Coenzyme Q in Thraustochytrium sp. RT2316-16: Effect of the Medium Composition
Abstract
:1. Introduction
2. Results
2.1. Identification and Quantification of CoQ10
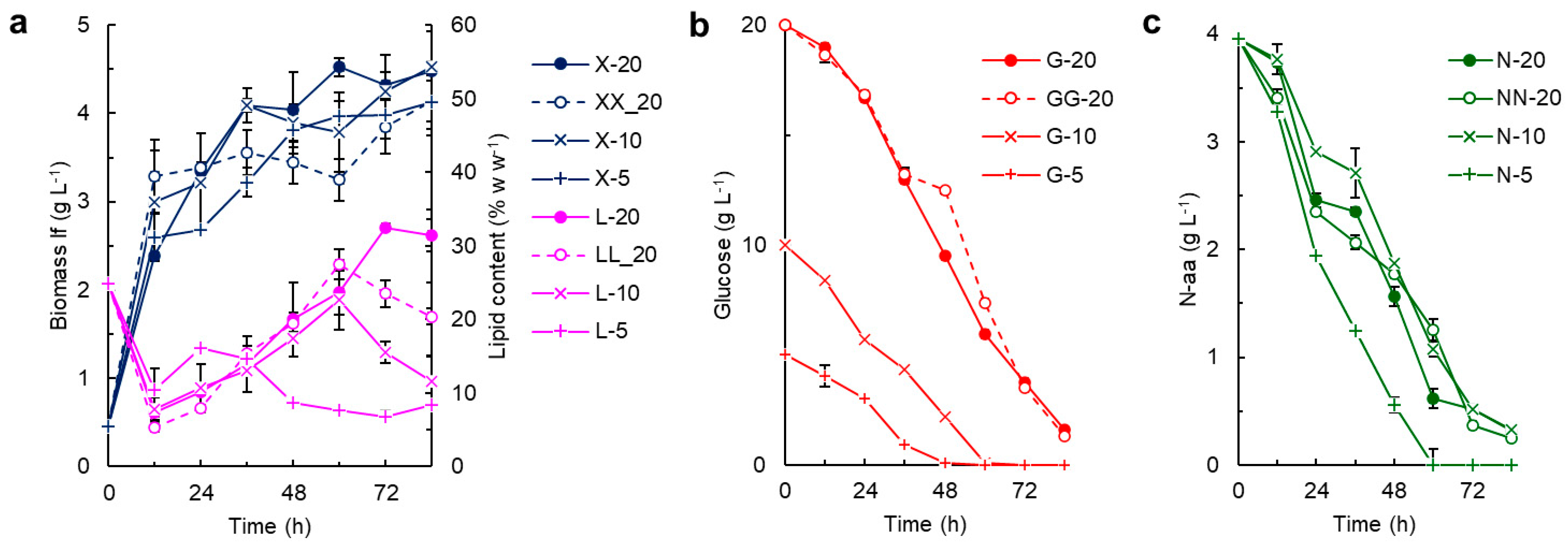
2.2. Effects of the Nitrogen Source on the CoQ Content in RT2316-16
2.3. Precursors of CoQ Ring Were Not Required for Its Biosynthesis in RT2316-16
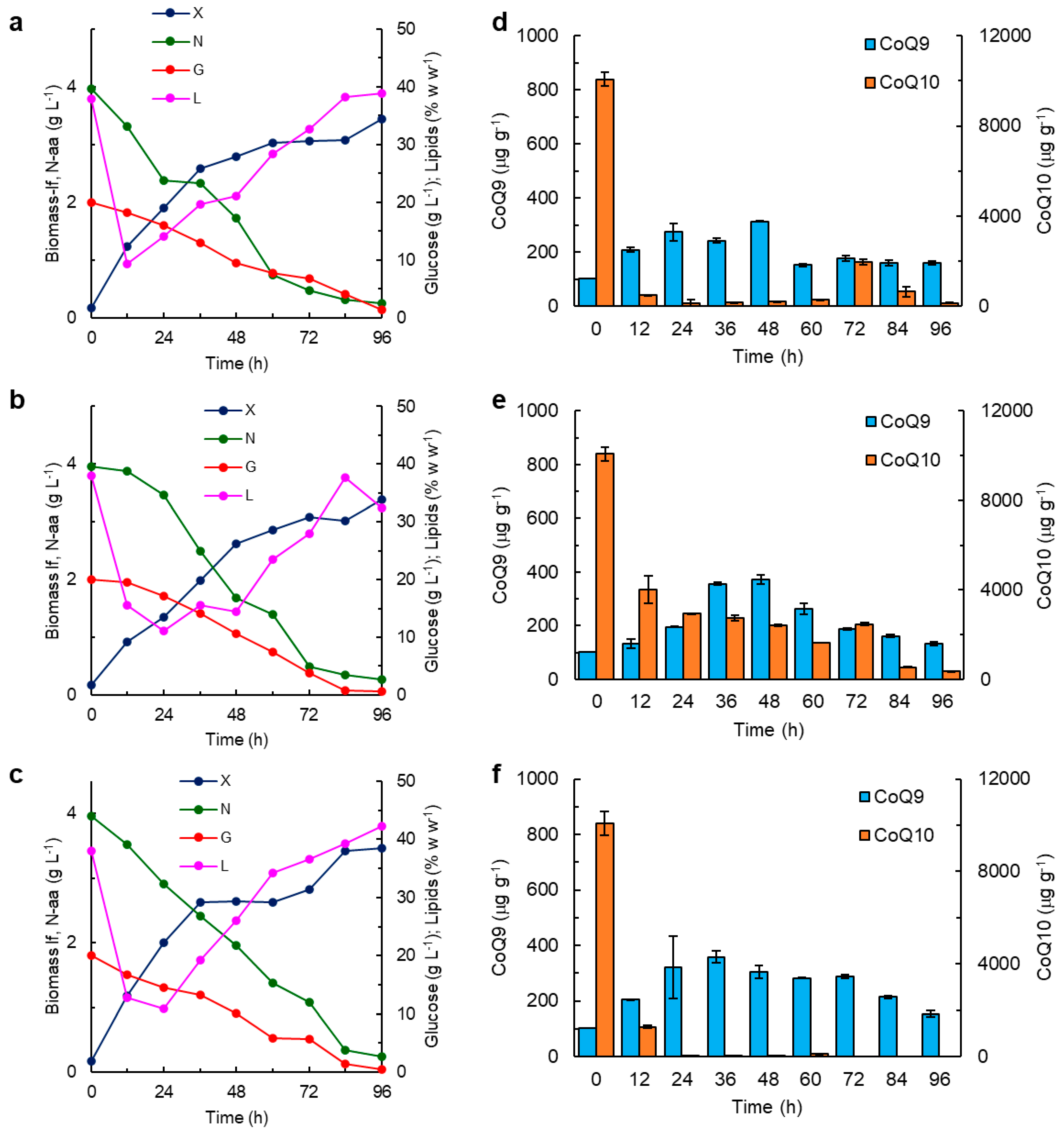
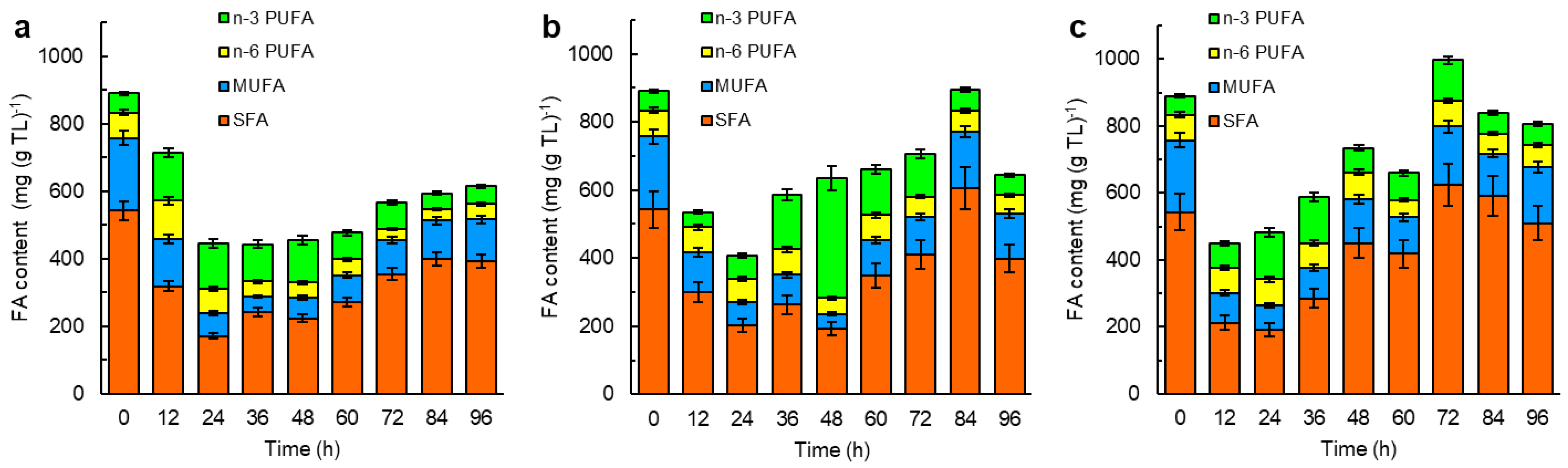
2.4. The CoQ Content in the Biomass: Influence of the Growth Stage and the Composition of the Culture Medium
3. Discussion
4. Materials and Methods
4.1. Microorganism and Inoculum Preparation
4.2. Effect of the Culture Medium Composition on the Production of CoQ10
4.3. Analyses
4.3.1. Cell Dry Weight
4.3.2. Residual Reducing Sugars
4.3.3. Residual Amino Acids
4.3.4. Extraction and Quantification of CoQ
4.3.5. UHPLC-MS/MS Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Cirilli, I.; Damiani, E.; Dludla, P.V.; Hargreaves, I.; Marcheggiani, F.; Millichap, L.E.; Orlando, P.; Silvestri, S.; Tiano, L. Role of coenzyme Q10 in health and disease: An update on the last 10 years (2010–2020). Antioxidants 2021, 10, 1325. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Díaz, C.; Barroso, M.P.; Navas, P. Plasma membrane coenzyme Q10 and growth control. Protoplasma 2000, 214, 19–23. [Google Scholar] [CrossRef]
- Fontaine, E.; Ichas, F.; Bernardi, P. A ubiquinone-binding site regulates the mitochondrial permeability transition pore. J. Biol. Chem. 1998, 273, 25734–25740. [Google Scholar] [CrossRef] [PubMed]
- Schniertshauer, D.; Gebhard, D.; Bergemann, J. Age-dependent loss of mitochondrial function in epithelial tissue can be reversed by coenzyme Q10. J. Aging Res. 2018, 2018, 6354680. [Google Scholar] [CrossRef] [PubMed]
- Bentinger, M.; Tekle, M.; Dallner, G. Coenzyme Q-biosynthesis and functions. Biochem. Biophys. Res. Commun. 2010, 396, 74–79. [Google Scholar] [CrossRef]
- Navas, P.; Cascajo, M.V.; Alcázar-Fabra, M.; Hernández-Camacho, J.D.; Sánchez-Cuesta, A.; Rodríguez, A.B.C.; Ballesteros-Simarro, M.; Arroyo-Luque, A.; Rodríguez-Aguilera, J.C.; Fernández-Ayala, D.J.M.; et al. Secondary CoQ10 deficiency, bioenergetics unbalance in disease and aging. Biofactors 2021, 47, 551–569. [Google Scholar] [CrossRef]
- Hernández-Camacho, J.D.; Bernier, M.; López-Lluch, G.; Navas, P. Coenzyme Q10 supplementation in aging and disease. Front. Physiol. 2018, 9, 44. [Google Scholar] [CrossRef]
- Hargreaves, I.; Heaton, R.A.; Mantle, D. Disorders of human coenzyme Q10 metabolism: An overview. Int. J. Mol. Sci. 2020, 21, 6695. [Google Scholar] [CrossRef]
- He, S.; Lu, H.; Zhang, G.; Ren, Z. Production of coenzyme Q10 by purple non-sulfur bacteria: Current development and future prospect. J. Clean. Prod. 2021, 307, 127326. [Google Scholar] [CrossRef]
- Burgardt, A.; Moustafa, A.; Persicke, M.; Sproß, J.; Patschkowski, T.; Risse, J.M.; Peters-Wendisch, P.; Lee, J.-H.; Wendisch, V.F. Coenzyme Q10 biosynthesis established in the non-ubiquinone containing Corynebacterium glutamicum by metabolic engineering. Front. Bioeng. Biotechnol. 2021, 9, 650961. [Google Scholar] [CrossRef]
- Siebert, M.; Severin, K.; Heide, L. Formation of 4-hydroxybenzoate in Escherichia coli: Characterization of the ubiC gene and its encoded enzyme chorismate pyruvate-lyase. Microbiology 1994, 140, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.E.; Dialamieh, G.H.; Bentley, R.; Springer, C.M.; Ramsey, V.G. Studies on coenzyme Q. Pattern of labeling in coenzyme Q9 after administration of isotopic acetate and aromatic amino acids to rats. J. Biol. Chem. 1965, 240, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.E. Biosynthesis of ubiquinones in animals. Vitam. Horm. 1967, 24, 551–574. [Google Scholar] [CrossRef]
- Pierrel, F.; Hamelin, O.; Douki, T.; Kieffer-Jaquinod, S.; Muhlenhoff, U.; Ozeir, M.; Lill, R.; Fontecave, M. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 2010, 17, 449–459. [Google Scholar] [CrossRef]
- Lombard, J.; Moreira, D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol. Biol. Evol. 2011, 28, 87–99. [Google Scholar] [CrossRef]
- Stefely, J.; Pagliarini, D. Biochemistry of mitochondrial coenzyme Q biosynthesis. Trends Biochem. Sci. 2017, 42, 824–843. [Google Scholar] [CrossRef]
- Rohmer, M.; Knani, M.; Simonin, P.; Sutter, B.; Sahm, H. Isoprenoid biosynthesis in bacteria: A novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 1993, 15, 517–524. [Google Scholar] [CrossRef]
- Li, W. Bringing bioactive compounds into membranes: The UbiA superfamily of intramembrane aromatic prenyltransferases. Trends Biochem. Sci. 2016, 41, 356–370. [Google Scholar] [CrossRef]
- González-Mariscal, I.; Martin-Montalvo, A.; Vázquez-Fonseca, L.; Pomares-Viciana, T.; Sánchez-Cuesta, A.; Fernández-Ayala, D.; Navas, P.; Santos-Ocana, C. The mitochondrial phosphatase PPTC7 orchestrates mitochondrial metabolism regulating coenzyme Q10 biosynthesis. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 1235–1248. [Google Scholar] [CrossRef]
- Gu, S.; Yao, J.; Yuan, Q.; Xue, P.; Zheng, Z.; Yu, Z. Kinetics of Agrobacterium tumefaciens ubiquinone-10 batch production. Process Biochem. 2006, 41, 1908–1912. [Google Scholar] [CrossRef]
- Kien, N.; Kong, I.; Lee, M.; Kim, J. Coenzyme Q10 production in a 150-l reactor by a mutant strain of Rhodobacter sphaeroides. J. Ind. Microbiol. Biotechnol. 2010, 37, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Tokdar, P.; Ranadive, P.; Deshmukh, S.; Khora, S. Optimization of fermentation process conditions for the production of CoQ10 using Paracoccus denitrificans ATCC 19367 fusant strain PF-P1. Int. J. Eng. Res. Technol. 2017, 6, 135–143. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, W.; Zhong, W.; Zhong, L.; Fang, J.; Li, X.; Wu, S.; Chen, J. Coenzyme Q10 production by Sphingomonas sp. ZUTE03 with novel precursors isolated from tobacco waste in a two-phase conversion system. J. Microbiol. Biotechnol. 2011, 21, 494–502. [Google Scholar] [CrossRef]
- Zhong, L.; Kong, Z.; Bian, T.; Cheng, C.; Shu, M.; Shi, Y.; Pan, J.; Therese, B.; Zhong, W. Identification and characterization of a novel coenzyme Q10-producing strain, Proteus penneri CA8, newly isolated from Ebolowa, Cameroon. J. Chem. Pharm. Res. 2013, 5, 201–209. [Google Scholar]
- Shukla, S.; Dubey, K. CoQ10 a super-vitamin: Review on application and biosynthesis. 3 Biotech 2018, 8, 249. [Google Scholar] [CrossRef]
- Kot, A.; Błażejak, S.; Kurcz, A.; Gientka, I.; Kieliszek, M. Rhodotorula glutinis potential source of lipids, carotenoids, and enzymes for use in industries. Appl. Microbiol. Biotechnol. 2016, 100, 6103–6117. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Kim, S.; Seo, J.; Sim, W.; Moon, H.; Lee, J. Lactate increases coenzyme Q10 production by Agrobacterium tumefacians. World J. Microbiol. Biotechnol. 2008, 24, 887–890. [Google Scholar] [CrossRef]
- He, M.; Rutledge, S.; Kelly, D.; Palmer, C.; Murdoch, G.; Majumder, N.; Nicholls, R.; Pei, Z.; Watkins, P.; Vockley, J. A new genetic disorder in mitochondrial fatty acid β-oxidation: ACAD9 deficiency. Am. J. Hum. Gen. 2007, 81, 87–103. [Google Scholar] [CrossRef]
- Chi, G.; Xu, Y.; Cao, X.; Li, Z.; Cao, M.; Chisti, Y.; He, H. Production of polyunsaturated fatty acids by Schizochytrium (Aurantiochytrium) spp. Biotechnol. Adv. 2022, 55, 107897. [Google Scholar] [CrossRef]
- Jakobsen, A.; Aasen, I.; Josefsen, K.; Strøm, A. Accumulation of docosahexaenoic acid-rich lipid in thraustochytrid Aurantiochytrium sp. strain T66: Effects of N and P starvation and O2 limitation. Appl. Microbiol. Biotechnol. 2008, 80, 297–306. [Google Scholar] [CrossRef]
- Li, J.; Liu, R.; Chang, G.; Li, X.; Chang, M.; Liu, Y.; Jin, Q.; Wang, X. A strategy for the highly efficient production of docosahexaenoic acid by Aurantiochytrium limacinum SR21 using glucose and glycerol as the mixed carbon sources. Bioresource Technol. 2015, 177, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Aasen, I.M.; Ertesvåg, H.; Heggeset, T.M.; Liu, B.; Brautaset, T.; Vadstein, O.; Ellingsen, T.E. Thraustochytrids as production organisms for docosahexaenoic acid (DHA), squalene, and carotenoids. Appl. Microbiol. Biotechnol. 2016, 100, 4309–4321. [Google Scholar] [CrossRef]
- Jain, R.; Raghukumar, S.; Tharanathan, R.; Bhosle, N.B. Extracellular polysaccharide production by thraustochytrid protists. Mar. Biotechnol. 2005, 7, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Leyton, A.; Flores, L.; Shene, C.; Chisti, Y.; Larama, G.; Asenjo, J.; Armenta, R. Antarctic thraustochytrids as sources of carotenoids and high-value fatty acids. Mar. Drugs 2021, 19, 386. [Google Scholar] [CrossRef] [PubMed]
- Valdebenito, D.; Urrutia, S.; Leyton, A.; Chisti, Y.; Asenjo, J.A.; Shene, C. Nitrogen sources affect the long-chain polyunsaturated fatty acids content in Thraustochytrium sp. RT2316-16. Mar. Drugs 2023, 21, 15. [Google Scholar] [CrossRef]
- Xu, W.; Yang, S.; Zhao, J.; Su, T.; Zhao, L.; Liu, J. Improving coenzyme Q8 production in Escherichia coli employing multiple strategies. J. Ind. Microbiol. Biotechnol. 2014, 41, 1297–1303. [Google Scholar] [CrossRef]
- Arenas-Jal, M.; Suñé-Negre, J.M.; García-Montoya, E. Coenzyme Q10 supplementation: Efficacy, safety, and formulation challenges. Compr. Rev. Food Sci. Food Saf. 2020, 19, 574–594. [Google Scholar] [CrossRef]
- Saini, R. Coenzyme Q10: The essential nutrient. J. Pharm. Bioallied Sci. 2011, 3, 466–467. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Y.; Du, Q.; Hou, X.; Wang, M.; Lu, S. Functional analysis of polyprenyl diphosphate synthase genes involved in plastoquinone and ubiquinone biosynthesis in Salvia miltiorrhiza. Front. Plant Sci. 2019, 10, 893. [Google Scholar] [CrossRef]
- Rousseau, G.; Varin, F. Determination of ubiquinone-9 and 10 levels in rat tissues and blood by high-performance liquid chromatography with ultraviolet detection. J. Chromatogr. Sci. 1998, 36, 247–252. [Google Scholar] [CrossRef]
- Saiki, R.; Nagata, A.; Kainou, T.; Matsuda, H.; Kawamukai, M. Characterization of solanesyl and decaprenyl diphosphate synthases in mice and humans. FEBS J. 2005, 272, 5606–5622. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.H.; Ko, T.P.; Wang, A.H. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 2002, 269, 3339–3354. [Google Scholar] [CrossRef]
- Armenta, R.E.; Burja, A.; Radianingtyas, H.; Barrow, C.J. Critical assessment of various techniques for the extraction of carotenoids and co-enzyme Q10 from the thraustochytrid strain ONC-T18. J. Agric. Food Chem. 2006, 54, 9752–9758. [Google Scholar] [CrossRef] [PubMed]
- Kawamukai, M. Biosynthesis and bioproduction of coenzyme Q10 by yeasts and other organisms. Biotechnol. Appl. Biochem. 2009, 53, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Araújo, W.L.; Ishizaki, K.; Nunes-Nesi, A.; Larson, T.R.; Tohge, T.; Krahnert, I.; Witt, S.; Obata, T.; Schauer, N.; Graham, I.A.; et al. Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell. 2010, 22, 1549–1563. [Google Scholar] [CrossRef]
- Yuan, Y.; Tian, Y.; Yue, T. Improvement of coenzyme Q10 production: Mutagenesis induced by high hydrostatic pressure treatment and optimization of fermentation conditions. J. Biomed. Biotechnol. 2012, 2012, 607329. [Google Scholar] [CrossRef]
- Knowles, C.; Redfearn, E. The effect of combined-nitrogen sources on the synthesis and function of the electron transport system of Azotobacter vinelandii. Biochim. Biophys. Acta Bioenerg. 1968, 162, 348–355. [Google Scholar] [CrossRef]
- Tian, Y.; Yue, T.; Yuan, Y.; Soma, P.; Lo, Y. Improvement of cultivation medium for enhanced production of coenzyme Q10 by photosynthetic Rhodospirillum rubrum. Biochem. Eng. J. 2010, 51, 160–166. [Google Scholar] [CrossRef]
- Winrow, M.J.; Rudney, H. The incorporation of p-hydroxybenzoic acid and isopentenyl pyrophosphate into ubiquinone precursors by cell-free preparations of rat tissues. Biochem. Biophys. Res. Comm. 1969, 37, 833–840. [Google Scholar] [CrossRef]
- Doyle, R.; Dixson, G. Coenzyme Q10 production using Sporidobolus johnsonii. U.S. Patent 20110269196A1, 3 November 2011. [Google Scholar]
- Marbois, B.; Xie, L.; Choi, S.; Hirano, K.; Hyman, K.; Clarke, C. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 27827–27838. [Google Scholar] [CrossRef]
- Alam, S.S.; Nambudiri, A.; Rudney, H. 4-Hydroxybenzoate: Polyprenyl transferase and the prenylation of 4-aminobenzoate in mammalian tissues. Arch. Biochem. Biophys. 1975, 171, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Forsgren, M.; Attersand, A.; Lake, S.; Grünler, J.; Swiezewska, E.; Dallner, G.; Climent, I. Isolation and functional expression of human COQ2, a gene encoding a polyprenyl transferase involved in the synthesis of CoQ. Biochem. J. 2004, 382, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Ohara, K.; Yazaki, K.; Nozaki, K.; Uchida, N.; Kawamukai, M.; Nojiri, H.; Yamane, H. The AtPPT1 gene encoding 4-hydroxybenzoate polyprenyl diphosphate transferase in ubiquinone biosynthesis is required for embryo development in Arabidopsis thaliana. Plant Mol. Biol. 2004, 55, 567–577. [Google Scholar] [CrossRef]
- Ohara, K.; Yamamoto, K.; Hamamoto, M.; Sasaki, K.; Yazaki, K. Functional characterization of OsPPT1, which encodes p-hydroxybenzoate polyprenyltransferase involved in ubiquinone biosynthesis in Oryza sativa. Plant Cell Physiol. 2006, 47, 581–590. [Google Scholar] [CrossRef]
- Qiu, X.; Xie, X.; Meesapyodsuk, D. Molecular mechanisms for biosynthesis and assembly of nutritionally important very long chain polyunsaturated fatty acids in microorganisms. Prog. Lipid Res. 2020, 79, 101047. [Google Scholar] [CrossRef] [PubMed]
- Wanke, M.; Swiezewska, E.; Dallner, G. Half-life of ubiquinone and plastoquinone in spinach cells. Plant Sci. 2000, 154, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Kainou, T.; Matsuda, H.; Kawamukai, M. Biological significance of the side chain length of ubiquinone in Saccharomyces cerevisiae. FEBS Lett. 1998, 431, 241–244. [Google Scholar] [CrossRef]
- Yoshida, H.; Kotani, Y.; Ochiai, K.; Araki, K. Production of ubiquione-10 using bacteria. J. Gen. Appl. Microbiol. 1998, 44, 19–26. [Google Scholar] [CrossRef]
- Ha, S.J.; Kim, S.Y.; Seo, J.H.; Jeya, M.; Zhang, Y.W.; Ramu, T.; Kim, I.W.; Lee, J.K. Ca2+ increases the specific coenzyme Q10 content in Agrobacterium tumefaciens. Bioprocess Biosyst. Eng. 2009, 32, 697–700. [Google Scholar] [CrossRef]
- Colella, A.; Patamia, M.; Galtieri, A.; Giardina, B. Cold adaptation and oxidative metabolism of Antarctic fish. Ital. J. Zool. 2000, 67 (Suppl. 1), 33–36. [Google Scholar] [CrossRef]
- Leyton, A.; Shene, C.; Chisti, Y.; Asenjo, J.A. Production of carotenoids and phospholipids by Thraustochytrium sp. in batch and repeated-batch culture. Mar. Drugs 2022, 20, 416. [Google Scholar] [CrossRef] [PubMed]
- Shene, C.; Leyton, A.; Rubilar, M.; Pinelo, M.; Acevedo, F.; Morales, E. Production of lipids and docosahexasaenoic acid (DHA) by a native Thraustochytrium strain. Eur. J. Lipid Sci. Technol. 2013, 115, 890–900. [Google Scholar] [CrossRef]
- Nielsen, P.; Petersen, D.; Dambmann, C. Improved method for determining food protein degree of hydrolysis. J. Food Sci. 2006, 66, 642–646. [Google Scholar] [CrossRef]
- Yi Wu, H.; Biqin Chen, H.; Guanghuang Zhen, H.; Youpao Zhu, H. Fermentation method for producing co-enzyme Q10. U.S. Patent 9689008B22017, 27 June 2017. [Google Scholar]


| Factor | Biomass (g L−1) | CoQ9 (µg g−1) | CoQ10 (µg g−1) |
|---|---|---|---|
| pHB £ | 3.8 ± 0.5 (72 h) | 22 ± 18 | ND |
| pHB £ | 4.9 ± 0.7 (96 h) | 3 ± 2 | 158 ± 19 |
| pHB ¥ | 2.4 ± 0.3 (72 h) | 4 ± 15 | 800 ± 12 |
| pHB ¥ | 2.4 ± 0.5 (96 h) | 2 ± 19 | 151 ± 3 |
| Control | 3.0 ± 0.4 (72 h) | 163 ± 39 | 100 ± 22 |
| No pABA † | 1.8 ± 0.3 (72 h) | 174 ± 37 | 489 ± 55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores, L.; Shene, C.; Asenjo, J.A.; Chisti, Y. Coenzyme Q in Thraustochytrium sp. RT2316-16: Effect of the Medium Composition. Mar. Drugs 2023, 21, 586. https://doi.org/10.3390/md21110586
Flores L, Shene C, Asenjo JA, Chisti Y. Coenzyme Q in Thraustochytrium sp. RT2316-16: Effect of the Medium Composition. Marine Drugs. 2023; 21(11):586. https://doi.org/10.3390/md21110586
Chicago/Turabian StyleFlores, Liset, Carolina Shene, Juan A. Asenjo, and Yusuf Chisti. 2023. "Coenzyme Q in Thraustochytrium sp. RT2316-16: Effect of the Medium Composition" Marine Drugs 21, no. 11: 586. https://doi.org/10.3390/md21110586
APA StyleFlores, L., Shene, C., Asenjo, J. A., & Chisti, Y. (2023). Coenzyme Q in Thraustochytrium sp. RT2316-16: Effect of the Medium Composition. Marine Drugs, 21(11), 586. https://doi.org/10.3390/md21110586








