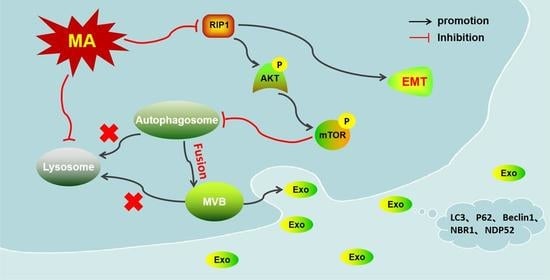
RIP1 Mediates Manzamine-A-Induced Secretory Autophagy in Breast Cancer
Abstract
:1. Introduction
2. Results
2.1. MA Inhibits Breast Cancer Cell Proliferation
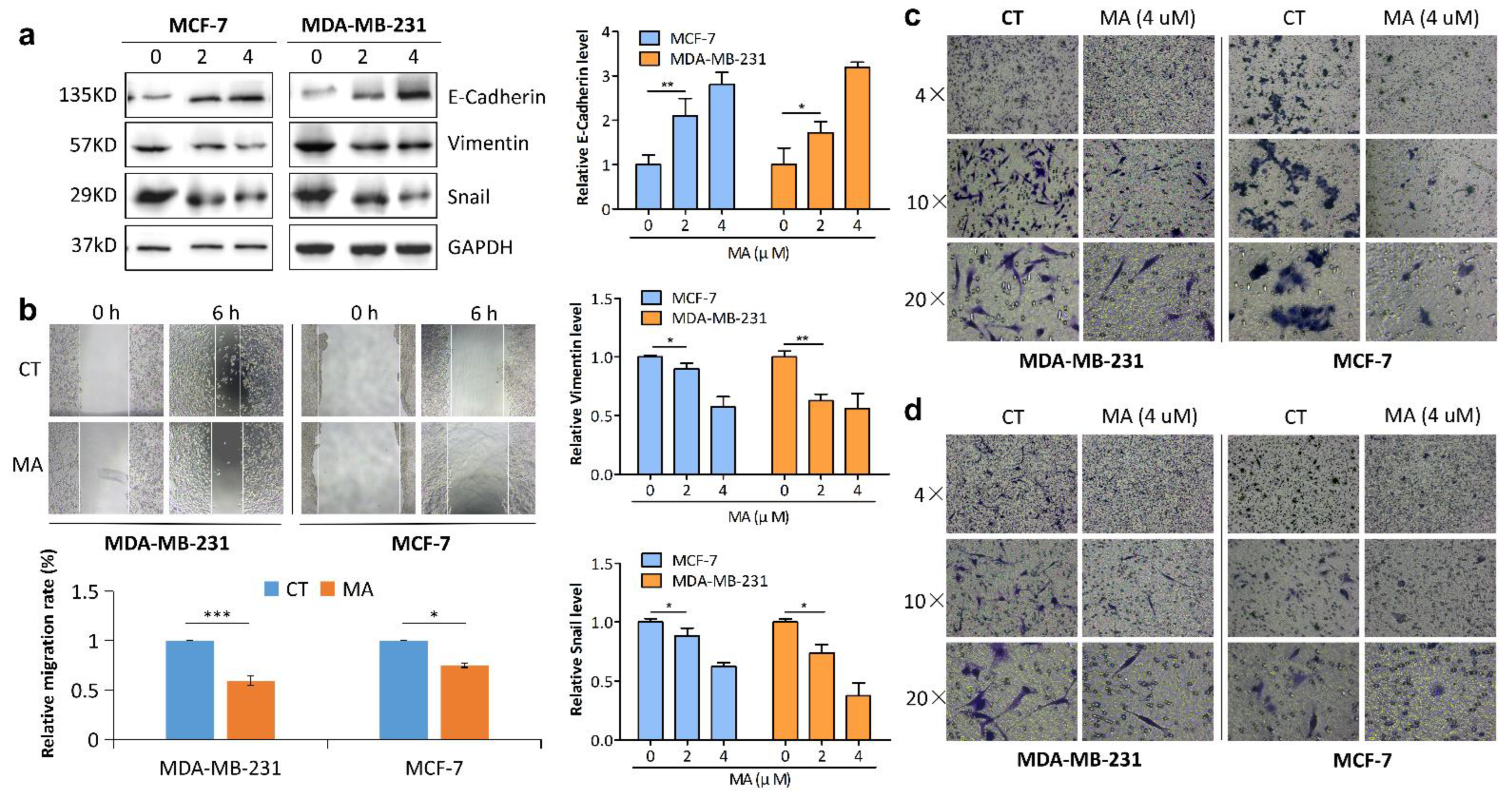
2.2. MA Inhibits Breast Cancer Cell Migration and Invasion
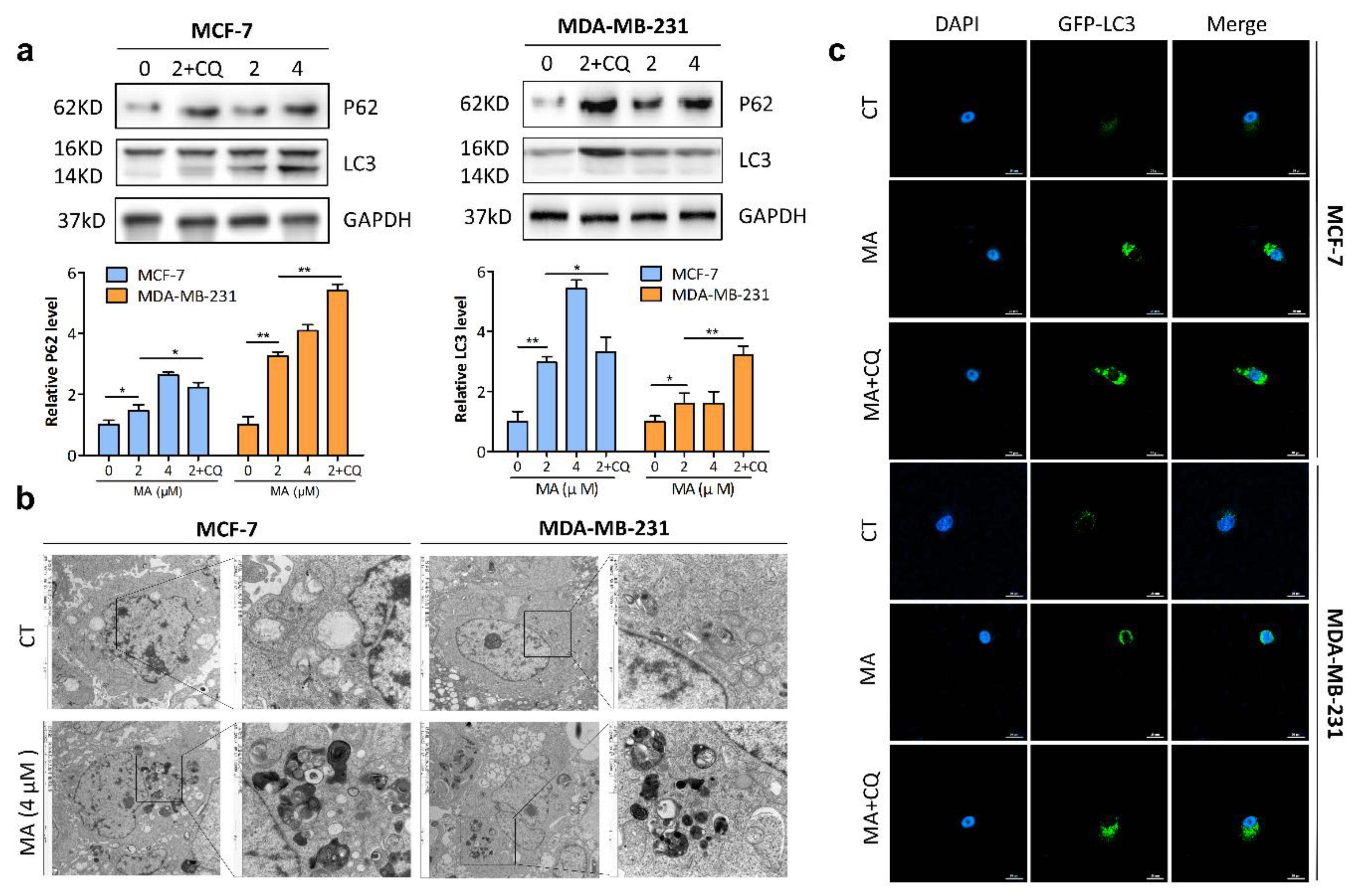
2.3. MA Promotes Autophagosome Formation and Inhibits Autophagosome Degradation in Breast Cancer Cells
2.4. RIP1 Mediates MA-Induced Autophagy through Akt/mTOR Pathway
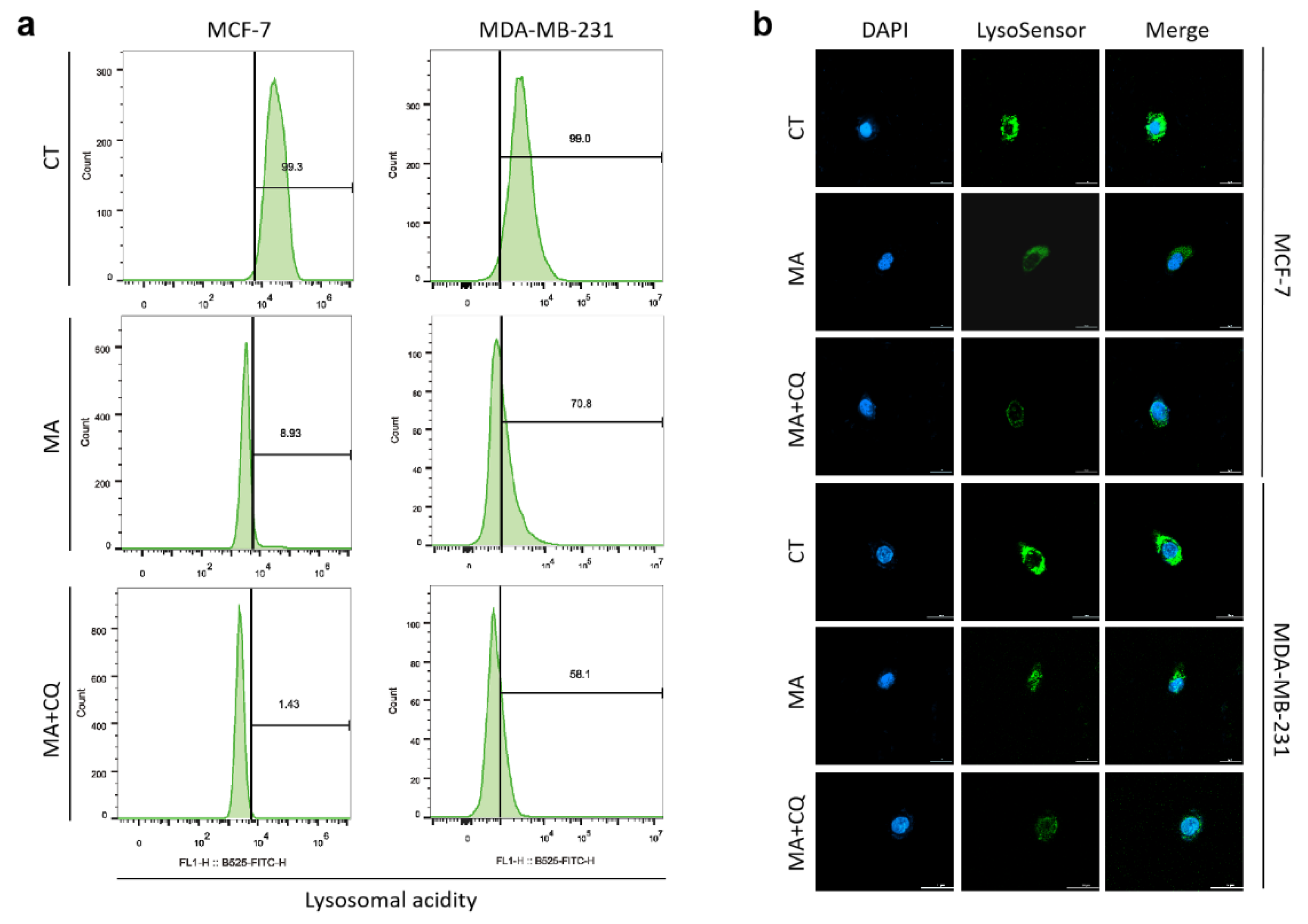
2.5. MA Reduces Lysosomal Acidity in Breast Cancer Cells
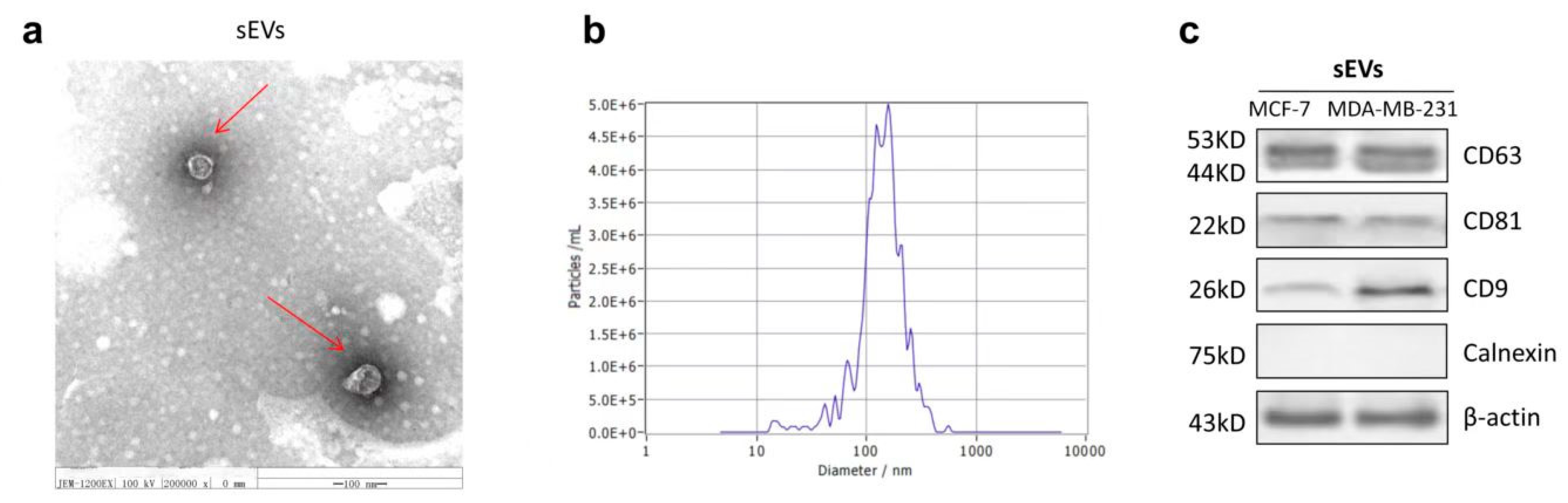
2.6. The Characterization of sEVs
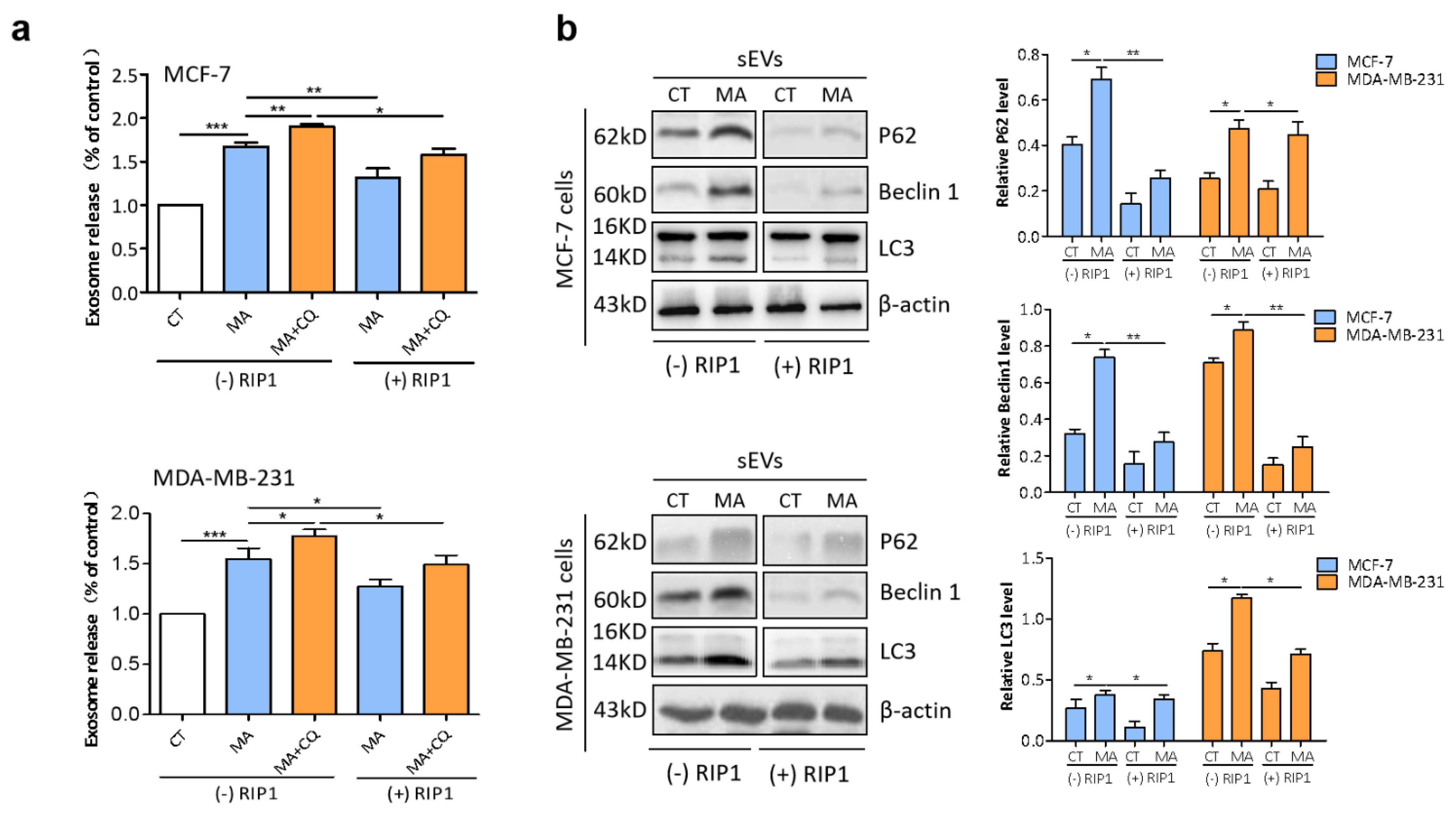
2.7. RIP1 Mediates MA-Induced Secretory Autophagy
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Reagents
4.2. Cell Viability Assay
4.3. Cell Proliferation Assay
4.4. Colony Formation Assay
4.5. Scratch Wound Assay
4.6. Transwell Assay
4.7. Western Blot Analysis
4.8. Transmission Electron Microscopy (TEM)
4.9. Confocal Microscropy
4.10. Lysosensor Assay
4.11. sEVs Isolation and Purification
4.12. Nanoparticle Tracking Analysis (NTA)
4.13. sEVs Quantification
4.14. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Das, R.; Rauf, A.; Mitra, S.; Bin Emran, T.; Hossain, J.; Khan, Z.; Naz, S.; Ahmad, B.; Meyyazhagan, A.; Pushparaj, K.; et al. Therapeutic potential of marine macrolides: An overview from 1990 to 2022. Chem. Interact. 2022, 365, 110072. [Google Scholar] [CrossRef] [PubMed]
- Ashok, P.; Lathiya, H.; Murugesan, S. Manzamine alkaloids as antileishmanial agents: A review. Eur. J. Med. Chem. 2015, 97, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Ashok, P.; Kumar, B.K.; Chander, S.; Sekhar, C.; Kondapalli, V.G.; Sankaranarayanan, M. Anti-infective Potential of Manzamine Alkaloids—A Review. Med. Chem. 2022, 18, 629–654. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, K.; Fujiwara, Y.; Hayashida, A.; Horlad, H.; Kato, H.; Rotinsulu, H.; Losung, F.; Mangindaan, R.E.; de Voogd, N.J.; Takeya, M.; et al. Manzamine A, a marine-derived alkaloid, inhibits accumulation of cholesterol ester in macrophages and suppresses hyperlipidemia and atherosclerosis in vivo. Bioorg. Med. Chem. 2013, 21, 3831–3838. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, M.; Hammond, N.L.; Peng, J.; Wahyuono, S.; McIntosh, K.A.; Charman, W.N.; Mayer, A.M.S.; Hamann, M.T. New Manzamine Alkaloids from an Indo-Pacific Sponge. Pharmacokinetics, Oral Availability, and the Significant Activity of Several Manzamines against HIV-I, AIDS Opportunistic Infections, and Inflammatory Diseases. J. Med. Chem. 2004, 47, 3512–3517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maglioni, S.; Arsalan, N.; Hamacher, A.; Afshar, S.; Schiavi, A.; Beller, M.; Ventura, N. High-Content C. elegans Screen Identifies Natural Compounds Impacting Mitochondria-Lipid Homeostasis and Promoting Healthspan. Cells 2021, 11, 100. [Google Scholar] [CrossRef]
- Karan, D.; Dubey, S.; Pirisi, L.; Nagel, A.; Pina, I.; Choo, Y.-M.; Hamann, M.T. The Marine Natural Product Manzamine A Inhibits Cervical Cancer by Targeting the SIX1 Protein. J. Nat. Prod. 2020, 83, 286–295. [Google Scholar] [CrossRef]
- Kallifatidis, G.; Hoepfner, D.; Jaeg, T.; Guzmán, E.A.; Wright, A.E. The Marine Natural Product Manzamine A Targets Vacuolar ATPases and Inhibits Autophagy in Pancreatic Cancer Cells. Mar. Drugs 2013, 11, 3500–3516. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.-C.; Kuo, T.-T.; Chang, H.-Y.; Liu, W.-S.; Hsia, S.-M.; Huang, T.-C. Manzamine A Exerts Anticancer Activity against Human Colorectal Cancer Cells. Mar. Drugs 2018, 16, 252. [Google Scholar] [CrossRef] [Green Version]
- Yadav, A.K.; Vashishta, V.; Joshi, N.; Taneja, P. AR-A 014418 Used against GSK3beta Downregulates Expression of hnRNPA1 and SF2/ASF Splicing Factors. J. Oncol. 2014, 2014, 695325. [Google Scholar] [CrossRef] [Green Version]
- Choezom, D.; Gross, J.C. Neutral sphingomyelinase 2 controls exosome secretion by counteracting V-ATPase-mediated endosome acidification. J. Cell Sci. 2022, 135, jcs259324. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [Green Version]
- Negahdaripour, M.; Owji, H.; Eskandari, S.; Zamani, M.; Vakili, B.; Nezafat, N. Small extracellular vesicles (sEVs): Discovery, functions, applications, detection methods and various engineered forms. Expert Opin. Biol. Ther. 2020, 21, 371–394. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Teixeira, A.F.; Zhu, H.-J.; Dijke, P.T. Cancer associated-fibroblast-derived exosomes in cancer progression. Mol. Cancer 2021, 20, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Raudenska, M.; Balvan, J.; Masarik, M. Crosstalk between autophagy inhibitors and endosome-related secretory pathways: A challenge for autophagy-based treatment of solid cancers. Mol. Cancer 2021, 20, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Pleet, M.L.; Branscome, H.; DeMarino, C.; Pinto, D.O.; Zadeh, M.A.; Rodriguez, M.; Sariyer, I.K.; El-Hage, N.; Kashanchi, F. Autophagy, EVs, and Infections: A Perfect Question for a Perfect Time. Front. Cell. Infect. Microbiol. 2018, 8, 362. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Ren, D. Lysosomal physiology. Annu. Rev. Physiol. 2015, 77, 57–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, L.; Peng, M.; Liu, S.; Liu, Y.; Wan, X.; Hou, Y.; Qin, Y.; Yang, L.; Chen, S.; Zeng, H.; et al. Hypoxia-stimulated ATM activation regulates autophagy-associated exosome release from cancer-associated fibroblasts to promote cancer cell invasion. J. Extracell. Vesicles 2021, 10, e12146. [Google Scholar] [CrossRef] [PubMed]
- Khuanjing, T.; Ongnok, B.; Maneechote, C.; Siri-Angkul, N.; Prathumsap, N.; Arinno, A.; Chunchai, T.; Arunsak, B.; Chattipakorn, S.C.; Chattipakorn, N. Acetylcholinesterase inhibitor ameliorates doxorubicin-induced cardiotoxicity through reducing RIP1-mediated necroptosis. Pharmacol. Res. 2021, 173, 105882. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jiang, H.; Chen, S.; Du, F.; Wang, X. The Mitochondrial Phosphatase PGAM5 Functions at the Convergence Point of Multiple Necrotic Death Pathways. Cell 2012, 148, 228–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wawruszak, A.; Luszczki, J.; Czerwonka, A.; Okon, E.; Stepulak, A. Assessment of Pharmacological Interactions between SIRT2 Inhibitor AGK2 and Paclitaxel in Different Molecular Subtypes of Breast Cancer Cells. Cells 2022, 11, 1211. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in Mammalian Autophagy Research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Joshi, K.; Denning, M.F.; Zhang, J. RIPK3 signaling and its role in the pathogenesis of cancers. Cell Mol. Life Sci. 2021, 78, 7199–7217. [Google Scholar] [CrossRef] [PubMed]
- Mijaljica, D.; Prescott, M.; Devenish, R.J. V-ATPase engagement in autophagic processes. Autophagy 2011, 7, 666–668. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Wei, Y.; Liu, W.; Liu, L.; Guo, Z.; Fan, C.; Wang, L.; Hu, J.; Li, B. Mechanical Stress-Dependent Autophagy Component Release via Extracellular Nanovesicles in Tumor Cells. ACS Nano 2019, 13, 4589–4602. [Google Scholar] [CrossRef] [PubMed]
- Fader, C.M.; I Colombo, M. Autophagy and multivesicular bodies: Two closely related partners. Cell Death Differ. 2008, 16, 70–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulet, C.R.; Champagne, A.; Bernard, G.; Vandal, D.; Chabaud, S.; Pouliot, F.; Bolduc, S. Cancer-associated fibroblasts induce epithelial–mesenchymal transition of bladder cancer cells through paracrine IL-6 signalling. BMC Cancer 2019, 19, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, R.; Karp, C.M.; Beaudoin, B.; Vuong, N.; Chen, G.; Chen, H.-Y.; Bray, K.; Reddy, A.; Bhanot, G.; Gelinas, C.; et al. Autophagy Suppresses Tumorigenesis through Elimination of p62. Cell 2009, 137, 1062–1075. [Google Scholar] [CrossRef] [Green Version]
- Bao, L.; Chandra, P.K.; Moroz, K.; Zhang, X.; Thung, S.N.; Wu, T.; Dash, S. Impaired autophagy response in human hepatocellular carcinoma. Exp. Mol. Pathol. 2013, 96, 149–154. [Google Scholar] [CrossRef] [Green Version]
- Jiang, T.-X.; Zou, J.-B.; Zhu, Q.-Q.; Liu, C.H.; Wang, G.-F.; Du, T.-T.; Luo, Z.-Y.; Guo, F.; Zhou, L.-M.; Liu, J.-J.; et al. SIP/CacyBP promotes autophagy by regulating levels of BRUCE/Apollon, which stimulates LC3-I degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 13404–13413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaleağasıoğlu, F.; Ali, D.M.; Berger, M.R. Multiple Facets of Autophagy and the Emerging Role of Alkylphosphocholines as Autophagy Modulators. Front. Pharmacol. 2020, 11, 547. [Google Scholar] [CrossRef]
- Mauthe, M.; Orhon, I.; Rocchi, C.; Zhou, X.; Luhr, M.; Hijlkema, K.J.; Coppes, R.P.; Engedal, N.; Mari, M.; Reggiori, F.; et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018, 14, 1435–1455. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yin, X. Panobinostat inhibits breast cancer progression via Vps34-mediated exosomal pathway. Hum. Cell 2022, 36, 366–376. [Google Scholar] [CrossRef]
- Wang, X.; Qian, C.; Yang, Y.; Liu, M.-Y.; Ke, Y.; Qian, Z.-M. Phosphorylated Rasal2 facilitates breast cancer progression. Ebiomedicine 2019, 50, 144–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salazar, G.; Cullen, A.; Huang, J.; Zhao, Y.; Serino, A.; Hilenski, L.; Patrushev, N.; Forouzandeh, F.; Hwang, H.S. SQSTM1/p62 and PPARGC1A/PGC-1alpha at the interface of autophagy and vascular senescence. Autophagy 2020, 16, 1092–1110. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yin, X.; Yang, Y. Rasal2 suppresses breast cancer cell proliferation modulated by secretory autophagy. Mol. Cell Biochem. 2019, 462, 115–122. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Liu, Y.; Qin, H.; Qi, G.; Chen, X.; Lyu, Y.; Han, Y. RIP1 Mediates Manzamine-A-Induced Secretory Autophagy in Breast Cancer. Mar. Drugs 2023, 21, 151. https://doi.org/10.3390/md21030151
Wang X, Liu Y, Qin H, Qi G, Chen X, Lyu Y, Han Y. RIP1 Mediates Manzamine-A-Induced Secretory Autophagy in Breast Cancer. Marine Drugs. 2023; 21(3):151. https://doi.org/10.3390/md21030151
Chicago/Turabian StyleWang, Xuan, Yuanpeng Liu, Huan Qin, Guocui Qi, Xuehong Chen, Yi Lyu, and Yantao Han. 2023. "RIP1 Mediates Manzamine-A-Induced Secretory Autophagy in Breast Cancer" Marine Drugs 21, no. 3: 151. https://doi.org/10.3390/md21030151
APA StyleWang, X., Liu, Y., Qin, H., Qi, G., Chen, X., Lyu, Y., & Han, Y. (2023). RIP1 Mediates Manzamine-A-Induced Secretory Autophagy in Breast Cancer. Marine Drugs, 21(3), 151. https://doi.org/10.3390/md21030151