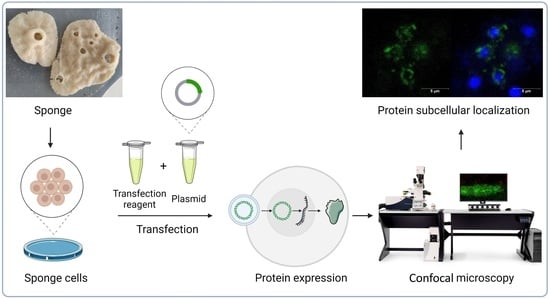
Transfection of Sponge Cells and Intracellular Localization of Cancer-Related MYC, RRAS2, and DRG1 Proteins
Abstract
1. Introduction
2. Results
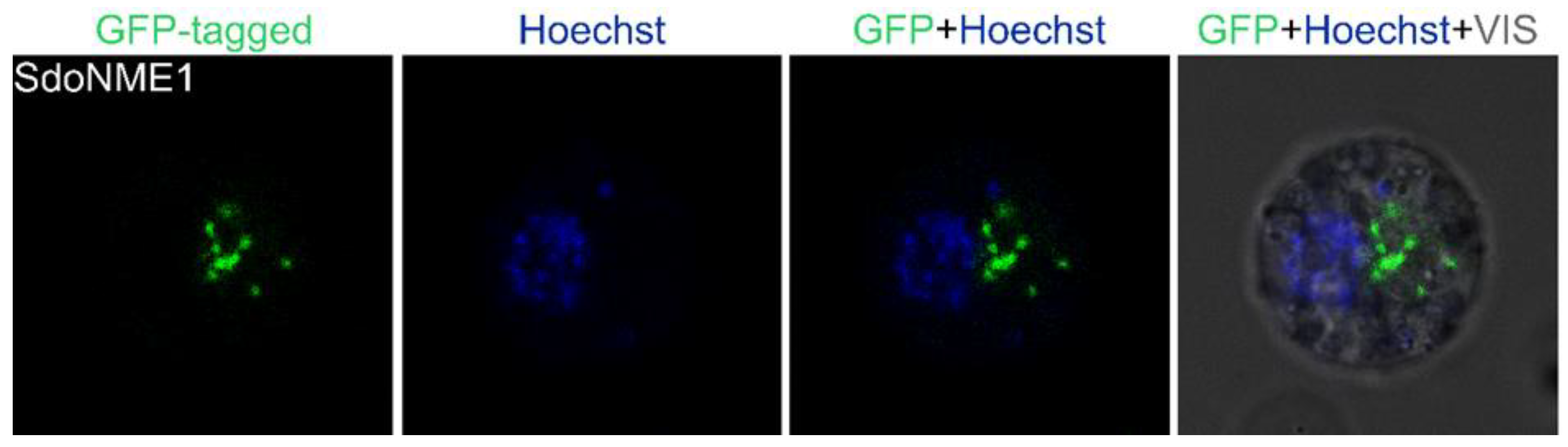
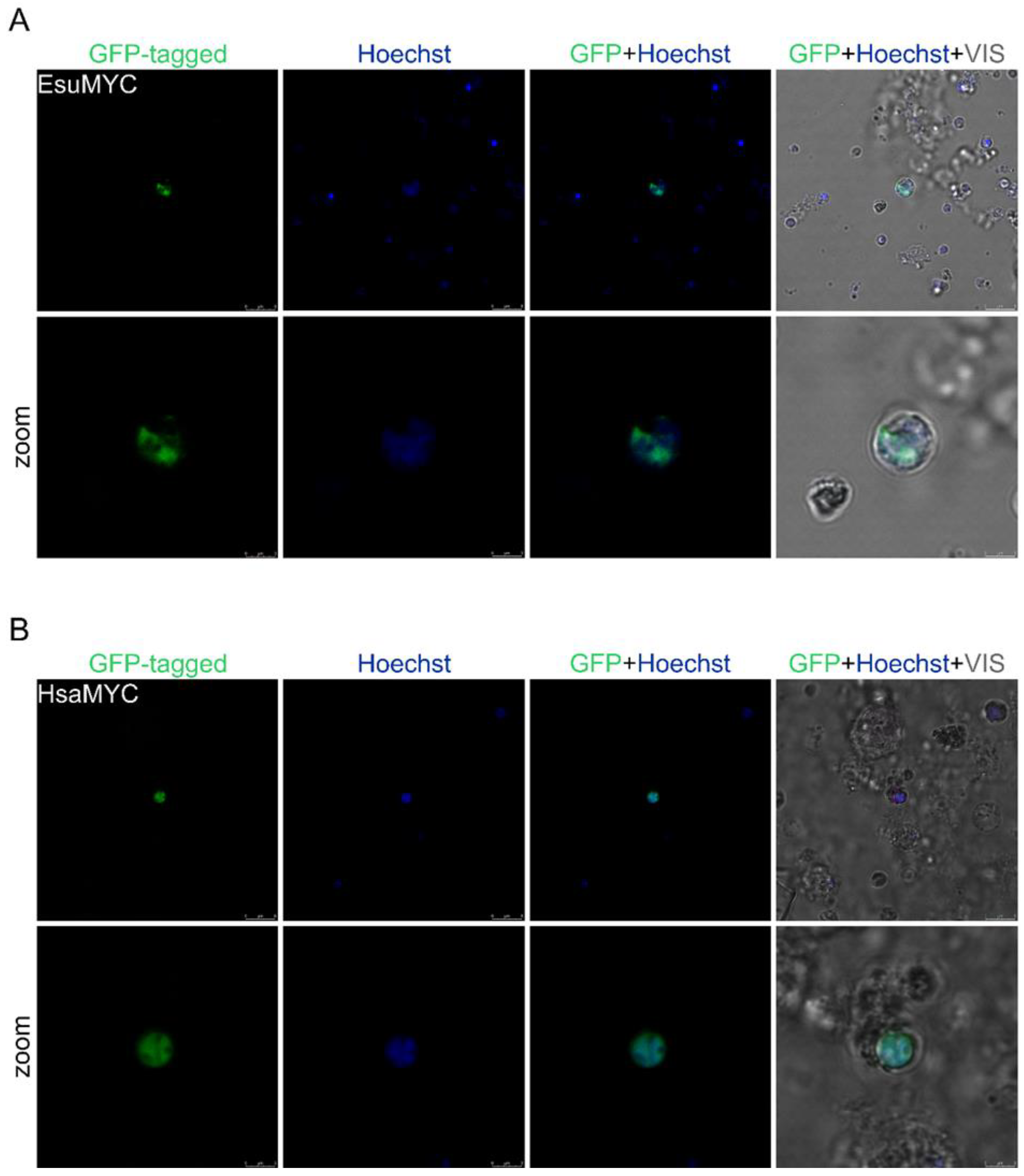
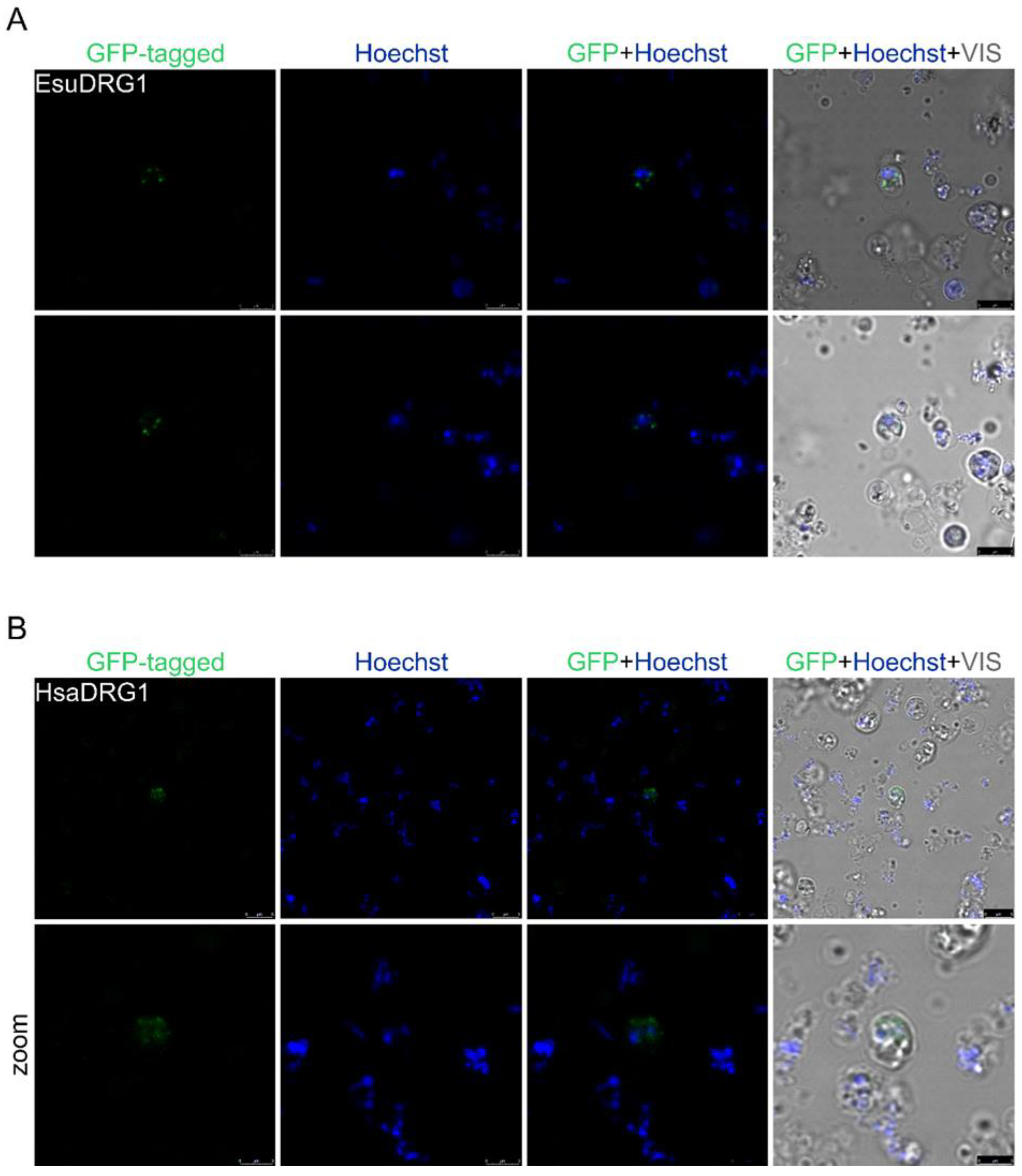
2.1. Nuclear Protein Localization
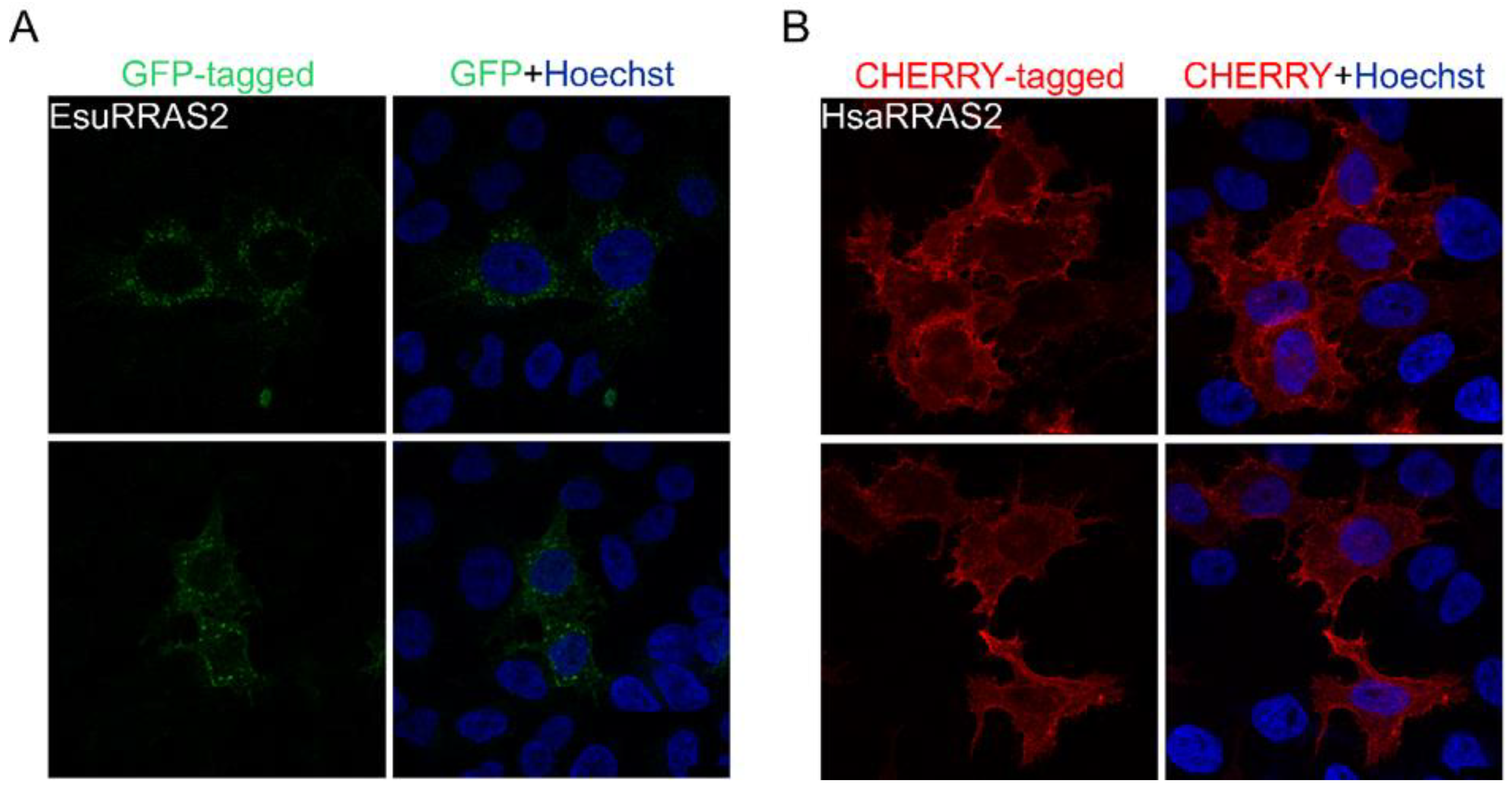
2.2. Membrane Protein Localization
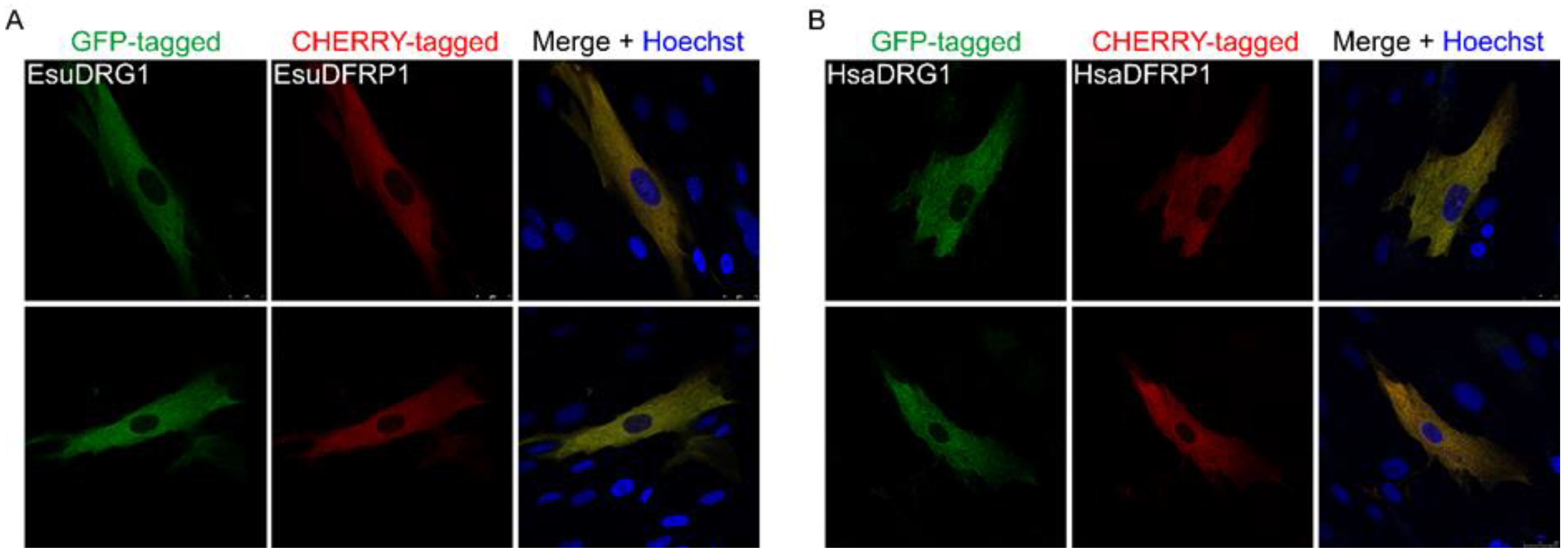
2.3. Cytoplasmic Protein Localization
3. Discussion
4. Materials and Methods
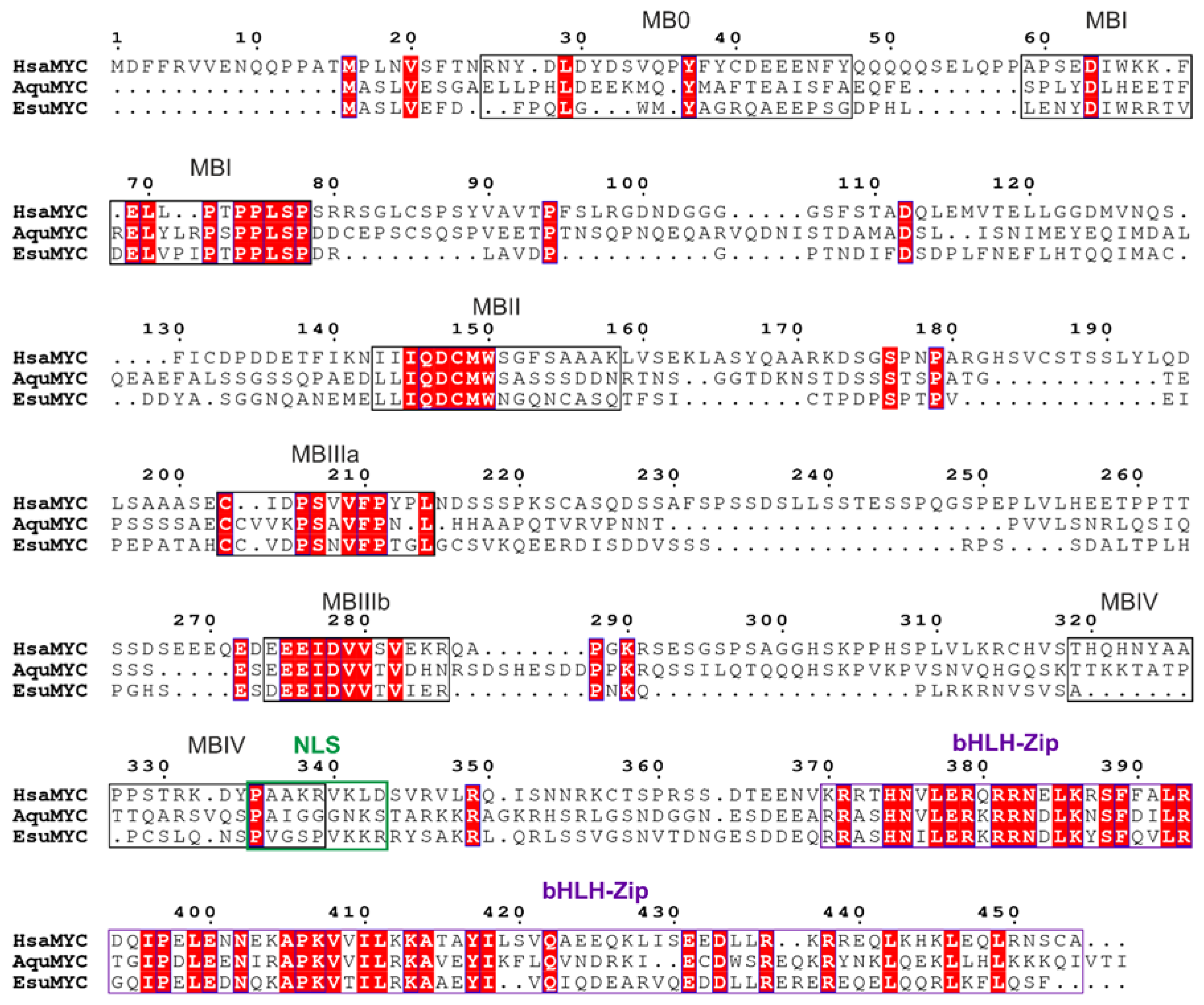
4.1. Sequence Analysis
4.2. Cell Culture
4.3. Plasmids
4.4. Transfection
4.5. Detection of Fluorescent Proteins and Confocal Microscopy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Barela Hudgell, M.A.; Smith, L.C. Lipofection mediated transfection fails for sea urchin coelomocytes. PLoS ONE 2022, 17, e0267911. [Google Scholar] [CrossRef]
- Kim, T.K.; Eberwine, J.H. Mammalian cell transfection: The present and the future. Anal. Bioanal. Chem. 2010, 397, 3173–3178. [Google Scholar] [CrossRef]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef]
- Ogay, I.D.; Lihoradova, O.A.; Azimova, S.S.; Abdukarimov, A.A.; Slack, J.M.; Lynn, D.E. Transfection of insect cell lines using polyethylenimine. Cytotechnology 2006, 51, 89–98. [Google Scholar] [CrossRef]
- Trotter, K.M.; Wood, H.A. Transfection Techniques for Producing Recombinant Baculoviruses. Mol. Biotechnol. 1996, 6, 329–334. [Google Scholar] [CrossRef]
- Recillas-Targa, F. Multiple Strategies for Gene Transfer, Expression, Knockdown, and Chromatin Influence in Mammalian Cell Lines and Transgenic. Mol. Biotechnol. 2006, 34, 337–354. [Google Scholar] [CrossRef]
- Carter, M.; Shieh, J. Gene Delivery Strategies. In Guide to Research Techniques in Neuroscience; Elsevier Inc.: Amsterdam, The Netherlands, 2015; pp. 239–252. [Google Scholar]
- Jordan, E.T.; Collins, M.; Terefe, J.; Ugozzoli, L.; Rubio, T. Optimizing Electroporation Conditions in Primary and Other Difficult-to-Transfect Cells. J. Biomol. Tech. 2008, 19, 328–334. [Google Scholar]
- Horibe, T.; Torisawa, A.; Akiyoshi, R.; Hatta-Ohashi, Y.; Suzuki, H.; Kawakami, K. Transfection efficiency of normal and cancer cell lines and monitoring of promoter activity by single-cell bioluminescence imaging. Luminescence 2014, 29, 96–100. [Google Scholar] [CrossRef]
- Kucharski, M.; Mrowiec, P.; Ocłoń, E. Current standards and pitfalls associated with the transfection of primary fibroblast cells. Biotechnol. Prog. 2021, 37, e3152. [Google Scholar] [CrossRef]
- Segeritz, C.-P.; Vallier, L. Cell Culture: Growing Cells as Model Systems In Vitro. Basic Sci. Methods Clin. Res. 2017, 151–172. [Google Scholar]
- HeLa Transfection Information and Resources | HeLa Cell Line. Available online: https://www.hela-transfection.com/transfection-information/ (accessed on 23 December 2022).
- Theodoulou, F.L.; Miller, A.J. Xenopus oocytes as a heterologous expression system for plant proteins. Mol. Biotechnol. 1995, 3, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Gaitán-Peñas, H.; Pusch, M.; Estévez, R. Expression of LRRC8/VRAC Currents in Xenopus Oocytes: Advantages and Caveats. Int. J. Mol. Sci. Rev. 2018, 19, 719. [Google Scholar] [CrossRef] [PubMed]
- Lungu-Mitea, S.; Oskarsson, A.; Lundqvist, J. Development of an oxidative stress in vitro assay in zebrafish (Danio rerio) cell lines. Sci. Rep. 2018, 8, 12380. [Google Scholar] [CrossRef]
- Kim, D.; Seok, S.; Baek, M.; Lee, H.; Na, Y.; Park, S.; Lee, H.; Dutta, N.K.; Kawakami, K.; Park, J. Estrogen-Responsive Transient Expression Assay Using a Brain Aromatase-Based Reporter Gene in Zebrafish (Danio rerio). Comp. Med. 2009, 59, 416–423. [Google Scholar] [PubMed]
- Chen, L.C.; Wu, J.L.; Shiau, C.Y.; Chen, J.Y. Organization and promoter analysis of the zebrafish (Danio rerio) chemokine gene (CXC-64) promoter. Fish Physiol. Biochem. 2010, 36, 511–521. [Google Scholar] [CrossRef]
- Kunkel, G.R.; Tracy, J.A.; Jalufka, F.L.; Lekve, A.C. CHD8short, a naturally-occurring truncated form of a chromatin remodeler lacking the helicase domain, is a potent transcriptional coregulator. Gene 2017, 641, 303–309. [Google Scholar] [CrossRef]
- Jin, Y.L.; Chen, L.M.; Le, Y.; Li, Y.L.; Hong, Y.H.; Jia, K.T. Establishment of a cell line with high transfection efficiency from zebrafish Danio rerio embryos and its susceptibility to fish viruses. J. Fish Biol. 2017, 91, 1018–1031. [Google Scholar] [CrossRef]
- Seok, S.H.; Baek, M.W.; Lee, H.Y.; Kim, D.J.; Na, Y.R.; Noh, K.J.; Park, S.H.; Lee, H.K.; Lee, B.H.; Park, J.H. In vivo alternative testing with zebrafish in ecotoxicology. J. Vet. Sci. 2008, 9, 351–357. [Google Scholar] [CrossRef]
- Safina, D.R.; Selina, P.I.; Roschina, M.P.; Karaseva, M.A.; Komissarov, A.A.; Demidyuk, I.V.; Sverdlov, E.D.; Kostrov, S.V. Functional efficiency of PCR vectors in vitro and at the organism level. PLoS ONE 2020, 15, e0232045. [Google Scholar] [CrossRef]
- Manivannan, S.N.; Jacobsen, T.L.; Lyon, P.; Selvaraj, B.; Halpin, P.; Simcox, A. Targeted Integration of Single-Copy Transgenes in Drosophila melanogaster Tissue-Culture Cells Using Recombination-Mediated Cassette Exchange. Genetics 2015, 201, 1319–1328. [Google Scholar] [CrossRef]
- Lang, Y.; Pi, X.; Di, Z.; Zhang, Q.; Wang, H.; Shen, B.; Li, F.; Liu, G.; Yu, Y.; Li, X.; et al. Molecular characterization and expression analysis of CS αβ defensin genes from the scorpion Mesobuthus martensii. Biosci. Rep. 2017, 37, BSR20171282. [Google Scholar] [CrossRef]
- Grimaldi, G.; Di Nocera, P.P. Transient expression of Drosophila melanogaster rDNA promoter into cultured Drosophila cells. Nucleic Acids Res. 1986, 14, 6417–6432. [Google Scholar] [CrossRef] [PubMed]
- Di Nocera, P.P.; Dawid, I.B. Transient expression of genes introduced into cultured cells of Drosophila. Proc. Natl. Acad. Sci. USA 1983, 80, 7095–7098. [Google Scholar] [CrossRef] [PubMed]
- Suárez Patino, S.F.; Astray Mancini, R.; Pereira, C.A.; Torres Suazo, C.A.; Zucatelli Mendonca, R.; Calil Jorge, A.S.; Jorge, C. Transient expression of rabies virus glycoprotein (RVGP) in Drosophila melanogaster Schneider 2 (S2) cells. J. Biotechnol. 2014, 192, 255–262. [Google Scholar] [CrossRef]
- Leopold, R.A.; Hughes, K.J.; DeVault, J.D. Using electroporation and a slot cuvette to deliver plasmid DNA to insect embryos. Genet. Anal. Biomol. Eng. 1996, 12, 197–200. [Google Scholar] [CrossRef]
- Keith, M.B.A.; Patrick, J.; Iatrou, K.; Behie, A. Use of Flow Cytometry to Rapidly Optimize the Transfection of Animal Cells. Res. Rep. 2000, 28, 148–154. [Google Scholar] [CrossRef]
- Gundersen-Rindal, D.; Slack, J.M.; Lynn, D.E. Transfection of Lymantria dispar insect cell lines. Methods Cell Sci. 2001, 22, 257–263. [Google Scholar] [CrossRef]
- Gammon, D.B.; Duraffour, S.; Rozelle, D.K.; Hehnly, H.; Sharma, R.; Sparks, M.E.; West, C.C.; Chen, Y.; Moresco, J.J.; Andrei, G.; et al. A single vertebrate DNA virus protein disarms invertebrate immunity to RNA virus infection. Elife 2014, 25, e02910. [Google Scholar] [CrossRef]
- Corsaro, B.G.; DiRenzo, J.; Fraser, M.J. Transfection of cloned Heliothis zea cell lines with the DNA genome of the Heliothis zea nuclear polyhedrosis virus. J. Virol. Methods 1989, 25, 283–292. [Google Scholar] [CrossRef]
- Corsaro, B.G.; Fraser, M.J. Transfection of Lepidopteran insect cells with Baculovirus DNA. J. Tissue Cult. Methods 1989, 12, 7–11. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, Y. Marine invertebrate cell culture: A decade of development. J. Oceanogr. 2014, 70, 405–414. [Google Scholar] [CrossRef]
- Davy, S.K.; Allemand, D.; Weis, V.M. Cell Biology of Cnidarian-Dinoflagellate Symbiosis. Microbiol. Mol. Biol. Rev. 2012, 76, 229–261. [Google Scholar] [CrossRef] [PubMed]
- Huete-Stauffer, C.; Valisano, L.; Gaino, E.; Vezzulli, L.; Cerrano, C. Development of long-term primary cell aggregates from Mediterranean octocorals. Vitr. Cell. Dev. Biol.—Anim. 2015, 51, 815–826. [Google Scholar] [CrossRef]
- Rinkevich, B. Marine Invertebrate Cell Cultures: New Millennium Trends. Mar. Biotechnol. 2005, 7, 429–439. [Google Scholar] [CrossRef]
- Weis, V.M.; Davy, S.K.; Hoegh-Guldberg, O.; Rodriguez-Lanetty, M.; Pringle, J.R. Cell biology in model systems as the key to understanding corals. Trends Ecol. Evol. 2008, 23, 369–376. [Google Scholar] [CrossRef]
- Vandepas, L.E.; Warren, K.J.; Amemiya, C.T.; Browne, W.E. Establishing and maintaining primary cell cultures derived from the ctenophore Mnemiopsis leidyi. J. Exp. Biol. 2017, 220, 1197–1201. [Google Scholar] [CrossRef]
- Rinkevich, B. Cell Cultures from Marine Invertebrates: New Insights for Capturing Endless Stemness. Mar. Biotechnol. 2011, 13, 345–354. [Google Scholar] [CrossRef]
- Shi, H.; Ruan, L.; Söderhäll, I.; Söderhäll, K.; Xu, X. Transfection of crayfish hematopoietic tissue cells. Dev. Comp. Immunol. 2018, 88, 70–76. [Google Scholar] [CrossRef]
- Tracy, A.N.; Yadavalli, R.; Reed, K.S.; Parnaik, R.; Poulton, N.J.; Bishop-Bailey, D.; Fernández Robledo, J.A. Genome to phenome tools: In vivo and in vitro transfection of Crassostrea virginica hemocytes. Fish Shellfish Immunol. 2020, 103, 438–441. [Google Scholar] [CrossRef]
- Miljkovic, M.; Galliot, B. Cnidarian and Bilaterian Promoters Can Direct GFP Expression in Transfected Hydra. Dev. Biol. 2002, 246, 377–390. [Google Scholar] [CrossRef]
- Bosch, T.C.G.; Khalturin, K. Patterning and cell differentiation in Hydra: Novel genes and the limits to conservation. Can. J. Zool. 2002, 80, 1670–1677. [Google Scholar] [CrossRef]
- Brennecke, T.; Gellner, K.; Bosch, T.C.G. The lack of a stress response in Hydra oligactis is due to reduced hsp70 mRNA stability. Eur. J. Biochem. 1998, 255, 703–709. [Google Scholar] [CrossRef]
- Lohmann, J.U.; Endl, I.; Bosch, T.C.G. Silencing of Developmental Genes in Hydra. Dev. Biol. 1999, 214, 211–214. [Google Scholar] [CrossRef]
- Böttger, A.; Alexandrova, O.; Cikala, M.; Schade, M.; Herold, M.; David, C.N. GFP expression in Hydra: Lessons from the particle gun. Dev. Genes Evol. 2002, 212, 302–305. [Google Scholar] [CrossRef]
- Suga, H.; Ruiz-Trillo, I. Development of ichthyosporeans sheds light on the origin of metazoan multicellularity. Dev. Biol. 2013, 377, 284–292. [Google Scholar] [CrossRef]
- Parra-Acero, H.; Ros-Rocher, N.; Perez-Posada, A.; Kożyczkowska, A.; Sánchez-Pons, N.; Nakata, A.; Suga, H.; Najle, S.R.; Ruiz-Trillo, I. Transfection of Capsaspora owczarzaki, a close unicellular relative of animals. Development 2018, 145, dev162107. [Google Scholar] [CrossRef]
- Nellen, W.; Silan, C.; Firtel, R.A. DNA-Mediated Transformation in Dictyostelium discoideum: Regulated Expression of an Actin Gene Fusion. Mol. Cell. Biol. 1984, 4, 2890–2898. [Google Scholar] [CrossRef]
- Gaudet, P.; Pilcher, K.E.; Fey, P.; Chisholm, R.L. Transformation of Dictyostelium discoideum with plasmid DNA. Nat. Protoc. 2007, 2, 1317–1324. [Google Scholar] [CrossRef]
- Grasela, J.J.; Pomponi, S.A.; Rinkevich, B.; Grima, J. Efforts to develop a cultured sponge cell line: Revisiting an intractable problem. Vitr. Cell. Dev. Biol. Anim. 2012, 48, 12–20. [Google Scholar] [CrossRef]
- Revilla-i-Domingo, R.; Schmidt, C.; Zifko, C.; Raible, F. Establishment of Transgenesis in the Demosponge. Genetics 2018, 210, 435–443. [Google Scholar] [CrossRef]
- Rivera, A.S.; Hammel, J.U.; Haen, K.M.; Danka, E.S.; Cieniewicz, B.; Winters, I.P.; Posfai, D.; Wörheide, G.; Lavrov, D.V.; Knight, S.W.; et al. RNA interference in marine and freshwater sponges: Actin knockdown in Tethya wilhelma and Ephydatia muelleri by ingested dsRNA expressing bacteria. BMC Biotechnol. 2011, 11, 67. [Google Scholar] [CrossRef]
- Pomponi, S.A. Biology of the Porifera: Cell culture. Can. J. Zool. 2006, 84, 167–174. [Google Scholar] [CrossRef]
- Rocher, C.; Vernale, A.; Fierro-Constaín, L.; Séjourné, N.; Chenesseau, S.; Marschal, C.; Le Goff, E.; Dutilleul, M.; Matthews, C.; Marschal, F.; et al. The buds of Oscarella lobularis (Porifera): A new convenient model for sponge cell and developmental biology. bioRxiv 2020. bioRxiv:2020.06.23.167296. [Google Scholar] [CrossRef]
- Pfannkuchen, M.; Brümmer, F. Heterologous expression of DsRed2 in young sponges (Porifera). Int. J. Dev. Biol. 2009, 53, 1113–1117. [Google Scholar] [CrossRef]
- Reid, P.J.W.; Matveev, E.; Mcclymont, A.; Posfai, D.; Hill, A.L.; Leys, S.P. Wnt signaling and polarity in freshwater sponges. BMC Ecol. Evol. 2018, 18, 12. [Google Scholar] [CrossRef]
- Love, G.D.; Grosjean, E.; Stalvies, C.; Fike, D.A.; Grotzinger, J.P.; Bradley, A.S.; Kelly, A.E.; Bhatia, M.; Meredith, W.; Snape, C.E.; et al. Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature 2008, 457, 718–721. [Google Scholar] [CrossRef]
- Harcet, M.; Roller, M.; Ćetković, H.; Perina, D.; Wiens, M.; Müller, W.E.G.; Vlahoviček, K. Demosponge EST sequencing reveals a complex genetic toolkit of the simplest metazoans. Mol. Biol. Evol. 2010, 27, 2747–2756. [Google Scholar] [CrossRef]
- Srivastava, M.; Simakov, O.; Chapman, J.; Fahey, B.; Gauthier, M.E.A.; Mitros, T.; Richards, G.S.; Conaco, C.; Dacre, M.; Hellsten, U.; et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 2010, 466, 720–726. [Google Scholar] [CrossRef]
- Ćetković, H.; Harcet, M.; Roller, M.; Herak Bosnar, M. A survey of metastasis suppressors in Metazoa. Lab. Investig. 2018, 98, 554–570. [Google Scholar] [CrossRef]
- Ćetkovic, H.; Halasz, M.; Bosnar, M.H. Sponges: A Reservoir of Genes Implicated in Human Cancer. Mar. Drugs 2018, 16, 20. [Google Scholar] [CrossRef]
- Beljan, S.; Herak Bosnar, M.; Ćetković, H. Rho Family of Ras-Like GTPases in Early-Branching Animals. Cells 2020, 9, 2279. [Google Scholar] [CrossRef]
- Ćetkovic, H.; Grebenjuk, V.A.; Müller, W.E.G.; Gamulin, V. Src proteins/src genes: From sponges to mammals. Gene 2004, 342, 251–261. [Google Scholar] [CrossRef]
- Ćetković, H.; Müller, W.E.G.; Gamulin, V. Bruton tyrosine kinase-like protein, BtkSD, is present in the marine sponge Suberites domuncula. Genomics 2004, 83, 743–745. [Google Scholar] [CrossRef]
- Ćetkovic, H.; Mikoč, A.; Müller, W.E.G.; Gamulin, V. Ras-like small GTPases form a large family of proteins in the marine sponge Suberites domuncula. J. Mol. Evol. 2007, 64, 332–341. [Google Scholar] [CrossRef]
- Ćetkovic, H.; Müller, I.M.; Müller, W.E.G.; Gamulin, V. Characterization and phylogenetic analysis of a cDNA encoding the Fes/FER related, non-receptor protein-tyrosine kinase in the marine sponge Sycon raphanus. Gene 1998, 216, 77–84. [Google Scholar] [CrossRef]
- Kruse, M.; Gamulin, V.; Ćetković, H.; Pancer, Z.; Müller, I.M.; Müller, W.E.G. Molecular evolution of the metazoan protein kinase C multigene family. J. Mol. Evol. 1996, 43, 374–383. [Google Scholar] [CrossRef]
- Perina, D.; Ćetković, H.; Harcet, M.; Premzl, M.; Lukić-Bilela, L.; Müller, W.E.G.; Gamulin, V. The complete set of ribosomal proteins from the marine sponge Suberites domuncula. Gene 2006, 366, 275–284. [Google Scholar] [CrossRef]
- Perina, D.; Bosnar, M.H.; Bago, R.; Mikoč, A.; Harcet, M.; Deželjin, M.; Ćetković, H. Sponge non-metastatic Group i Nme gene/protein—Structure and function is conserved from sponges to humans. BMC Evol. Biol. 2011, 11, 87. [Google Scholar] [CrossRef]
- Perina, D.; Korolija, M.; Hadžija, M.P.; Grbeša, I.; Belužić, R.; Imešek, M.; Morrow, C.; Marjanović, M.P.; Bakran-Petricioli, T.; Mikoč, A.; et al. Functional and structural characterization of FAU gene/protein from marine sponge Suberites domuncula. Mar. Drugs 2015, 13, 4179–4196. [Google Scholar] [CrossRef]
- Perina, D.; Mikoč, A.; Harcet, M.; Imešek, M.; Sladojević, D.; Brcko, A.; Ćetković, H. Characterization of Bruton’s tyrosine kinase gene and protein from marine sponge Suberites domuncula. Croat. Chem. Acta 2012, 85, 223–229. [Google Scholar] [CrossRef]
- Beljan, S.; Dominko, K.; Talajić, A.; Kasun, A.H.; Vidaček, N.Š; Bosnar, M.H.; Vlahoviček, K.; Ćetković, H. Structure and function of cancer—Related developmentally regulated GTP—Binding protein 1 (DRG1) is conserved between sponges and humans. Sci. Rep. 2022, 12, 11379. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Song, X.C.; Yang, X.; Peng, T.; Wang, Y.; Zhou, W.G. Protein Subcellular Localization Profiling of Breast Cancer Cells by Dissociable Antibody MicroArray (DAMA) Staining. J. Mol. Evol. 2011, 10, 1536–1544. [Google Scholar] [CrossRef]
- Curtis, P.D.; Quardokus, E.M.; Lawler, M.L.; Guo, X.; Klein, D.; Chen, J.C.; Arnold, R.J.; Brun, Y.V. The scaffolding and signaling functions of a localization factor impact polar development. Mol. Microbiol. 2012, 84, 712–735. [Google Scholar] [CrossRef]
- Ahmadi, S.E.; Rahimi, S.; Zarandi, B.; Chegeni, R.; Safa, M. MYC: A multipurpose oncogene with prognostic and therapeutic implications in blood malignancies. J. Hematol. Oncol. 2021, 14, 121. [Google Scholar] [CrossRef] [PubMed]
- Hartl, M.; Mitterstiller, A.-M.; Valovka, T.; Breuker, K.; Hobmayer, B.; Bister, K. Stem cell-specific activation of an ancestral myc protooncogene with conserved basic functions in the early metazoan Hydra. Proc. Natl. Acad. Sci. USA 2010, 107, 4051–4056. [Google Scholar] [CrossRef] [PubMed]
- Popov, N.; Wanzel, M.; Madiredjo, M.; Zhang, D.; Beijersbergen, R.; Bernards, R.; Moll, R.; Elledge, S.J.; Eilers, M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 2007, 9, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Madden, S.K.; De Araujo, A.D.; Gerhardt, M.; Fairlie, D.P.; Mason, J.M. Taking the Myc out of cancer: Toward therapeutic strategies to directly inhibit c-Myc. Mol. Cancer 2021, 20, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gao, X.; Yuan, M.; Yang, B.; He, Q.; Cao, J. Targeting Myc Interacting Proteins as a Winding Path in Cancer Therapy. Front. Pharmacol. 2021, 12, 748852. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; Mcwilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Xavier, R.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef]
- Wang, K.; Peng, K. RRAS2 knockdown suppresses osteosarcoma progression by inactivating the MEK/ERK signaling pathway. Anticancer Drugs 2019, 30, 933–939. [Google Scholar] [CrossRef]
- Movilla, N.; Crespo, P.; Bustelo, X.R. Signal transduction elements of TC21, an oncogenic member of the R-Ras subfamily of GTP-binding proteins. Oncogene 1999, 18, 5860–5869. [Google Scholar] [CrossRef] [PubMed]
- Rosário, M.; Paterson, H.F.; Marshall, C.J. Activation of the Raf/MAP kinase cascade by the Ras-related protein TC21 is required for the TC21-mediated transformation of NIH 3T3 cells. EMBO J. 1999, 18, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Hao, X.; Ge, C.; Zhao, F.; Zhu, M.; Chen, T.; Yao, M.; He, X.; Li, J. TC21 promotes cell motility and metastasis by regulating the expression of E-cadherin and N-cadherin in hepatocellular carcinoma. Int. J. Oncol. 2010, 37, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Pereira, A.M.; Sims, D.; Dexter, T.; Fenwick, K.; Assiotis, I.; Kozarewa, I.; Mitsopoulos, C.; Hakas, J.; Zvelebil, M.; Lord, C.J.; et al. Genome-wide functional screen identi fi es a compendium of genes affecting sensitivity to tamoxifen. Proc. Natl. Acad. Sci. USA 2012, 109, 2730–2735. [Google Scholar] [CrossRef]
- Larive, R.M.; Abad, A.; Cardaba, C.M.; Hernández, T.; Cañamero, M.; Gutkind, J.S. The Ras-like protein R-Ras2/TC21 is important for proper mammary gland development. Mol. Biol. Cell 2012, 23, 2373–2387. [Google Scholar] [CrossRef] [PubMed]
- Capri, Y.; Flex, E.; Krumbach, O.H.F.; Carpentieri, G.; Cecchetti, S.; Lißewski, C.; Adariani, S.R.; Schanze, D.; Brinkmann, J.; Piard, J.; et al. Activating Mutations of RRAS2 Are a Rare Cause of Noonan Syndrome. Am. J. Hum. Genet. 2019, 104, 1223–1232. [Google Scholar] [CrossRef]
- Zhang, X.; Spiegelman, N.A.; Nelson, O.D.; Jing, H.; Lin, H. SIRT6 regulates Ras-related protein R-Ras2 by lysine defatty-acylation. Elife 2017, 6, e25158. [Google Scholar] [CrossRef]
- Zeng, F.; Li, X.; Pires-Alves, M.; Chen, X.; Hawk, C.W.; Jin, H. Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells. Cell Rep. 2021, 37, 109877. [Google Scholar] [CrossRef]
- Schellhaus, A.K.; Moreno-Andrés, D.; Chugh, M.; Yokoyama, H.; Moschopoulou, A.; De, S.; Bono, F.; Hipp, K.; Schäffer, E.; Antonin, W. Developmentally Regulated GTP binding protein 1 (DRG1) controls microtubule dynamics. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Westrip, C.A.E.E.; Zhuang, Q.; Hall, C.; Eaton, C.D.; Coleman, M.L. Developmentally regulated GTPases: Structure, function and roles in disease. Cell. Mol. Life Sci. 2021, 78, 7219–7235. [Google Scholar] [CrossRef]
- Sazuka, T.; Kinoshita, M.; Tomooka, Y.; Ikawa, Y.; Noda, M.; Kumar, S. Expression of DRG during murine embryonic development. Biochem. Biophys. Res. Commun. 1992, 189, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Sommer, K.A.; Petersen, G.; Bautz, E.K.F. The gene upstream of DmRP128 codes for a novel GTP-binding protein of Drosophila melanogaster. MGG Mol. Gen. Genet. 1994, 242, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Azuma, S.; Ikawa, S.; Semba, K.; Inoue, J. Identification of DRG family regulatory proteins (DFRPs): Specific regulation of DRG1 and DRG2. Genes Cells 2005, 10, 139–150. [Google Scholar] [CrossRef]
- Wilson, H.V. On some phenomena of coalescence and regeneration in sponges. J. Exp. Zool. 1907, 5, 245–258. [Google Scholar] [CrossRef]
- Custodio, M.R.; Prokic, I.; Steffen, R.; Koziol, C.; Borojevic, R.; Brümmer, F.; Nickel, M.; Müller, W.E.G. Primmorphs generated from dissociated cells of the sponge Suberites domuncula: A model system for studies of cell proliferation and cell death. Mech. Ageing Dev. 1998, 105, 45–59. [Google Scholar] [CrossRef]
- Schippers, K.J. Sponge Cell Culture; Wageningen University: Wageningen, The Netherlands, 2013. [Google Scholar]
- Pomponi, S.A.; Willoughby, R.; Kelly-Borges, M. Sponge Cell Culture. In Molecular Approaches to the Study of the Ocean; Cooksey, K.E., Ed.; Springer International Publishing: Dordrech, The Netherlands, 1998; pp. 423–433. ISBN 9789461734419. [Google Scholar]
- Wittlieb, J.; Khalturin, K.; Lohmann, J.U.; Anton-Erxleben, F.; Bosch, T.C.G. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 6208–6211. [Google Scholar] [CrossRef]
- Herak Bosnar, M.; De Gunzburg, J.; Weber, I.; Pavelic, J. Subcellular localization of A and B Nm23/NDPK subunits. Exp. Cell Res. 2004, 298, 275–284. [Google Scholar] [CrossRef]
- Radić, M.; Šoštar, M.; Weber, I.; Ćetković, H.; Slade, N.; Bosnar, M.H. The Subcellular Localization and Oligomerization Preferences of NME1/NME2 upon Radiation-Induced DNA Damage. Int. J. Mol. Sci. 2020, 21, 2363. [Google Scholar] [CrossRef]
- Proust, B.; Radić, M.; Vidaček, N.Š; Cottet, C.; Attia, S.; Lamarche, F.; Ačkar, L.; Mikulčić, V.G.; Tokarska-Schlattner, M.; Bosnar, M.H. NME6 is a phosphotransfer-inactive, monomeric NME/NDPK family member and functions in complexes at the interface of mitochondrial inner membrane and matrix. Cell Biosci. 2021, 11, 195. [Google Scholar] [CrossRef] [PubMed]
- Perina, D.; Herak Bosnar, M.; Mikoč, A.; Müller, W.E.G.; Ćetković, H. Characterization of Nme6-like gene/protein from marine sponge Suberites domuncula. Naunyn. Schmiedeberg’s Arch. Pharmacol. 2011, 384, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Schlattner, U. The complex functions of the nme family—A matter of location and molecular activity. Int. J. Mol. Sci. 2021, 22, 13083. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.J. Sponging off Nature for New Drug Leads. Biochem. Pharmacol. 2017, 139, 3–14. [Google Scholar] [CrossRef]
- Varijakzhan, D.; Loh, J.Y.; Yap, W.S.; Yusoff, K.; Seboussi, R.; Lim, S.H.E.; Lai, K.S.; Chong, C.M. Bioactive compounds from marine sponges: Fundamentals and applications. Mar. Drugs 2021, 19, 246. [Google Scholar] [CrossRef]
- Munro, M.H.G.; Blunt, J.W.; Dumdei, E.J.; Hickford, S.J.H.; Lill, R.E.; Li, S.; Battershill, C.N.; Duckworth, A.R. The discovery and development of marine compounds with pharmaceutical potential. J. Biotechnol. 1999, 70, 15–25. [Google Scholar] [CrossRef]
- Osinga, R.; Tramper, J.; Wijffels, H. Cultivation of Marine Sponges. J. Mar. Biotechnol. 1999, 1, 509–532. [Google Scholar] [CrossRef]
- Schippers, K.J.; Sipkema, D.; Osinga, R.; Smidt, H.; Pomponi, S.A.; Martens, D.E.; Wijffels, R.H. Cultivation of Sponges, Sponge Cells and Symbionts. Achievements and Future Prospects. Adv. Mar. Biol. 2012, 62, 273–337. [Google Scholar] [CrossRef]
- BLAST: Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 23 December 2022).
- Chernogor, L.I.; Denikina, N.N.; Belikov, S.I.; Ereskovsky, A.V. Long-Term Cultivation of Primmorphs from Freshwater Baikal Sponges Lubomirskia baikalensis. Mar. Biotechnol. 2011, 13, 782–792. [Google Scholar] [CrossRef]
- Dominko, K.; Rastija, A.; Sobocanec, S.; Vidatic, L.; Meglaj, S.; Babic Lovincic, A.; Hutter-Paier, B.; Colombo, A.; Lichtenthaler, S.F.; Tahirovic, S.; et al. Impaired Retromer Function in Niemann-Pick Type C Disease Is Dependent on Intracellular Cholesterol Accumulation. Int. J. Mol. Sci. 2021, 22, 13256. [Google Scholar] [CrossRef]





| Construct Name/Organism | Origin | Cloned In | Primers/Restriction Site |
|---|---|---|---|
| EsuMYC-GFP E. subterraneus | Esu cDNA | pEGFP-N1 | XhoI 5′-GTCTAGCTCGAGATGGCGTCGTTGGTAGAGTTC-3′ BamHI 5′-CTAGACGAATTCCGGGAAAAACTTTGCAGAAACTTC-3′ |
| HsaMYC-GFP H. sapiens | HG11346-UT SinoBiological | pEGFP-N1 | NdeI 5′-GTCTAGGAATTCATGCCCCTCAACGTTAGC-3′ BamHI 5′-CTAGACGGATCCGCGGACGCACAAGAGTTCCG-3′ |
| EsuRRAS-GFP E. subterraneus | Esu cDNA | pEGFP-C1 | XhoI 5′-GTCTAGCTCGAGGCATGGCGGCCAACAAAGAC-3′ BamHI 5′-CTAGACGGATCCTCACAGAATTACACATTTCTTC-3′ |
| HsaRRAS2-CHERRY H. sapiens | RC204591 OriGene | pmCherry-C1 | XhoI 5′-GTCTAGCTCGAGGCATGGCCGCGGCCGGCTGGCG-3′ BamHI 5′-CTAGACGGATCCTTAGAAAATGACACAATGGCAG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dominko, K.; Talajić, A.; Radić, M.; Vidaček, N.Š.; Vlahoviček, K.; Bosnar, M.H.; Ćetković, H. Transfection of Sponge Cells and Intracellular Localization of Cancer-Related MYC, RRAS2, and DRG1 Proteins. Mar. Drugs 2023, 21, 119. https://doi.org/10.3390/md21020119
Dominko K, Talajić A, Radić M, Vidaček NŠ, Vlahoviček K, Bosnar MH, Ćetković H. Transfection of Sponge Cells and Intracellular Localization of Cancer-Related MYC, RRAS2, and DRG1 Proteins. Marine Drugs. 2023; 21(2):119. https://doi.org/10.3390/md21020119
Chicago/Turabian StyleDominko, Kristina, Antea Talajić, Martina Radić, Nikolina Škrobot Vidaček, Kristian Vlahoviček, Maja Herak Bosnar, and Helena Ćetković. 2023. "Transfection of Sponge Cells and Intracellular Localization of Cancer-Related MYC, RRAS2, and DRG1 Proteins" Marine Drugs 21, no. 2: 119. https://doi.org/10.3390/md21020119
APA StyleDominko, K., Talajić, A., Radić, M., Vidaček, N. Š., Vlahoviček, K., Bosnar, M. H., & Ćetković, H. (2023). Transfection of Sponge Cells and Intracellular Localization of Cancer-Related MYC, RRAS2, and DRG1 Proteins. Marine Drugs, 21(2), 119. https://doi.org/10.3390/md21020119