Generation of Bioactive Peptides from Porphyridium sp. and Assessment of Their Potential for Use in the Prevention of Hypertension, Inflammation and Pain
Abstract
:1. Introduction
2. Results
2.1. Proximate Compositional Analysis of Porphyridium sp. and Generated Hydrolysates
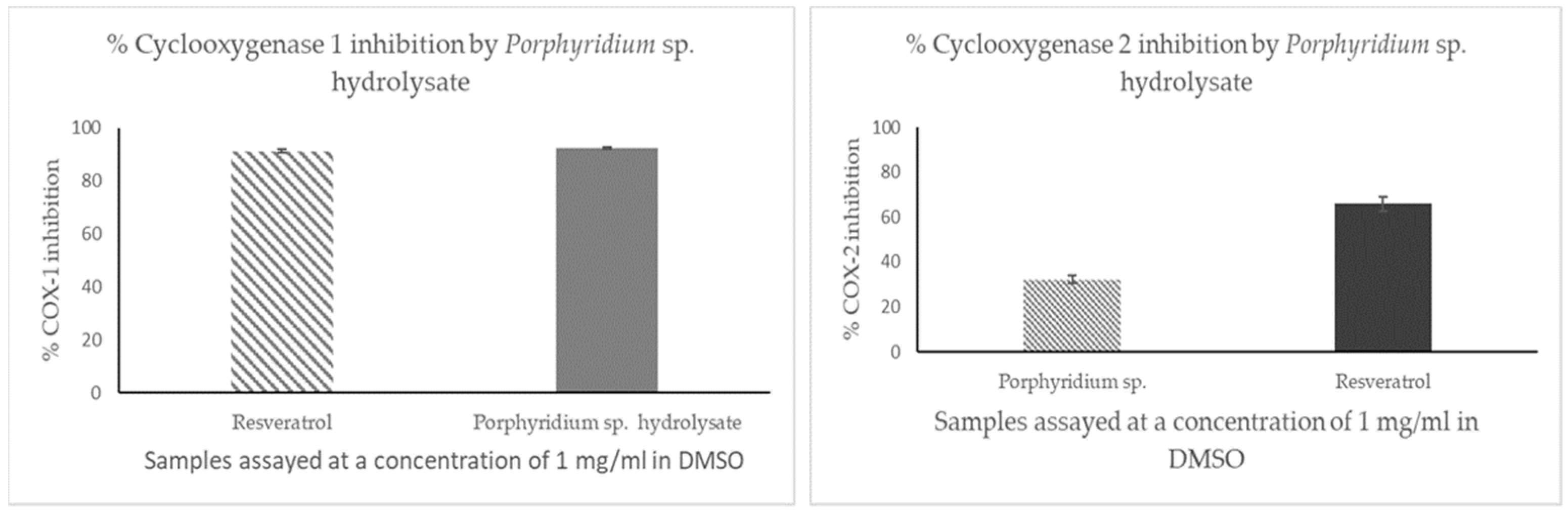
2.2. In Vitro Screening for Cyclooxygenase 1 and 2 Inhibition
2.3. Mass Spectrometry and In Silico Analysis of Peptides
2.3.1. Peptides Identified Using Mass Spectrometry
2.3.2. Determination of the PeptideRanker Scores, Novelty, and Potential Bioactivities of Peptides
2.3.3. Chemical Synthesis of Selected Peptides
2.4. Product Development–Production of Jelly Candies
2.5. In Vivo Antihypertensive Trial
2.6. Statistical Analysis
3. Discussion
4. Materials and Methods
4.1. Porphyridium sp. Hydrolysate and Permeate Generation
4.2. In Vitro Bioactivities Assessment
4.2.1. In Vitro Screening for Cyclooxygenase (COX-1 and COX-2) Inhibitory Activities
4.2.2. Mass Spectrometry in Tandem and In Silico Analysis
4.3. Chemical Synthesis of Peptides
4.4. Product Formulation
4.5. In Vivo Trial in SHRs
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2017, 9, 7204–7218. [Google Scholar] [CrossRef] [Green Version]
- Parisien, M.; Lima, L.V.; Dagostino, C.; El-Hachem, N.; Drury, G.L.; Grant, A.V.; Huising, J.; Verma, V.; Meloto, C.B.; Silva, J.R.; et al. Acute inflammatory response via neutrophil activation protects against the development of chronic pain. Sci. Transl. Med. 2022, 14, eabj9954. [Google Scholar] [CrossRef] [PubMed]
- Süleyman, H.; Demircan, B.; Karagöz, Y. Anti-inflammatory and side effects of cyclooxygenase inhibitors. Pharmacol. Rep. 2007, 59, 247–258. [Google Scholar] [PubMed]
- Patrono, C. Cardiovascular effects of cyclooxygenase-2 inhibitors: A mechanistic and clinical perspective. Br. J. Clin. Pharmacol. 2016, 82, 957–964. [Google Scholar] [CrossRef] [Green Version]
- Attiq, A.; Jalil, J.; Husain, K.; Ahmad, W. Raging the War Against Inflammation With Natural Products. Front. Pharmacol. 2018, 9, 976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Wang, C.; Shi, Q.; Ma, C. Preparation of different molecular weight polysaccharides from Porphyridium cruentum and their antioxidant activities. Int. J. Biol. Macromol. 2009, 45, 42–47. [Google Scholar] [CrossRef]
- Dillon, H.; Sumanchi, A.; Rao, K. US20070167398A1. In Methods and Compositions for Reducing Inflammation and Preventing Oxidative Damage; Current Holdrs TerraVia Holdings Inc.: Dallas, TX, USA, 2007. [Google Scholar]
- Sekar, S.; Chandramohan, M. Phycobiliproteins as a commodity: Trends in applied research, patents and commercialization. J. Appl. Phycol. 2008, 20, 113–136. [Google Scholar] [CrossRef]
- Risjani, Y.; Mutmainnah, N.; Manurung, P.; Wulan, S.N.; Yunianta. Exopolysaccharide from Porphyridium cruentum (purpureum) is Not Toxic and Stimulates Immune Response against Vibriosis: The Assessment Using Zebrafish and White Shrimp Litopenaeus vannamei. Mar. Drugs. 2021, 19, 133. [Google Scholar] [CrossRef]
- Kavitha, M.D.; Gouda, K.G.M.; Aditya Rao, S.J.; Shilpa, T.S.; Shetty, N.P.; Sarada, R. Atheroprotective effect of novel peptides from Porphyridium purpureum in RAW 264.7 macrophage cell line and its molecular docking study. Biotechnol. Lett. 2019, 41, 91–106. [Google Scholar] [CrossRef]
- Mora, L.; Gallego, M.; Toldrá, F. ACEI-Inhibitory Peptides Naturally Generated in Meat and Meat Products and Their Health Relevance. Nutrients 2018, 10, 1259. [Google Scholar] [CrossRef] [Green Version]
- Di Bernardini, R.; Mullen, A.M.; Bolton, D.; Kerry, J.; O’Neill, E.; Hayes, M. Assessment of the angiotensin-I-converting enzyme (ACE-I) inhibitory and antioxidant activities of hydrolysates of bovine brisket sarcoplasmic proteins produced by papain and characterisation of associated bioactive peptidic fractions. Meat Sci. 2012, 90, 226–235. [Google Scholar] [CrossRef]
- Hayes, M.; Mora, L.; Lucakova, S. Identification of Bioactive Peptides from Nannochloropsis oculata Using a Combination of Enzymatic Treatment, in Silico Analysis and Chemical Synthesis. Biomolecules 2022, 12, 1806. [Google Scholar] [CrossRef]
- Bleakley, S.; Hayes, M.; O’ Shea, N.; Gallagher, E.; Lafarga, T. Predicted Release and Analysis of Novel ACE-I, Renin, and DPP-IV Inhibitory Peptides from Common Oat (Avena sativa) Protein Hydrolysates Using in Silico Analysis. Foods 2017, 6, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, E.; Conlon, M.; Hayes, M. In vitro enzyme inhibitory effects of green and brown Australian seaweeds and potential impact on metabolic syndrome. J. Appl. Phycol. 2023, 35, 893–910. [Google Scholar] [CrossRef]
- Hayes, M.; Stanton, C.; Slattery, H.; O’Sullivan, O.; Hill, C.; Fitzgerald, G.F.; Ross, R.P. Casein fermentate of Lactobacillus animalis DPC6134 contains a range of novel propeptide Angiotensin-Converting Enzyme inhibitors. Appl. Environ. Microbiol. 2007, 73, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira-Lemos, E.; Almeida, A.R.; Vouga, B.; Morais, C.; Correia, I.; Pereira, P.; Guiné, R.P. Development and characterization of healthy gummy jellies containing natural fruits. Open Agric. 2021, 6, 466–478. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.; Pollastri, G.; Shields, D.C. Towards the improved discovery and design of functional peptides: Common features of diverse classes permit generalised prediction of bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [Green Version]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the Expasy Server; Walker, J.M., Ed.; The Proteomics Protocols Handbook, Humana Press; Full text—Copyright Humana Press: Totowa, NJ, USA, 2005. [Google Scholar]
- Gomez-Ruiz, J.A.; Ramos, M.; Recio, I. Identification of novel angiotensin-converting enzyme inhibitory peptides from ovine milk proteins by CE-MS and chromatographic techniques. Electrophoresis 2007, 28, 4202–4211. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Inhibition of dipeptidyl peptidase IV (DPP-IV) by proline containing casein derived peptides. J. Funct. Foods 2013, 5, 1909–1917. [Google Scholar] [CrossRef] [Green Version]
- Lan, V.T.T.; Ito, K.; Ohno, M.; Motoyama, T.; Ito, S.; Kawarasaki, Y. Analyzing a dipeptide library to identify human dipeptidyl peptidase IV inhibitor. Food Chem. 2015, 175, 66–73. [Google Scholar] [CrossRef]
- Dhanda, S.; Singh, H.; Singh, J. Hydrolysis of various bioactive peptides by goat brain dipeptidyl peptidase III. Cell Biochem. Funct. 2008, 23, 339–345. [Google Scholar] [CrossRef]
- Hatakenaka, T.; Kato, T.; Okamoto, K. In Vitro and In Silico Studies on Angiotensin I-Converting Enzyme Inhibitory Peptides Found in Hydrophobic Domains of Porcine Elastin. Molecules 2023, 28, 3337. [Google Scholar] [CrossRef] [PubMed]
- Naseri, A.; Jacobsen, C.; Sejberg, J.J.P.; Pedersen, T.E.; Larsen, J.; Hansen, K.M.; Holdt, S.L. Multi-Extraction and Quality of Protein and Carrageenan from Commercial Spinosum (Eucheuma denticulatum). Foods 2020, 9, 1072. [Google Scholar] [CrossRef]
- Blanco-Llamero, C.; García-García, P.; Señoráns, F.J. Combination of Synergic Enzymes and Ultrasounds as an Effective Pretreatment Process to Break Microalgal Cell Wall and Enhance Algal Oil Extraction. Foods 2021, 10, 1928. [Google Scholar] [CrossRef] [PubMed]
- Safi, C.; Rodriguez, L.C.; Mulder, W.; Engelen-Smit, N.; Spekking, W.; Broek, L.V.D.; Olivieri, G.; Sijtsma, L. Energy consumption and water-soluble protein release by cell wall disruption of Nannochloropsis gaditana. Bioresour. Technol. 2017, 239, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Toldrá, F.; Mora, L. Proteins and Bioactive Peptides in High Protein Content Foods. Foods 2021, 10, 1186. [Google Scholar] [CrossRef]
- Purcell, D.; Packer, M.A.; Hayes, M. Angiotensin-I-Converting Enzyme Inhibitory Activity of Protein Hydrolysates Generated from the Macroalga Laminaria digitata (Hudson) JV Lamouroux 1813. Foods 2022, 11, 1792. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, C.; Mora-Soler, L.; Gallagher, E.; O’ Connor, P.; Prieto, J.; Soler-Vila, A.; Hayes, M. Isolation and Characteri zation of Bioactive Pro-Peptides with in Vitro Renin Inhibitory Activities from the Macroalga Palmaria palmata. J. Agric. Food Chem. 2012, 60, 7421–7427. [Google Scholar] [CrossRef]
- Admassu, H.; Abdalbasit, M.; Gasmalla, A.; Yang, R.; Zhao, W. Bioactive peptides derived from seaweed protein and their health benefits: Antihypertensive, antioxidant and antidiabetic properties. J. Food Sci. 2018, 83, 6–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, T.; He, S.; Ni, C.; Wu, Y.; Xu, Z.; Chen, M.L.; Li, H.; Cheng, Y.; Wen, L. In Vitro Anti-Inflammatory Activity of Three Peptides Derived from the Byproduct of Rice Processing. Plant Foods Hum Nutr. 2022, 77, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Dullius, A.; Fassina, P.; Giroldi, M.; Goettert, M.I.; Volken de Souza, C.F. A biotechnological approach for the production of branched chain amino acid containing bioactive peptides to improve human health: A review. Food Res. Int. 2020, 131, 109002. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, S.; Shuib, A.S.; Abdullah, N. Structural characteristics and antihypertensive effects of angiotensin-1-converting enzyme inhibitory peptides in the Renin-Angiotensin and Kallikrein Kinin systems. Afr. J. Tradit. Complement. Altern. Med. 2017, 14, 383–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, R.; Aluko, R.E.; Ju, X.-R. Evaluating Molecular Mechanism of Hypotensive Peptides Interactions with Renin and Angiotensin Converting Enzyme. PLoS ONE 2014, 9, e91051. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural requirements of Angiotensin-I-converting enzyme inhibitory activity—Structure activity relationship study of di- and tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef]
- Torkova, A.; Kononikhin, A.; Bugrova, A.; Khotchenkov, V.; Tsentalovich, M.; Medvedeva, U. Effect of in vitro gastrointestinal digestion on bioactivity of poultry protein hydrolysate. Curr. Res. Nutr. Food Sci. 2016, 4 (Suppl. S2), 77–78. [Google Scholar]
- Chi, C.F.; Wang, B.; Wang, Y.M.; Zhang, B.; Deng, S.J. Isolation and characterization of three antioxidant peptides from protein hydrolysate of bluefin leatherjacket (Navodon septentrionalis) heads. J. Funct. Foods 2015, 12, 1–10. [Google Scholar] [CrossRef]
- He, J.; Guo, H.; Zhang, M.; Wang, M.; Sun, L.; Zhuang, Y. Purification and characterization of a novel calcium-binding heptapeptide from the hydrolysate of tilapia bone with its osteogenic activity. Foods 2022, 11, 468. [Google Scholar] [CrossRef]
- Dogne, J.M.; Supuran, C.T.; Pratico, D. Adverse cardiovascular effects of the coxibs. J. Med. Chem. 2005, 48, 2251–2257. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Złotek, U.; Jakubczyk, A.; Rybczyńska-Tkaczyk, K.; Ćwiek, P.; Baraniak, B.; Lewicki, S. Characteristics of New Peptides GQLGEHGGAGMG, GEHGGAGMGGGQFQPV, EQGFLPGPEESGR, RLARAGLAQ, YGNPVGGVGH, and GNPVGGVGHGTTGT as Inhibitors of Enzymes Involved in Metabolic Syndrome and Antimicrobial Potential. Molecules 2020, 25, 2492. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Yoshioka, K.; Mizutani, K.; Miyakoshi, M.; Murakami, T.; Akizawa, T. Blood pressure-depressing activity of a peptide derived from silkworm fibroin in spontaneously hypertensive rats. Biosci. Biotechnol. Biochem. 2006, 70, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Marczak, E.D.; Usui, H.; Fujita, H.; Yang, Y.; Yokoo, M.; Lipkowski, A.W.; Yoshikawa, M. New antihypertensive peptides isolated from rapeseed. Peptides 2003, 24, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Takaki-Doi, S.; Hashimoto, K.; Yamamura, M.; Kamei, C. Antihypertensive activities of royal jelly protein hydrolysate and its fractions in spontaneously hypertensive rats. Acta Med. Okayama. 2009, 63, 57–64. [Google Scholar] [CrossRef]
- Cao, S.; Wang, Z.; Xing, L.; Zhou, L.; Zhang, W. Bovine Bone Gelatin-Derived Peptides: Food Processing Characteristics and Evaluation of Antihypertensive and Antihyperlipidemic Activities. J. Agric. Food Chem. 2022, 70, 9877–9887. [Google Scholar] [CrossRef]
- Hoyle, N.T.; Merritt, J.H. Quality of fish protein hydrolysates from herring (Clupea harengus). J. Food Sci. 1994, 59, 76–79. [Google Scholar] [CrossRef]
- Association of Official Analytical Chemists. Official Methods of Analysis of AOAC International; AOAC International: Rockville, MD, USA, 1998. [Google Scholar]
- Willenberg, I.; Meschede, A.K.; Gueler, F.; Jang, M.-S.; Shushakova, N.; Schebb, N.H. Food Polyphenols Fail to Cause a Biologically Relevant Reduction of COX-2 Activity. PLoS ONE 2015, 10, e0139147. [Google Scholar] [CrossRef]
- Khatun, M.S.; Hasan, M.M.; Kurata, H. PreAIP: Computational prediction of anti-inflammatory peptides by integrating multiple complementary features. Front. Genet. 2019, 10, 129. [Google Scholar] [CrossRef] [Green Version]
- Purwaningtyas, H.P.; Suhartakik, N.; Mustofa, A. Formulation of jelly from candy Betel (Piper betle L.)—Suji (Pleomele angustofolia) leaf extract. J. Teknol. Dan Ind. Pangan 2017, 2, 25–30. (In Indonesian) [Google Scholar]
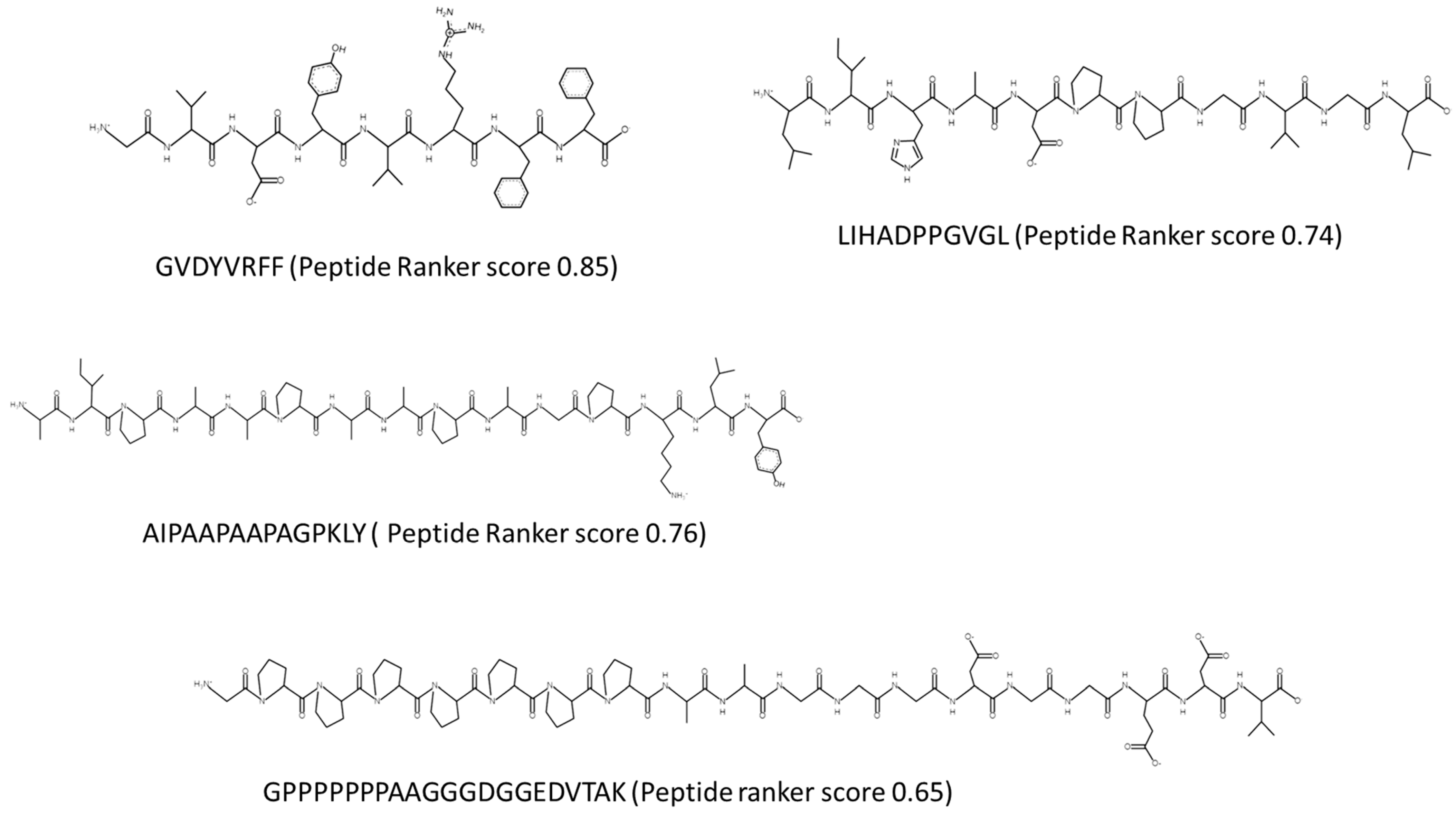

| Porphyridium sp. Peptides Generated from Hydrolysis | ||||||||
|---|---|---|---|---|---|---|---|---|
| Peptide Amino Acid Sequence (Single Letter Code) | Protein of Origin | Accession Code | Peptide Ranker Value * | PreAIP RF *† Combined Values | BIOPEP Search ** | Expasy Peptide Cutter *** | Bioactivities Associated with Peptide Fragments | Peptide Charge |
| GVDYVRFF | Uncharacterized protein OS = Porphyridium purpureum OX = 35,688 GN = FVE85_6371 PE = 4 SV = 1 | tr|A0A5J4Z524|A0A5J4Z524_PORPP | 0.857 | 0.374 Low confidence | Novel | GVD, Y, VR, F, F | GVD-alpha amylase inhibitor; VR-DPP-IV and ACE inhibitor peptide | 0 |
| AIPAAPAAPAGPKLY | Putative transporter YrhG OS = Porphyridium purpureum OX = 35,688 GN = FVE85_0107 PE = 4 SV = 1 | tr|A0A5J4YZ45|A0A5J4YZ45_PORPP | 0.768 | 0.262 Negative AIP | Novel | AIPAAPAAPAGPK; L, Y | All novel but GPK occurs at the C-terminal end of antioxidative, antibacterial, osteoanabolic and calcium binding peptides found in BIOPEP | 1 |
| LIHADPPGVGL | Uncharacterized protein OS = Porphyridium purpureum OX = 35,688 GN = FVE85_6371 PE = 4 SV = 1 | tr|A0A5J4Z524|A0A5J4Z524_PORPP | 0.743 | 0.390 Medium confidence AIP | Novel | L, IH, ADPPGVG, L | IH-DPP-IV inhibitor & DPP-III inhibitor; GVG occurs in alpha amylase and ACE-I inhibitors | −1 |
| GLDAGLSHCGVVNVCIP | UDP-glucose/GDP-mannose dehydrogenase OS = Ectocarpus siliculosus OX = 2880 GN = Esi_0052_0113 PE = 4 SV = 1 | tr|D8LPB4|D8LPB4_ECTSI | 0.729 | 0.500 High confidence AIP | Novel | G, L, DAG, L, SH, CGVVNVCIP | DAG-found in ACE-1 and antioxidative peptides; SH is a DPP-IV inhibitory peptide | −1 |
| LIHADPPGVGLTGF | Uncharacterized protein OS = Porphyridium purpureum OX = 35,688 GN = FVE85_6371 PE = 4 SV = 1 | tr|A0A5J4Z524|A0A5J4Z524_PORPP | 0.716 | 0.369 low confidence AIP | Novel | L, IH, ADPPGVG, L, TG, F | IH- DPP-IV inhibitor and DPP-III inhibitor, GVG occurs in alpha amylase and ACE-1 inhibitor peptides, TG-DPP-IV inhibitor and ACE-1 inhibitor | −1 |
| AIPAAPAAPAGPK | Putative transporter YrhG OS = Porphyridium purpureum OX = 35,688 GN = FVE85_0107 PE = 4 SV = 1 | tr|A0A5J4YZ45|A0A5J4YZ45_PORPP | 0.709 | Novel | AIPAAPAAPAGPK | All novel but GPK occurs at the C-terminal end of antioxidative, antibacterial, osteoanabolic and calcium binding peptides found in BIOPEP | 1 | |
| GPPPPPPPAASGGDGGEDVTAK | Adenylyl cyclase-associated protein 2 OS = Porphyridium purpureum OX = 35,688 GN = FVE85_5192 PE = 3 SV = 1 | tr|A0A5J4Z4T9|A0A5J4Z4T9_PORPP | 0.659 | 0.469 High confidence AIP | Novel | Resistance to cleavage by Trypsin, Chymotrypsin and Pepsin | TAK found in the immunomodulator peptide RTAKV | −2 |
| AAGGSLFEEYMR | Protein PYP1 OS = Porphyridium purpureum OX = 35,688 GN = FVE85_6364 PE = 4 SV = 1 | tr|A0A5J4Z470|A0A5J4Z470_PORPP | 0.648 | 0.461 Medium confidence AIP | Novel | AAGGS, L, F, EE, Y, M, R | GGS found in the sequence of an ACE-1 inhibitory peptide YAGGS | −1 |
| LFDGKVCTMLIIIT | NADH-ubiquinone oxidoreductase chain 5 OS = Rhodogorgon sp. OX = 2,485,824 GN = ND5 PE = 3 SV = 1 | tr|A0A3G3MIM0|A0A3G3MIM0_9FLOR | 0.641 | 0.545 High confidence AIP | Novel | L, F, DGK, VCTM, L, IIIT | all novel peptide fragments- no assigned bioactivities but IT is found in the ACE-1 inhibitory peptides ITT and ITK | 0 |
| LDSHLPINLPQGL | Ribulose bisphosphate carboxylase large chain (Fragment) OS = Porphyridium purpureum OX = 35,688 GN = rbcL PE = 3 SV = 1 | tr|Q1WFR3|Q1WFR3_PORPP | 0.619 | 0.428 Medium confidence AIP | Novel | L, DSH, L, PINL, PQG, L | PQG occurs at the end of the PEP inhibitory peptide PPPPGGPQPRPPQG | −1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayes, M.; Aluko, R.E.; Aurino, E.; Mora, L. Generation of Bioactive Peptides from Porphyridium sp. and Assessment of Their Potential for Use in the Prevention of Hypertension, Inflammation and Pain. Mar. Drugs 2023, 21, 422. https://doi.org/10.3390/md21080422
Hayes M, Aluko RE, Aurino E, Mora L. Generation of Bioactive Peptides from Porphyridium sp. and Assessment of Their Potential for Use in the Prevention of Hypertension, Inflammation and Pain. Marine Drugs. 2023; 21(8):422. https://doi.org/10.3390/md21080422
Chicago/Turabian StyleHayes, Maria, Rotimi E. Aluko, Elena Aurino, and Leticia Mora. 2023. "Generation of Bioactive Peptides from Porphyridium sp. and Assessment of Their Potential for Use in the Prevention of Hypertension, Inflammation and Pain" Marine Drugs 21, no. 8: 422. https://doi.org/10.3390/md21080422
APA StyleHayes, M., Aluko, R. E., Aurino, E., & Mora, L. (2023). Generation of Bioactive Peptides from Porphyridium sp. and Assessment of Their Potential for Use in the Prevention of Hypertension, Inflammation and Pain. Marine Drugs, 21(8), 422. https://doi.org/10.3390/md21080422








