine Drugs. 2023; 21(11):570. https://doi.org/10.3390/md21110570
Chicago/Turabian StyleGrinchenko, Andrei, Ivan Buriak, and Vadim Kumeiko. 2023. "Invertebrate C1q Domain-Containing Proteins: Molecular Structure, Functional Properties and Biomedical Potential" Marine Drugs 21, no. 11: 570. https://doi.org/10.3390/md21110570
APA StyleGrinchenko, A., Buriak, I., & Kumeiko, V. (2023). Invertebrate C1q Domain-Containing Proteins: Molecular Structure, Functional Properties and Biomedical Potential. Marine Drugs, 21(11), 570. https://doi.org/10.3390/md21110570





